Abstract
Epstein-Barr virus (EBV) is associated with the development of a variety of malignancies, including Hodgkin lymphoma. One of the few viral transcripts expressed in EBV-positive Hodgkin/Reed-Sternberg (HRS) cells of Hodgkin lymphoma is latent membrane protein 2A (LMP2A). This viral protein blocks B-cell receptor (BCR)-signaling in vitro. Furthermore, expression of LMP2A in developing B cells in vivo induces a global down-regulation of genes necessary for proper B-cell development. In this study we have analyzed gene transcription in primary B cells from LMP2A transgenic mice, LMP2A-expressing human B-cell lines, and LMP2A-positive and -negative EBV-infected lymphoblastoid cell lines (LCLs). We demonstrate that LMP2A increases the expression of genes associated with cell cycle induction and inhibition of apoptosis, alters the expression of genes involved in DNA and RNA metabolism, and decreases the expression of B-cell-specific factors and genes associated with immunity. Furthermore, many alterations in gene expression induced by LMP2A are similar to those recently described in HRS cells of Hodgkin lymphoma and activated, proliferating germinal center centroblasts/centrocytes. These correlations suggest that LMP2A expression in EBV-infected B cells may lead to the induction and maintenance of an activated, proliferative state that could ultimately result in the development of Hodgkin lymphoma. (Blood. 2003;102: 4166-4178)
Introduction
Epstein-Barr virus (EBV) infection is implicated in the development of a variety of malignancies of both lymphoid and epithelial origin, including Hodgkin lymphoma, Burkitt lymphoma, nasopharyngeal carcinoma, and various lymphoproliferative disorders arising in immunocompromised individuals.1,2 EBV establishes a lifelong latent infection in B lymphocytes by limiting viral gene expression in order to evade immune recognition. Independent studies report limited gene expression in B cells from healthy individuals harboring a latent EBV infection. Expressed transcripts in these cells include EBV nuclear antigen 1 (EBNA1), latent membrane protein 2A (LMP2A), EBV-encoded RNAs (EBERs), and BamHI-A-rightward transcripts (BARTS).2-6
Classical Hodgkin lymphoma is characterized by the presence of multinucleated, malignant Hodgkin/Reed-Sternberg (HRS) cells, which constitute only a minority of the tumor cells.7 HRS cells are presumed to derive from germinal center B cells that have lost expression of immunoglobulin (Ig) and other B-cell-specific characteristics.7-9 It has been estimated that nearly half of all Hodgkin lymphomas contain EBV DNA, and viral gene expression appears to be limited to EBNA1, LMP1, and LMP2A.2,10-12 The consistent detection of LMP2A transcripts in both tumor cells and latently infected B cells in vivo suggests that LMP2A plays an important role in viral persistence and in the development of EBV-associated diseases such as Hodgkin lymphoma.
In latently infected lymphocytes, LMP2A localizes to small glycolipid-enriched microdomains in the plasma membrane, where it is thought to mimic an activated B-cell receptor (BCR).13,14 Studies demonstrate that BCR activation in LMP2A-expressing cells fails to induce calcium mobilization and activation of the downstream signaling molecules Lyn, Syk, phosphatidylinositol 3-kinase (PI3-K), phospholipase C-γ2 (PLCγ2), Vav, Shc, and mitogen-activated protein kinase (MAPK).15,16 The amino-terminal domain of LMP2A is tyrosine phosphorylated and associates with Src family protein tyrosine kinases (PTKs) as well as Syk.16,17 Mutational analyses indicate that phosphotyrosines at positions 74 and 85 (an immunoreceptor tyrosine-based motif [ITAM]) in LMP2A bind Syk, while tyrosine 112 binds Lyn. All 3 residues are essential for the LMP2A-mediated block in BCR signal transduction.18,19
Expression of an LMP2A transgene in vivo interferes with normal B-cell development, allowing BCR-negative cells to exit the bone marrow and colonize peripheral lymphoid organs.20 Bone marrow B cells from these mice undergo Ig light chain, but not heavy chain, gene rearrangement, indicating that LMP2A signaling bypasses the requirement for Ig recombination and allows IgM-negative cells, which would normally undergo apoptosis, to colonize peripheral lymphoid organs.20 Furthermore, bone marrow cells from LMP2A transgenic mice form colonies in interleukin-7 (IL-7)-containing methylcellulose, and the B cells in these colonies lack IgM expression, indicating that LMP2A bypasses the requirement for Ig rearrangement and allows for IL-7-driven B-cell proliferation.20,21 The LMP2A-mediated effects on B-cell development and survival in vivo have been shown to require the ITAM (Syk binding).22 In addition, studies using mice deficient for the downstream B-cell signaling components BLNK (SLP-65) or Bruton tyrosine kinase (Btk) have demonstrated that LMP2A uses these molecules for its effects on B-cell development and survival.23,24
In a recent study, we used DNA microarray technology to identify alterations in gene expression in B cells from mice expressing the LMP2A transgene. We identified not only decreased expression of genes associated with normal B-cell development, but also reduced levels of the transcription factors that regulate expression of those genes.25 These results suggest that LMP2A induces a global down-regulation of gene transcription necessary for proper B-cell development. In this study, we present the complete results of our microarray analyses of B cells from LMP2A transgenic mice and include a comparison of gene transcription in human B-cell lines expressing LMP2A as well as LMP2A-positive and -negative EBV-infected lymphoblastoid cell lines (LCLs). These experiments have identified common pathways affected by LMP2A in both murine in vivo and human in vitro systems and have provided further relevance with regard to the role of LMP2A in the development of EBV-associated Hodgkin lymphoma.
Materials and methods
Isolation of primary B cells from mice
Construction and characterization of the Eμ TgE LMP2A transgenic mice have been described previously.20 All animals were housed at the Northwestern University Center for Experimental Animal Resources in accordance with university animal welfare guidelines. To isolate bone marrow B cells, femurs and tibias were flushed with 1 × phosphate-buffered saline containing 1% penicillin/streptomycin. Red blood cells were lysed in 155 mM ammonium chloride and 2 × 106 cells were placed in 3 mL Methocult M3630 methylcellulose cultures containing 10 ng/mL recombinant mouse IL-7 (Stemcell Technologies, Vancouver, BC, Canada). Colonies formed in 7 to 10 days were routinely more than 95% CD19+ B cells as demonstrated by flow cytometry as previously described,20 and these cells were used for microarray analyses. Spleens were dissociated between frosted slides in RPMI to prepare single cell suspensions. Red blood cells were lysed and B cells were purified at 4°C on magnetic cell sorting (MACS) columns using magnetic beads coated with CD19 antibodies (Miltenyi Biotec, Auburn, CA). B cells were tested for purity by fluorescent-activated cell sorter analysis, and cells shown to be more than 95% CD19+ B cells by flow cytometry were used for microarray analysis.
Cell lines
All cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 1000 U/mL penicillin, and 1000 μg/mL streptomycin. The BJAB cell line is an established EBV-negative B-lymphoma line (American Type Culture Collection, Rockville, MD). BJAB cell lines expressing LMP2A were generated by retrovirus infection as previously described.26 Gene expression was compared in 2 individual BJAB clones that contain vector alone (C1 and F6) and 2 clones that express LMP2A (C12 and F11). The EBV-infected LCLs have been described previously.27 Lines LCL1 and LCL2 are EBV+LMP2A+ and lines ES2 and ES5 are EBV+LMP2A- LCLs.
RNA preparation and microarray experiments
Total RNA was extracted from CD19+ murine B cells and human cell lines according to the Trizol reagent protocol (Invitrogen, Carlsbad, CA). RNA was then subjected to cleanup using the Qiagen Rneasy Mini Kit according to the RNA cleanup protocol (Qiagen, Valencia, CA). Aliquots of RNA were then removed and run on 1% agarose/formaldehyde gels to verify that no degradation occurred. Additionally, optical density (OD) readings were taken, and 20 μg total RNA having OD A260/A280 readings between 1.9 and 2.1 was used for reverse transcription. Double-stranded cDNA was then generated according to the Superscript Double-Stranded cDNA Synthesis Kit (Invitrogen) using an oligo dT/T7 primer (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24-3′). Following phenol/chloroform extraction and ethanol precipitation, cRNA was in vitro transcribed and labeled with biotin using the Enzo Bioarray RNA Transcription Labeling Kit (Affymetrix, Santa Clara, CA). The cRNA was purified using CHROMA SPIN 100 columns (Clontech, Palo Alto, CA) and ethanol was precipitated. cRNA having OD A260/A280 readings between 1.9 and 2.1 were again run on agarose/formaldehyde gels to check for transcripts ranging between 35 and 200 bases, and 20 μg of these products was fragmented in fragmentation buffer containing 200 mM Tris (tris(hydroxymethyl)aminomethane)-acetate, pH 8.1, 500 mM KOAc, and 150 mM MgOAc, at 95°C for 35 minutes. Fragmented, biotinylated cRNA was then submitted for hybridization by the Children's Memorial Institute for Education and Research (CMIER) microarray facility at Northwestern University. The murine genome U74A and human U133A chips were purchased from Affymetrix and used for analysis.
Statistical significance of differential expression
Affymetrix microarrays were used to quantify expression in all experiments. The Affymetrix system of expression quantification using perfect and mismatched oligonucleotides was used in conjunction with the system of background detection. At least 2 separate, identical experiments comparing gene transcription in LMP2A-positive versus LMP2A-negative cells were performed for each cell type, including murine bone marrow and splenic B cells, human BJABs, and human LCLs. Patricia Dyck, a bioinformaticist for Northwestern University, performed statistical analyses of the arrays. A t-distribution was applied to the difference between LMP2A-expressing and control cells. The t test quantifies the probability of differential expression through calculation of the magnitude and consistency of changes. Genes with probability values greater than .01 were deemed to be inconsistent, therefore statistically insignificant. For Tables 1, 2, S1, and S2, expressed sequence tags with no known homology or function were excluded, and the selected data were placed into categories based on protein function.
List of genes and gene categories induced by LMP2A
Murine bone marrow CD19+ . | Murine spleen CD19+ . | Human BJAB + LMP2A . | Human LCL + LMP2A . |
|---|---|---|---|
| Cell cycle/apoptosis | |||
| Baculoviral IAP repeat-containing 5 (survivin) | CT-2, cell cycle/apoptosis inhibitor 6 (Api6) | Nuclear mitotic apparatus protein 1 (Numa 1) | Cyclin F |
| Cyclin D 1 | Bcl-xL | Programmed cell death 6 (Pdcd6) | Cyclin I |
| Cyclin-dependent kinase 5 (cdk5) | Baculoviral IAP repeat-containing 5 (survivin) | Cysteine-rich protein, nucleolar (Hsa6591) | |
| Cyclin-dependent kinase 9 (Cdk9) | B-cell translocation gene 3 (Btg3) | Polo-like kinase (Plk) | |
| Inhibin beta E (Inhbe) | Caspase-11 | Rcl, c-myc responsive | |
| Leukemia inhibitory factor receptor (Lifr) | Caspase-3 | Circadian oscillary protein (Scop) | |
| Proliferating cell nuclear antigen (PCNA) | Cdc20 | ||
| Prolactin-like protein A (Prlpa) | Cell division cycle 2 homolog A (Cdc2a) | ||
| Prothymosin alpha (Ptma) | Cell division cycle 6 homolog (Cdc6) | ||
| TOB3 | CDC28 protein kinase 1 (Cks 1) | ||
| Cytochrome c-1 (Cyc 1) | |||
| Cyclin A2 | |||
| Cyclin B2 | |||
| Geminin (Gmnn) | |||
| MAD2 (mitotic arrest deficient)-like 1 (mad211) | |||
| Antigen identified by monoclonal antibody Ki 67 | |||
| Protein regulator of cytokinesis 1-like (Prc 1) | |||
| Neural stem cell-derived neuronal survival (Sdnsf) | |||
| Serine (or cysteine) proteinase inhibitor (Serpinb6) | |||
| Cd27 binding protein (Hindu God of destruction, Siva) | |||
| DNA/RNA metabolism | |||
| Apoptotic chromatin condensation ind in nucleus (Acinus) | Brain abundant, membrane-attached signal protein 1 (Basp 1) | Eukaryotic translation initiation factor 4 gamma (elF4g1) | BAF53A, chromatin remodeling |
| Absent, small, or homeotic discs 1 (Ash 1) | Eukaryotic translation elongation factor 1 delta (Eefld) | Histone family member H2B/S | Centromere protein A (Cenpa) |
| D-E-A-D box polypeptide 6 (Dbp6) | Eukaryotic translation initiation factor 3, subunit 7 (elF3s7) | H2B histone family, member H2BFS | elF-3s8 |
| Excision repair cross-comp repair deficiency (Ercc2) | ELL-related RNA polymerase II (E112) | H4 histone family, member H4FG | Glycyl-tRNA synthetase (Gars) |
| General control of amino acid synthesis-like 2 (Gen512) | High-mobility group box 2 nuclear protein (Hmgb2) | Polymerase (DNA directed), mu (Polm) | Histone H1x (H1fx) |
| General transcription factor III A (Gtf3a) | High-mobility group nucleosomal binding domain 2 (Hmgn2) | RNA binding motif, ss interacting protein (MSSP-1, Rbms 1) | IGF-II mRNA-binding protein 3 (Koc1) |
| H3 histone, family 3A (H3f3a) | Hn ribonucleoproteins methyltransferase-like 2 (Hhmr112) | Ribosomal protein S11 | Mitochondrial ribosomal protein S27 (Mrps27) |
| High mobility group nucleosomal binding domain 2 (Hmgn2) | Eukaryotic translation initiation factor 1A (IFly) | 40S ribosomal protein SA (Rsp4) | 8-oxoguanine DNA glycosylase (Oggl) |
| Mini chromosome maintenance-deficient 4 homolog (Mcmd4) | Mini chromosome maintenance deficient (Mcmd) | Splicing factor 3a, subunit 2 (Sf3a2) | DNA polymerase delta small subunit (Polo2) |
| Polymerase (DNA directed) alpha 2 (Pola2) | MORF-related gene X (mrgx) | SWI/SNF related regulator of chromatin (Smarca4) | RNA polymerase II, polypeptide J (Polr2j) |
| Polymerase (DNA directed) mu (Polm) | Mitochondrial ribosomal protein L30 (Mrpl30) | SWI/SNF related regulator of chromatin (Smarcb 1) | Replication protein A3 (Rpa3) |
| RAD51-associated protein 1 (Rad51 ap 1) | Polymerase (DNA directed), alpha 2 (Pola2) | Transcription termination factor, RNA polymerase I (Ttf1) | Seryl-tRNA synthetase (Sars) |
| Ribosomal protein L32 | RNA binding motif protein 10 (Rbm 10) | Zinc-finger helicase (hZFH) | Small nuclear ribonucleoprotein polypeptides B and B1 (Snrpb) |
| Ribosomal protein L5 | RNA binding motif protein 3 (Rbm3) | Zinc finger protein 161 (Znf161) | Structure-specific recognition protein 1 (Ssrp 1) |
| Ribosomal protein L7 | RNA binding motif, ss interacting protein (MSSP-1, Rbms 1) | Zinc finger protein 220 (Znf220) | ATP-dependent mitochondrial RNA helicase (SUV3, Sars) |
| Ribosomal protein S26 | Ribosomal protein L22 | Transcription elongation factor A (SII, Tceal) | |
| Ribosomal protein S4, X-linked (Rps4x) | Ribosomal protein S6 kinase polypeptide 1 (Rps6ka1) | Topoisomerase (DNA) II alpha (Top2a) | |
| Suppressor of Ty 4 homolog (Spt4h) | Ribosome binding protein 1 (Rrbp 1) | UMP synthase (Umps) | |
| DNA/RNA metabolism (continued) | |||
| Xlr-related, meiosis regulated (Xmr) | Ribonucleotide reductase M1 (Rrm1) | Tyrosyl-tRNA synthetase (Yars) | |
| Tyrosyl-tRNA synthetase (Yars) | Ribonucleotide reductase M2 subunit (Rrm2) | Zinc finger protein 9 (Znf9) | |
| Zinc finger protein 125, (ZT2, Zfp 125) | Topoisomerase (DNA) II alpha (Top2a) | Eukaryotic translation termination factor 1 (Etf1) | |
| Zinc finger protein, multitype (FOG Friend of GATA, Zfpm1) | |||
| Inflammation/immunity | |||
| Allograft inflammatory factor 1 (lbal, Aif1) | Lipocortin 1, annexin A1 (Anxa1) | Ig alpha chain C region (Alc) | |
| Complement component 9 (C9) | B-cell-receptor gene (Bcr) | CD86 antigen (CD28 antigen ligand 2, B7-2) | |
| Calumenin (Calu) | Complement Clq B chain (Clqb) | C-type lectin, superfamily member 2 (Clecsf2) | |
| CD84 antigen | Complement component 1, q subcomponent, c (Clqc) | HCGII-7 hemochromatosis candidate genes | |
| Fibroblast growth factor 1 (Fgf1) | Calumenin, secretory pathway (Calu) | Regulatory factor X, 2 (Rfx2) | |
| Fibroblast growth factor 5 (Fgf5) | CD68 antigen | ||
| Interferon activated gene 204 (lfi204) | CD7 antigen | ||
| Germ line immunoglobulin V(H)II gene H8 | CD9 antigen | ||
| Interferon gamma-inducing factor binding protein (lgifbp) | Cathelin-like protein (CLP, CRAMP, Cnlp) | ||
| Immunoglobuling J chain | Cytokine receptor-like molecule (EB13), EBV-ind gene 3 | ||
| Immunoglobulin kappa chain variable 20 (V20 family) | Endoglin (Eng) | ||
| Interleukin-13 receptor, alpha 2 (IL-13a2) | Ficolin A (Fena) | ||
| Interleukin-5 (IL-5) | Histocompatibility 13 (H13) | ||
| Immunity-associated protein (Imap) | Histocompatibility 47 (H47) | ||
| Lymphocyte-activation gene 3 (Lag 3) | H-2 class I histocompatibility antigen (HA1D) | ||
| Lymphocyte antigen 6 complex, locus D (Ly-6d) | Ig heavy chain V region (MPC 11, HV01) | ||
| Reg factor X-ass ankyrin-containing protein (Rfxank) | Single chain antibody scFv (scFvCPSQ) | ||
| TNF receptor superfamily, member 8 (Tnfrsf8) | mRNA (IB5) for IgA V-D-J-heavy chain | ||
| Immunoglobulin heavy chain V region mRNA | |||
| Clone WS2a43 mutated immunoglobulin heavy chain | |||
| Immunoglobulin heavy chain 8 (heavy chain of IgG3) | |||
| Ig active joining chain (J chain) of the b allele | |||
| Immunoglobulin kappa light chain variable region | |||
| Immunoglobulin variable gene V kappa | |||
| Immunoglobulin light chain V region | |||
| Immunoglobulin light chain IgG | |||
| Anti-DNA immunoglobulin light chain IgG | |||
| Extrachromosomal DNA for V kappa and J k coding joint | |||
| Ig rearranged truncated mu-chain | |||
| IFN-stimulated protein, 20 kDa (Isg207) | |||
| Integral membrane protein 3 (Itm3) | |||
| Immunoglobulin J chain | |||
| Lymphocyte antigen 6 complex, locus C (Ly-6c) | |||
| Mannose receptor, C type 1 (Mrc1) | |||
| Intracellular ca-binding prot (MRP 14, calgranulin B, s100a9) | |||
| Secretory leukoprotease inhibitor gene (Slpi) | |||
| TGF beta 1 induced transcript 4 (Tgfbli4) | |||
| TNF receptor superfamily, member 17 (Tnfrsf17) | |||
| Vascular cell adhesion molecule 1 (Vcam1) | |||
| Signaling | |||
| Adrenergic receptor, beta 3 (Adrb3) | rhoB gene, ras homolog (Arhb) | Ras homolog gene family, member G (rho G Arhg) | Cornichon-like (Cnil) |
| Cadherin EGF LAG 7-pass G-type rec 2 (flamingo 1, Celsr2) | Arsenate resistance protein 2 (Acr2, Ars2) | Rho GDP dissociation inhibitor (GDI) alpha (Arhgdia) | Dual-specificity tyr-phosphorylation reg kinase 1A (Dyrk 1a) |
| Calcium and integrin binding 1 (calmyrin, KIP, Cib 1) | GP1-anchored membrane protein 1 (Gpiap 1) | GTP binding protein overexpressed in skeletal muscle (Gem) | FK506 binding protein 2 (Fkbp2) |
| Cytokine inducible SH2-containing protein (SOCS-1, Cis) | Gene trap locus 3, Delta-like Dlk1 (Gtl3) | 5-hydroxytryptamine (serotonin) receptor 1E (Htr1e) | Guanylate cyclase activator (Guca1b) |
| Creatine kinase, mitochondrial 1 (Ckmt 1) | Potentially oncogenic Ras (Ki-Ras) | Inositol 1,4,5-triphosphate receptor, type 1 (ltpr 1) | Hyaluronan-mediated motility receptor (RHAMM CD168, Hmmr) |
| Tyrosine protein kinase (Csk) | Kit oncogene | Mitogen-activated protein kinase kinase 2 (Map2k2) | Mitogen-act protein kinase kinase kinase kinase 1 (Map4k1) |
| Casein kinase 1, delta (Csnk 1d) | Membrane-spanning 4-domains (Ms4a6b) | Pecanex-like 3 (Penxl3) | Adaptor protein p 150, P13-kinase regulatory subunit 4 (Pik3r4) |
| Delta-like 1 (Dll 1) | Phosphatidylinositol-4-phosphate 5-kinase (Pip5k2a) | 3-phosphoinositide-dependent protein kinase-1 (Pdpk 1) | Protein phosphatase 1G (formerly 2C) (PPM1G) |
| Ectodermal-neural cortex 1 (Enc1) | Protein tyrosine phosphatase, nonreceptor type 8 (Ptpn8) | Protein phosphatase 3 (formerly 2B)(calcineurin A alpha) | Protein phosphatase 2 (formerly 2A)(Ppp2r2a) |
| FK506 binding protein 4 (Fkbp4) | Protein tyr pptase, nonreceptor type (SHPS, BIT, Ptpns 1) | RaP2 interacting protein 8, small GTPase int prot (Rpip8) | |
| Fyn-related kinase (Frk) | Sell (suppressor of lin-12) 1 homolog (Sel1h) | Sac domain-containing inositol phosphatase 3 (Sac3) | |
| Frizzled homolog 2 (Fzd2) | Signal-transducing adaptor molecule 1 (Stam 1) | Tyrosine kinase 2 (Tyk2) | |
| Frizzled homolog 8 (Fzd8) | WW domain binding protein 2 (Wbp2) | Unc-51-like kinase 1 (Ulk 1) | |
| Ras GTPase activating protein binding protein 1 (G3BP) | Zeta-chain (TCR)-associated protein kinase (Zap-70) | ||
| Growth hormone-inducible transmembrane protein (Ghitm) | |||
| Protein phosphatase 3, catalytic subunit (Ppp3ca) | |||
| Protein tyrosine phosphatase, receptor type (Ftp-1, Ptprl) | |||
| GTP binding nuclear protein (Ran) | |||
| Wingless related MMTV integration site 10b (Wnt10b) | |||
| Wingless-related MMTV integration site 6 (Wnt6) | |||
| Transcription factors | |||
| BarH-like homeobox 1 (Barx1) | Carbon catabolite repression 4 homolog (Ccr4) | Bromodomain-containing 3 (Brd3) | Leucine-zipper protein, repressor of cAMP, MAPK (Atf5) |
| Distal-less homeobox 2 (Dlx2) | CCAAT/enhancer binding protein (C/EBP) | BTB (POZ) domain-containing 3 (Btbd3) | Cellular repressor of E1A-stimulated genes (Creg) |
| GLI-Kruppel family member (Gli1) | Cellular repressor of E1A-stimulated genes (Creg) | Bithorax Bx1 homeobox (Bx1) | Alternatively spliced cut-like 1 (Cutl1) |
| H6 homeobox 3 repressor (Hmx3) | dbpA homologue, cold shock domain protein A (Csda6) | CCR4-NOT transcription complex, subunit 8 (Cnot8) | Forkhead protein (FKHRLI, Fox03a) |
| LIM homeobox protein 1 (Lhx1) | Transcription factor (Dp 1) | NFk light polypeptide enhancer in B cells inhibitor (nfkbia) | |
| Similar to zinc finger protein pMLZ-4 (Mlz-4) | ets variant gene 6 (TEL, Etv6) | PR domain-containing 2, with ZNF domain (Prdm2) | |
| Myogenin (Myog) | Ewing sarcoma homolog (Ews) | Sirtuin silent mating type information reg 2 homolog 6 (Sirt6) | |
| Y-box binding protein (oxyR) | Enhancer of zeste homolog 2 (Ezh2) | SP140 nuclear body protein | |
| Paired box gene 3 (Pax3) | HIV type 1 enhancer binding protein 1 (Hivep 1) | ||
| Norepinephrine transporter (SLC6A5) | Inhibitor protein (Id2) | ||
| T-cell factor-1 (Tcf-1) | Makorin, ring finger protein, (Mkm1) | ||
| Transcription factor AP-2 beta (Tcfap2b) | T-cell acute leukemia 1 (Tal1) | ||
| Transcription factor 7 (Tcf7) | |||
| X-box binding protein 1 (Xbp1) | |||
| Tumor-associated | |||
| ElaC homolog 2 (Elac2) | Arginine-rich, mutated in early stage tumors (Armet) | HLA-B-associated transcript 3 (Bat3) | |
| Leukemia-associated gene (Pr22, Lag) | Ecotropic viral integration site 2 (Evi2) | Prostate tumor overexpressed gene 1 (Ptov 1) | |
| Neoplastic progression 2 (Npn2) | Immunoglobulin superfamily, member 4 (TSLC1, Igsf4) | ||
| Gene overexpressed in astrocytoma (Goa, Oea) | Leukemia-associated gene (Lag) | ||
| Proviral integration site 1 (Pim 1) | Expressed in nonmetastatic cells 3 (Nme3) | ||
| T-cell leukemia/lymphoma 1B (Tcl 1 bl) | Protein-serine/threonine kinase (Pim2) | ||
| Secreted acidic cys-rich glycoprotein/osteonectin (Sparc) | |||
| Ubiquitination | |||
| Proteasome (prosome, macropain) 26S subunit (psmd3) | Proteasome (prosome, macropain) subunit (Psma4) | F-box and leucine-rich repeat protein 7 (Fb17) | Proteasome (prosome, macropain) 26S subunit (Psmd8) |
| WW domain-containing protein 4 (Wwp2) | Proteasome (prosome, macropain) 26S subunit (psmc2) | Proteasome (prosome, macropain) subunit (Psmb9) | Ubiquitin-conjugating enzyme E2C (Ube2c) |
| Proteasome (prosome, macropain) 26S subunit (Psmd11) | Ubiquitin-conjugating enzyme E21 (UBC9, Ube21) | ||
| Ubiquitin fusion degradation 1-like (Ufd 11) | Ubiquitin-specific protease 22 (Usp22) |
Murine bone marrow CD19+ . | Murine spleen CD19+ . | Human BJAB + LMP2A . | Human LCL + LMP2A . |
|---|---|---|---|
| Cell cycle/apoptosis | |||
| Baculoviral IAP repeat-containing 5 (survivin) | CT-2, cell cycle/apoptosis inhibitor 6 (Api6) | Nuclear mitotic apparatus protein 1 (Numa 1) | Cyclin F |
| Cyclin D 1 | Bcl-xL | Programmed cell death 6 (Pdcd6) | Cyclin I |
| Cyclin-dependent kinase 5 (cdk5) | Baculoviral IAP repeat-containing 5 (survivin) | Cysteine-rich protein, nucleolar (Hsa6591) | |
| Cyclin-dependent kinase 9 (Cdk9) | B-cell translocation gene 3 (Btg3) | Polo-like kinase (Plk) | |
| Inhibin beta E (Inhbe) | Caspase-11 | Rcl, c-myc responsive | |
| Leukemia inhibitory factor receptor (Lifr) | Caspase-3 | Circadian oscillary protein (Scop) | |
| Proliferating cell nuclear antigen (PCNA) | Cdc20 | ||
| Prolactin-like protein A (Prlpa) | Cell division cycle 2 homolog A (Cdc2a) | ||
| Prothymosin alpha (Ptma) | Cell division cycle 6 homolog (Cdc6) | ||
| TOB3 | CDC28 protein kinase 1 (Cks 1) | ||
| Cytochrome c-1 (Cyc 1) | |||
| Cyclin A2 | |||
| Cyclin B2 | |||
| Geminin (Gmnn) | |||
| MAD2 (mitotic arrest deficient)-like 1 (mad211) | |||
| Antigen identified by monoclonal antibody Ki 67 | |||
| Protein regulator of cytokinesis 1-like (Prc 1) | |||
| Neural stem cell-derived neuronal survival (Sdnsf) | |||
| Serine (or cysteine) proteinase inhibitor (Serpinb6) | |||
| Cd27 binding protein (Hindu God of destruction, Siva) | |||
| DNA/RNA metabolism | |||
| Apoptotic chromatin condensation ind in nucleus (Acinus) | Brain abundant, membrane-attached signal protein 1 (Basp 1) | Eukaryotic translation initiation factor 4 gamma (elF4g1) | BAF53A, chromatin remodeling |
| Absent, small, or homeotic discs 1 (Ash 1) | Eukaryotic translation elongation factor 1 delta (Eefld) | Histone family member H2B/S | Centromere protein A (Cenpa) |
| D-E-A-D box polypeptide 6 (Dbp6) | Eukaryotic translation initiation factor 3, subunit 7 (elF3s7) | H2B histone family, member H2BFS | elF-3s8 |
| Excision repair cross-comp repair deficiency (Ercc2) | ELL-related RNA polymerase II (E112) | H4 histone family, member H4FG | Glycyl-tRNA synthetase (Gars) |
| General control of amino acid synthesis-like 2 (Gen512) | High-mobility group box 2 nuclear protein (Hmgb2) | Polymerase (DNA directed), mu (Polm) | Histone H1x (H1fx) |
| General transcription factor III A (Gtf3a) | High-mobility group nucleosomal binding domain 2 (Hmgn2) | RNA binding motif, ss interacting protein (MSSP-1, Rbms 1) | IGF-II mRNA-binding protein 3 (Koc1) |
| H3 histone, family 3A (H3f3a) | Hn ribonucleoproteins methyltransferase-like 2 (Hhmr112) | Ribosomal protein S11 | Mitochondrial ribosomal protein S27 (Mrps27) |
| High mobility group nucleosomal binding domain 2 (Hmgn2) | Eukaryotic translation initiation factor 1A (IFly) | 40S ribosomal protein SA (Rsp4) | 8-oxoguanine DNA glycosylase (Oggl) |
| Mini chromosome maintenance-deficient 4 homolog (Mcmd4) | Mini chromosome maintenance deficient (Mcmd) | Splicing factor 3a, subunit 2 (Sf3a2) | DNA polymerase delta small subunit (Polo2) |
| Polymerase (DNA directed) alpha 2 (Pola2) | MORF-related gene X (mrgx) | SWI/SNF related regulator of chromatin (Smarca4) | RNA polymerase II, polypeptide J (Polr2j) |
| Polymerase (DNA directed) mu (Polm) | Mitochondrial ribosomal protein L30 (Mrpl30) | SWI/SNF related regulator of chromatin (Smarcb 1) | Replication protein A3 (Rpa3) |
| RAD51-associated protein 1 (Rad51 ap 1) | Polymerase (DNA directed), alpha 2 (Pola2) | Transcription termination factor, RNA polymerase I (Ttf1) | Seryl-tRNA synthetase (Sars) |
| Ribosomal protein L32 | RNA binding motif protein 10 (Rbm 10) | Zinc-finger helicase (hZFH) | Small nuclear ribonucleoprotein polypeptides B and B1 (Snrpb) |
| Ribosomal protein L5 | RNA binding motif protein 3 (Rbm3) | Zinc finger protein 161 (Znf161) | Structure-specific recognition protein 1 (Ssrp 1) |
| Ribosomal protein L7 | RNA binding motif, ss interacting protein (MSSP-1, Rbms 1) | Zinc finger protein 220 (Znf220) | ATP-dependent mitochondrial RNA helicase (SUV3, Sars) |
| Ribosomal protein S26 | Ribosomal protein L22 | Transcription elongation factor A (SII, Tceal) | |
| Ribosomal protein S4, X-linked (Rps4x) | Ribosomal protein S6 kinase polypeptide 1 (Rps6ka1) | Topoisomerase (DNA) II alpha (Top2a) | |
| Suppressor of Ty 4 homolog (Spt4h) | Ribosome binding protein 1 (Rrbp 1) | UMP synthase (Umps) | |
| DNA/RNA metabolism (continued) | |||
| Xlr-related, meiosis regulated (Xmr) | Ribonucleotide reductase M1 (Rrm1) | Tyrosyl-tRNA synthetase (Yars) | |
| Tyrosyl-tRNA synthetase (Yars) | Ribonucleotide reductase M2 subunit (Rrm2) | Zinc finger protein 9 (Znf9) | |
| Zinc finger protein 125, (ZT2, Zfp 125) | Topoisomerase (DNA) II alpha (Top2a) | Eukaryotic translation termination factor 1 (Etf1) | |
| Zinc finger protein, multitype (FOG Friend of GATA, Zfpm1) | |||
| Inflammation/immunity | |||
| Allograft inflammatory factor 1 (lbal, Aif1) | Lipocortin 1, annexin A1 (Anxa1) | Ig alpha chain C region (Alc) | |
| Complement component 9 (C9) | B-cell-receptor gene (Bcr) | CD86 antigen (CD28 antigen ligand 2, B7-2) | |
| Calumenin (Calu) | Complement Clq B chain (Clqb) | C-type lectin, superfamily member 2 (Clecsf2) | |
| CD84 antigen | Complement component 1, q subcomponent, c (Clqc) | HCGII-7 hemochromatosis candidate genes | |
| Fibroblast growth factor 1 (Fgf1) | Calumenin, secretory pathway (Calu) | Regulatory factor X, 2 (Rfx2) | |
| Fibroblast growth factor 5 (Fgf5) | CD68 antigen | ||
| Interferon activated gene 204 (lfi204) | CD7 antigen | ||
| Germ line immunoglobulin V(H)II gene H8 | CD9 antigen | ||
| Interferon gamma-inducing factor binding protein (lgifbp) | Cathelin-like protein (CLP, CRAMP, Cnlp) | ||
| Immunoglobuling J chain | Cytokine receptor-like molecule (EB13), EBV-ind gene 3 | ||
| Immunoglobulin kappa chain variable 20 (V20 family) | Endoglin (Eng) | ||
| Interleukin-13 receptor, alpha 2 (IL-13a2) | Ficolin A (Fena) | ||
| Interleukin-5 (IL-5) | Histocompatibility 13 (H13) | ||
| Immunity-associated protein (Imap) | Histocompatibility 47 (H47) | ||
| Lymphocyte-activation gene 3 (Lag 3) | H-2 class I histocompatibility antigen (HA1D) | ||
| Lymphocyte antigen 6 complex, locus D (Ly-6d) | Ig heavy chain V region (MPC 11, HV01) | ||
| Reg factor X-ass ankyrin-containing protein (Rfxank) | Single chain antibody scFv (scFvCPSQ) | ||
| TNF receptor superfamily, member 8 (Tnfrsf8) | mRNA (IB5) for IgA V-D-J-heavy chain | ||
| Immunoglobulin heavy chain V region mRNA | |||
| Clone WS2a43 mutated immunoglobulin heavy chain | |||
| Immunoglobulin heavy chain 8 (heavy chain of IgG3) | |||
| Ig active joining chain (J chain) of the b allele | |||
| Immunoglobulin kappa light chain variable region | |||
| Immunoglobulin variable gene V kappa | |||
| Immunoglobulin light chain V region | |||
| Immunoglobulin light chain IgG | |||
| Anti-DNA immunoglobulin light chain IgG | |||
| Extrachromosomal DNA for V kappa and J k coding joint | |||
| Ig rearranged truncated mu-chain | |||
| IFN-stimulated protein, 20 kDa (Isg207) | |||
| Integral membrane protein 3 (Itm3) | |||
| Immunoglobulin J chain | |||
| Lymphocyte antigen 6 complex, locus C (Ly-6c) | |||
| Mannose receptor, C type 1 (Mrc1) | |||
| Intracellular ca-binding prot (MRP 14, calgranulin B, s100a9) | |||
| Secretory leukoprotease inhibitor gene (Slpi) | |||
| TGF beta 1 induced transcript 4 (Tgfbli4) | |||
| TNF receptor superfamily, member 17 (Tnfrsf17) | |||
| Vascular cell adhesion molecule 1 (Vcam1) | |||
| Signaling | |||
| Adrenergic receptor, beta 3 (Adrb3) | rhoB gene, ras homolog (Arhb) | Ras homolog gene family, member G (rho G Arhg) | Cornichon-like (Cnil) |
| Cadherin EGF LAG 7-pass G-type rec 2 (flamingo 1, Celsr2) | Arsenate resistance protein 2 (Acr2, Ars2) | Rho GDP dissociation inhibitor (GDI) alpha (Arhgdia) | Dual-specificity tyr-phosphorylation reg kinase 1A (Dyrk 1a) |
| Calcium and integrin binding 1 (calmyrin, KIP, Cib 1) | GP1-anchored membrane protein 1 (Gpiap 1) | GTP binding protein overexpressed in skeletal muscle (Gem) | FK506 binding protein 2 (Fkbp2) |
| Cytokine inducible SH2-containing protein (SOCS-1, Cis) | Gene trap locus 3, Delta-like Dlk1 (Gtl3) | 5-hydroxytryptamine (serotonin) receptor 1E (Htr1e) | Guanylate cyclase activator (Guca1b) |
| Creatine kinase, mitochondrial 1 (Ckmt 1) | Potentially oncogenic Ras (Ki-Ras) | Inositol 1,4,5-triphosphate receptor, type 1 (ltpr 1) | Hyaluronan-mediated motility receptor (RHAMM CD168, Hmmr) |
| Tyrosine protein kinase (Csk) | Kit oncogene | Mitogen-activated protein kinase kinase 2 (Map2k2) | Mitogen-act protein kinase kinase kinase kinase 1 (Map4k1) |
| Casein kinase 1, delta (Csnk 1d) | Membrane-spanning 4-domains (Ms4a6b) | Pecanex-like 3 (Penxl3) | Adaptor protein p 150, P13-kinase regulatory subunit 4 (Pik3r4) |
| Delta-like 1 (Dll 1) | Phosphatidylinositol-4-phosphate 5-kinase (Pip5k2a) | 3-phosphoinositide-dependent protein kinase-1 (Pdpk 1) | Protein phosphatase 1G (formerly 2C) (PPM1G) |
| Ectodermal-neural cortex 1 (Enc1) | Protein tyrosine phosphatase, nonreceptor type 8 (Ptpn8) | Protein phosphatase 3 (formerly 2B)(calcineurin A alpha) | Protein phosphatase 2 (formerly 2A)(Ppp2r2a) |
| FK506 binding protein 4 (Fkbp4) | Protein tyr pptase, nonreceptor type (SHPS, BIT, Ptpns 1) | RaP2 interacting protein 8, small GTPase int prot (Rpip8) | |
| Fyn-related kinase (Frk) | Sell (suppressor of lin-12) 1 homolog (Sel1h) | Sac domain-containing inositol phosphatase 3 (Sac3) | |
| Frizzled homolog 2 (Fzd2) | Signal-transducing adaptor molecule 1 (Stam 1) | Tyrosine kinase 2 (Tyk2) | |
| Frizzled homolog 8 (Fzd8) | WW domain binding protein 2 (Wbp2) | Unc-51-like kinase 1 (Ulk 1) | |
| Ras GTPase activating protein binding protein 1 (G3BP) | Zeta-chain (TCR)-associated protein kinase (Zap-70) | ||
| Growth hormone-inducible transmembrane protein (Ghitm) | |||
| Protein phosphatase 3, catalytic subunit (Ppp3ca) | |||
| Protein tyrosine phosphatase, receptor type (Ftp-1, Ptprl) | |||
| GTP binding nuclear protein (Ran) | |||
| Wingless related MMTV integration site 10b (Wnt10b) | |||
| Wingless-related MMTV integration site 6 (Wnt6) | |||
| Transcription factors | |||
| BarH-like homeobox 1 (Barx1) | Carbon catabolite repression 4 homolog (Ccr4) | Bromodomain-containing 3 (Brd3) | Leucine-zipper protein, repressor of cAMP, MAPK (Atf5) |
| Distal-less homeobox 2 (Dlx2) | CCAAT/enhancer binding protein (C/EBP) | BTB (POZ) domain-containing 3 (Btbd3) | Cellular repressor of E1A-stimulated genes (Creg) |
| GLI-Kruppel family member (Gli1) | Cellular repressor of E1A-stimulated genes (Creg) | Bithorax Bx1 homeobox (Bx1) | Alternatively spliced cut-like 1 (Cutl1) |
| H6 homeobox 3 repressor (Hmx3) | dbpA homologue, cold shock domain protein A (Csda6) | CCR4-NOT transcription complex, subunit 8 (Cnot8) | Forkhead protein (FKHRLI, Fox03a) |
| LIM homeobox protein 1 (Lhx1) | Transcription factor (Dp 1) | NFk light polypeptide enhancer in B cells inhibitor (nfkbia) | |
| Similar to zinc finger protein pMLZ-4 (Mlz-4) | ets variant gene 6 (TEL, Etv6) | PR domain-containing 2, with ZNF domain (Prdm2) | |
| Myogenin (Myog) | Ewing sarcoma homolog (Ews) | Sirtuin silent mating type information reg 2 homolog 6 (Sirt6) | |
| Y-box binding protein (oxyR) | Enhancer of zeste homolog 2 (Ezh2) | SP140 nuclear body protein | |
| Paired box gene 3 (Pax3) | HIV type 1 enhancer binding protein 1 (Hivep 1) | ||
| Norepinephrine transporter (SLC6A5) | Inhibitor protein (Id2) | ||
| T-cell factor-1 (Tcf-1) | Makorin, ring finger protein, (Mkm1) | ||
| Transcription factor AP-2 beta (Tcfap2b) | T-cell acute leukemia 1 (Tal1) | ||
| Transcription factor 7 (Tcf7) | |||
| X-box binding protein 1 (Xbp1) | |||
| Tumor-associated | |||
| ElaC homolog 2 (Elac2) | Arginine-rich, mutated in early stage tumors (Armet) | HLA-B-associated transcript 3 (Bat3) | |
| Leukemia-associated gene (Pr22, Lag) | Ecotropic viral integration site 2 (Evi2) | Prostate tumor overexpressed gene 1 (Ptov 1) | |
| Neoplastic progression 2 (Npn2) | Immunoglobulin superfamily, member 4 (TSLC1, Igsf4) | ||
| Gene overexpressed in astrocytoma (Goa, Oea) | Leukemia-associated gene (Lag) | ||
| Proviral integration site 1 (Pim 1) | Expressed in nonmetastatic cells 3 (Nme3) | ||
| T-cell leukemia/lymphoma 1B (Tcl 1 bl) | Protein-serine/threonine kinase (Pim2) | ||
| Secreted acidic cys-rich glycoprotein/osteonectin (Sparc) | |||
| Ubiquitination | |||
| Proteasome (prosome, macropain) 26S subunit (psmd3) | Proteasome (prosome, macropain) subunit (Psma4) | F-box and leucine-rich repeat protein 7 (Fb17) | Proteasome (prosome, macropain) 26S subunit (Psmd8) |
| WW domain-containing protein 4 (Wwp2) | Proteasome (prosome, macropain) 26S subunit (psmc2) | Proteasome (prosome, macropain) subunit (Psmb9) | Ubiquitin-conjugating enzyme E2C (Ube2c) |
| Proteasome (prosome, macropain) 26S subunit (Psmd11) | Ubiquitin-conjugating enzyme E21 (UBC9, Ube21) | ||
| Ubiquitin fusion degradation 1-like (Ufd 11) | Ubiquitin-specific protease 22 (Usp22) |
At least 2 separate, identical experiments comparing gene transcription in LMP2A-positive versus LMP2A-negative cells were performed for each cell type. Statistical analyses of the arrays were performed as described in “Materials and methods.” Genes with probability values of less than .01 and differential ratios of greater than 1.1 in LMP2A-positive cells compared with LMP2A-negative cells are listed.
List of genes and gene categories repressed by LMP2A
Murine bone marrow CD 19+ . | Murine spleen CD19+ . | Human BJAB + LMP2A . | Human LCL + LMP2A . |
|---|---|---|---|
| Cell cycle/apoptosis | |||
| SH3P9 myc box dependent interacting (Bin1) | SH3P9 myc box-dependent interacting (Bin1) | BTG family, member 3 (Btg3) | ATM kinase |
| Caspase-6 | B-cell translocation gene 2 (Btg2) | Cyclin A2 | RBP-1-like protein (Bcaa) |
| EGL nine homolog 1 (Egln 1) | p53 variant | Cell division cycle 2 (Cdc2) | pptase and tensin (PTEN) |
| Ubiquinol-cytochrome c reductase (Ubcr) | Proline-rich protein expressed in brain (Prtb) | CHK 1 checkpoint homolog (Chek1) | Retinoblastomaa susceptibility (Rbl) |
| Signal-induced proliferation associated (Spa-1) | CDC28 protein kinase 2 (Cks2) | ||
| (tre-2/USP6, BUB2, cdc16) domain (Tbc 1d1) | Defender against cell death 1 (Dad 1) | ||
| Transgene insert site 737 (TgN737) | Programmed cell death 4 (Pdcd4) | ||
| Pituitary tumor-transforming 1 (Pttg 1) | |||
| Ubiquinol-cytochrome C reductase (Ucrq) | |||
| YME1-like 1 (Yme 1I1) | |||
| DNA/RNA metabolism | |||
| Heterogeneous nuclear riboprotein H2 | H1 histone family, member 2 | Eukaryotic translation initiation factor (eIF2s2) | eIF-1A |
| H2B histone family, member S | Polymerase (RNA) II (DNA directed) | Eukaryotic translation initiation factor (eIF4a1) | Histone H2A/g (H2AFG) |
| DNA-binding nuclear protein NP220 | Polymerase (RNA) II (DNA directed) | High-mobility group box 2 (Hmgb2) | Histone H2A.2 (H2AFO) |
| Polymerase (RNA) II (DNA directed) | RAD50 homolog | Heterogeneous nuclear ribonucleoprotein (Hnrph1) | Histone H2A.X (H2AFX) |
| RADI homolog | Rael RNA export 1 homolog | Mitochondrial ribosomal protein 63 | Histone H2B/h |
| Trf-proximal protein homolog (Trfp) | Ribosomal protein L21 | Mitochondrial ribosomal protein L3 | Histone H2B/e |
| Zinc finger protein 292 | Zinc finger protein 36 (TISII) | High mobility group-like nuclear protein (Nhp2) | Histone H2B1 S |
| Zinc finger protein, subfamily 1A, 4 | Polymerase (RNA) II (DNA directed) polypeptide J | Histone H2BFC | |
| DNA repair protein rad18 | Histone H2BFD | ||
| RAD1 homolog | Histone H2B/k (H2BFK) | ||
| RNA-binding region containing (RNPI, RRM) | Histone H2BFL | ||
| Ribosomal protein L26-like 1 | Histone H2BFR | ||
| Ribosomal protein L38 | H4 histone family G (H4FG) | ||
| Zinc finger protein 9 | |||
| Inflammation/immunity | |||
| Colony-stimulating factor 1 receptor (Csf 1r) | Adenosine A2a receptor (Adora2a) | Allograft inflammatory factor 1 (Aif1) | Butyrophilin (BTF4) |
| Deoxynucleotidyltransferase, terminal (TdT) | Complement component 1r | Complement component 1q receptor 1 | CD69 |
| Histocompatibility 2, class II antigen A | CD19 antigen | Complement component 4A | IFN-induced leucine zipper protein 35 (Ifi35) |
| Histocompatibility 2, Q region locus 7 | CD22 antigen | Integrin, beta 2 (LFA-1) | IFN-induced protein 44 (Ifi44) |
| Interferon (alpha and beta) receptor 2 | Histocompatibility 2, D region locus 1 | Ig kappa chain V-I region (Igk) | IgG lambda light chain V-J-C region |
| Germ line immunoglobulin V(H)II gene | Histocompatibility 2, class II, locus DMa | T-cell receptor alpha locus (TCR alpha) | Interleukin-7 (IL-7) |
| Immunoglobulin lambda chain, variable 1 | IFN-γ inducible protein 30 (Ifi30) | Pre-B lymphocyte gene 3 (vpreB) | Interferon regulatory factor 7B (IRF7b) |
| Interleukin-16 (IL-16) | IFN-γ inducible protein (Ifi47) | Interferon-induced protein (IRF4/Pip/Isg15) | |
| Integrin beta 4 binding protein (Itgb4bp) | Delta-immunoglobulin (IgD) | Melanoma antigen rec by T cells (MART-1, Mlana) | |
| Immunoglobulin lambda 5 | IgE Fc receptor, low affinity II, alpha | B-lymphocyte cell-surface antigen B1 (CD20) | |
| Ig B-cell antigen receptor gene (mb-1) | Germ line IgH chain gene, DJC region | 2,5 oligoadenylate synthetase 2 (Oas2) | |
| Q2-k gene for class I antigen (MHC I) | Immunoglobulin heavy chain 6 of IgM | HLA-E, transketolase (Tkt) | |
| Recombinase activating gene-2 (Rag2) | IL-4-induced gene 1 (IL4i1) | Tumor necrosis factor receptor 1b (Tnfrsf1b) | |
| TRAF-interacting protein (Traip) | TNF-beta mRNA, lymphotoxin A | TNFR7, CD27 | |
| Uromodulin (Umod) | Lymphotoxin B | ||
| VpreB | Lymphocyte antigen 9 (Ly9) | ||
| Immunoglobulin-associated alpha (mb-1) | |||
| Myeloid-associated differentiation marker (Myadm) | |||
| TNF receptor superfamily (tnfrsf5, CD40) vpreB | |||
| Signaling | |||
| Raf-related oncogene (A-raf) | Adrenergic receptor, beta 2 (b2ar) | Casein kinase II (CKII) | Calmodulin 1 |
| Cornichon homolog (Cnih) | B lymphoid kinase (Blk) | Developmentally regulated GTP binding protein (Drg1) | Dual-specificity phosphatase 5 (Dusp5) |
| FK506 binding protein 1a (Flb 1a) | Bruton tyrosine kinase (Btk) | Growth factor receptor-bound protein 2 (Grb2) | Inositol 1,4,5 trisphosphate receptor (Itpr1) |
| Guanine nucleotide binding protein, alpha inhibiting (Gnai2) | Adapter protein CIKS (Act 1) | Hyaluronan-mediated motility receptor (RHAMM) | Phosphoinositide-binding protein (Pip3e) |
| Protein phosphatase 1B (Ppm1b) | tyr-thr dual specificity phosphatase PAC-1 (Dusp2) | Protein phosphatase 1, catalytic subunit (PP1) | Cytohesin binding protein HE (Pscdbp) |
| Pleckstrin homology, Sec7 and coiled/coil domain (Pscd3) | Growth hormone (Gh) | Receptor (calcitonin) activity modifying protein (Ramp2) | Ras-associated domain family 2 (Ral GDS/AF-6) |
| Rho interacting protein 3 (Rhoip3) | Glia maturation factor, gamma (Gmfγ) | RAS guanyl releasing protein 1 (Rasgrp1) | Wingless-type MMTV integration site (Wnt6) |
| Sel 1 inhibitor of Notch | Hematopoietic cell phosphatase (SHP) | Ras suppressor protein 1 (Rsp1) | |
| Serine/threonine kinase 10 (LOK, Stk 10) | Interferon regulatory factor 1 (IRF1) | vav 3 oncogene (Vav3) | |
| Spleen tyrosine kinase Syk | Lymphocyte antigen 57 (BLNK) | WW domain binding protein 11 (Wbp11) | |
| TPR-containing, SH2-binding phosphoprotein (Tsp) | Membrane-bound C2 domain containing (Mbc2) | ||
| Membrane-spanning 4-domains (Ms4a4d) | |||
| Nuclear receptor binding protein (Nrbp) | |||
| RAS p21 protein activator 3 (Rasa3) | |||
| Regulator of G-protein signaling 19 (Rgs19) | |||
| Ras-like without CAAX 1 (Rit1) | |||
| Small protein effector 1 of Cdc42 (Spec1) | |||
| SWAP complex protein, 70 kDa (Swap70) | |||
| Tubby-like protein 3 (Tulp3) | |||
| Transcription factors | |||
| E2A transcription factor | B-cell leukemia/lymphoma 6 (Bcl-6) | C-terminal binding protein 1 (CTBP1) | Capicua homolog (Cic) |
| E4F transcription factor 1 (E4f1) | E2A transcription factor | Early growth response 4 (Egr4) | HB9 homeobox transcription factor (H1xb9) |
| Early B-cell factor (EBF) | Early B-cell factor (EBF) | Enhancer of rudimentary homolog (Erh) | Sex comb on midleg-like 2 (Scml2) |
| Early development regulator 2 (Edr2, Mph2) | Kruppel-like factor LKLF mRNA (Kif2) | Nuclear receptor subfamily 4 (Nur77, Nr4a1) | RP58, zinc finger protein (Znf238) |
| E26 avian leukemia oncogene 2 (Ets2) | LIM only 2 (Lmo2) | PBX/knotted 1 homeobox 2 (KN1, Pknox2) | |
| Host cell factor C1 transcription factor (Hcfc1) | Max dimerization protein 4 (Mad4) | Wilms tumor 1-associating protein (Wtap) | |
| Lymphoid enhancer factor-1 (LEF-1) | Nuclear factor of kappa in B cells 2 (Nfkb2) | ||
| Nuclear transcription factor-Y gamma (NF-Yγ) | pax-5 transcription factor | ||
| Pax-5 | SCAN-KRAB-zinc finger gene 1 (Skz1) | ||
| Ets transcription factor PU.1 | SpiB Ets family member | ||
| Transcription factor-like 1 (YL-1, Tcfl1) | Tripartite motif protein 21 (Trim21) | ||
| T-cell leukemia, homeobox 2, hox11L1 (Tlx2) | |||
| Tripartite motif protein 25 (Trim25) | |||
| Ubiquitination | |||
| Autophagy 12-like (Apg121) | WW domain-containing protein 4 (Wwp2) | ||
| Ubiquilin 1 (Ubqln1) | |||
| Ubiquitin-specific protease 15 (Usp15) |
Murine bone marrow CD 19+ . | Murine spleen CD19+ . | Human BJAB + LMP2A . | Human LCL + LMP2A . |
|---|---|---|---|
| Cell cycle/apoptosis | |||
| SH3P9 myc box dependent interacting (Bin1) | SH3P9 myc box-dependent interacting (Bin1) | BTG family, member 3 (Btg3) | ATM kinase |
| Caspase-6 | B-cell translocation gene 2 (Btg2) | Cyclin A2 | RBP-1-like protein (Bcaa) |
| EGL nine homolog 1 (Egln 1) | p53 variant | Cell division cycle 2 (Cdc2) | pptase and tensin (PTEN) |
| Ubiquinol-cytochrome c reductase (Ubcr) | Proline-rich protein expressed in brain (Prtb) | CHK 1 checkpoint homolog (Chek1) | Retinoblastomaa susceptibility (Rbl) |
| Signal-induced proliferation associated (Spa-1) | CDC28 protein kinase 2 (Cks2) | ||
| (tre-2/USP6, BUB2, cdc16) domain (Tbc 1d1) | Defender against cell death 1 (Dad 1) | ||
| Transgene insert site 737 (TgN737) | Programmed cell death 4 (Pdcd4) | ||
| Pituitary tumor-transforming 1 (Pttg 1) | |||
| Ubiquinol-cytochrome C reductase (Ucrq) | |||
| YME1-like 1 (Yme 1I1) | |||
| DNA/RNA metabolism | |||
| Heterogeneous nuclear riboprotein H2 | H1 histone family, member 2 | Eukaryotic translation initiation factor (eIF2s2) | eIF-1A |
| H2B histone family, member S | Polymerase (RNA) II (DNA directed) | Eukaryotic translation initiation factor (eIF4a1) | Histone H2A/g (H2AFG) |
| DNA-binding nuclear protein NP220 | Polymerase (RNA) II (DNA directed) | High-mobility group box 2 (Hmgb2) | Histone H2A.2 (H2AFO) |
| Polymerase (RNA) II (DNA directed) | RAD50 homolog | Heterogeneous nuclear ribonucleoprotein (Hnrph1) | Histone H2A.X (H2AFX) |
| RADI homolog | Rael RNA export 1 homolog | Mitochondrial ribosomal protein 63 | Histone H2B/h |
| Trf-proximal protein homolog (Trfp) | Ribosomal protein L21 | Mitochondrial ribosomal protein L3 | Histone H2B/e |
| Zinc finger protein 292 | Zinc finger protein 36 (TISII) | High mobility group-like nuclear protein (Nhp2) | Histone H2B1 S |
| Zinc finger protein, subfamily 1A, 4 | Polymerase (RNA) II (DNA directed) polypeptide J | Histone H2BFC | |
| DNA repair protein rad18 | Histone H2BFD | ||
| RAD1 homolog | Histone H2B/k (H2BFK) | ||
| RNA-binding region containing (RNPI, RRM) | Histone H2BFL | ||
| Ribosomal protein L26-like 1 | Histone H2BFR | ||
| Ribosomal protein L38 | H4 histone family G (H4FG) | ||
| Zinc finger protein 9 | |||
| Inflammation/immunity | |||
| Colony-stimulating factor 1 receptor (Csf 1r) | Adenosine A2a receptor (Adora2a) | Allograft inflammatory factor 1 (Aif1) | Butyrophilin (BTF4) |
| Deoxynucleotidyltransferase, terminal (TdT) | Complement component 1r | Complement component 1q receptor 1 | CD69 |
| Histocompatibility 2, class II antigen A | CD19 antigen | Complement component 4A | IFN-induced leucine zipper protein 35 (Ifi35) |
| Histocompatibility 2, Q region locus 7 | CD22 antigen | Integrin, beta 2 (LFA-1) | IFN-induced protein 44 (Ifi44) |
| Interferon (alpha and beta) receptor 2 | Histocompatibility 2, D region locus 1 | Ig kappa chain V-I region (Igk) | IgG lambda light chain V-J-C region |
| Germ line immunoglobulin V(H)II gene | Histocompatibility 2, class II, locus DMa | T-cell receptor alpha locus (TCR alpha) | Interleukin-7 (IL-7) |
| Immunoglobulin lambda chain, variable 1 | IFN-γ inducible protein 30 (Ifi30) | Pre-B lymphocyte gene 3 (vpreB) | Interferon regulatory factor 7B (IRF7b) |
| Interleukin-16 (IL-16) | IFN-γ inducible protein (Ifi47) | Interferon-induced protein (IRF4/Pip/Isg15) | |
| Integrin beta 4 binding protein (Itgb4bp) | Delta-immunoglobulin (IgD) | Melanoma antigen rec by T cells (MART-1, Mlana) | |
| Immunoglobulin lambda 5 | IgE Fc receptor, low affinity II, alpha | B-lymphocyte cell-surface antigen B1 (CD20) | |
| Ig B-cell antigen receptor gene (mb-1) | Germ line IgH chain gene, DJC region | 2,5 oligoadenylate synthetase 2 (Oas2) | |
| Q2-k gene for class I antigen (MHC I) | Immunoglobulin heavy chain 6 of IgM | HLA-E, transketolase (Tkt) | |
| Recombinase activating gene-2 (Rag2) | IL-4-induced gene 1 (IL4i1) | Tumor necrosis factor receptor 1b (Tnfrsf1b) | |
| TRAF-interacting protein (Traip) | TNF-beta mRNA, lymphotoxin A | TNFR7, CD27 | |
| Uromodulin (Umod) | Lymphotoxin B | ||
| VpreB | Lymphocyte antigen 9 (Ly9) | ||
| Immunoglobulin-associated alpha (mb-1) | |||
| Myeloid-associated differentiation marker (Myadm) | |||
| TNF receptor superfamily (tnfrsf5, CD40) vpreB | |||
| Signaling | |||
| Raf-related oncogene (A-raf) | Adrenergic receptor, beta 2 (b2ar) | Casein kinase II (CKII) | Calmodulin 1 |
| Cornichon homolog (Cnih) | B lymphoid kinase (Blk) | Developmentally regulated GTP binding protein (Drg1) | Dual-specificity phosphatase 5 (Dusp5) |
| FK506 binding protein 1a (Flb 1a) | Bruton tyrosine kinase (Btk) | Growth factor receptor-bound protein 2 (Grb2) | Inositol 1,4,5 trisphosphate receptor (Itpr1) |
| Guanine nucleotide binding protein, alpha inhibiting (Gnai2) | Adapter protein CIKS (Act 1) | Hyaluronan-mediated motility receptor (RHAMM) | Phosphoinositide-binding protein (Pip3e) |
| Protein phosphatase 1B (Ppm1b) | tyr-thr dual specificity phosphatase PAC-1 (Dusp2) | Protein phosphatase 1, catalytic subunit (PP1) | Cytohesin binding protein HE (Pscdbp) |
| Pleckstrin homology, Sec7 and coiled/coil domain (Pscd3) | Growth hormone (Gh) | Receptor (calcitonin) activity modifying protein (Ramp2) | Ras-associated domain family 2 (Ral GDS/AF-6) |
| Rho interacting protein 3 (Rhoip3) | Glia maturation factor, gamma (Gmfγ) | RAS guanyl releasing protein 1 (Rasgrp1) | Wingless-type MMTV integration site (Wnt6) |
| Sel 1 inhibitor of Notch | Hematopoietic cell phosphatase (SHP) | Ras suppressor protein 1 (Rsp1) | |
| Serine/threonine kinase 10 (LOK, Stk 10) | Interferon regulatory factor 1 (IRF1) | vav 3 oncogene (Vav3) | |
| Spleen tyrosine kinase Syk | Lymphocyte antigen 57 (BLNK) | WW domain binding protein 11 (Wbp11) | |
| TPR-containing, SH2-binding phosphoprotein (Tsp) | Membrane-bound C2 domain containing (Mbc2) | ||
| Membrane-spanning 4-domains (Ms4a4d) | |||
| Nuclear receptor binding protein (Nrbp) | |||
| RAS p21 protein activator 3 (Rasa3) | |||
| Regulator of G-protein signaling 19 (Rgs19) | |||
| Ras-like without CAAX 1 (Rit1) | |||
| Small protein effector 1 of Cdc42 (Spec1) | |||
| SWAP complex protein, 70 kDa (Swap70) | |||
| Tubby-like protein 3 (Tulp3) | |||
| Transcription factors | |||
| E2A transcription factor | B-cell leukemia/lymphoma 6 (Bcl-6) | C-terminal binding protein 1 (CTBP1) | Capicua homolog (Cic) |
| E4F transcription factor 1 (E4f1) | E2A transcription factor | Early growth response 4 (Egr4) | HB9 homeobox transcription factor (H1xb9) |
| Early B-cell factor (EBF) | Early B-cell factor (EBF) | Enhancer of rudimentary homolog (Erh) | Sex comb on midleg-like 2 (Scml2) |
| Early development regulator 2 (Edr2, Mph2) | Kruppel-like factor LKLF mRNA (Kif2) | Nuclear receptor subfamily 4 (Nur77, Nr4a1) | RP58, zinc finger protein (Znf238) |
| E26 avian leukemia oncogene 2 (Ets2) | LIM only 2 (Lmo2) | PBX/knotted 1 homeobox 2 (KN1, Pknox2) | |
| Host cell factor C1 transcription factor (Hcfc1) | Max dimerization protein 4 (Mad4) | Wilms tumor 1-associating protein (Wtap) | |
| Lymphoid enhancer factor-1 (LEF-1) | Nuclear factor of kappa in B cells 2 (Nfkb2) | ||
| Nuclear transcription factor-Y gamma (NF-Yγ) | pax-5 transcription factor | ||
| Pax-5 | SCAN-KRAB-zinc finger gene 1 (Skz1) | ||
| Ets transcription factor PU.1 | SpiB Ets family member | ||
| Transcription factor-like 1 (YL-1, Tcfl1) | Tripartite motif protein 21 (Trim21) | ||
| T-cell leukemia, homeobox 2, hox11L1 (Tlx2) | |||
| Tripartite motif protein 25 (Trim25) | |||
| Ubiquitination | |||
| Autophagy 12-like (Apg121) | WW domain-containing protein 4 (Wwp2) | ||
| Ubiquilin 1 (Ubqln1) | |||
| Ubiquitin-specific protease 15 (Usp15) |
At least 2 separate, identical experiments comparing gene transcription in LMP2A-positive versus LMP2A-negative cells were performed for each cell type. Statistical analyses of the arrays were performed as described in “Materials and methods.” Genes with probability values of less than .01 and differential ratios of less than 1.1 in LMP2A-positive cells compared with LMP2A-negative cells are listed.
Real-time PCR
Total RNA was extracted from CD19+ murine bone marrow and splenic B cells using Trizol reagent (Invitrogen). RNA (5 μg) was reverse transcribed according to the Superscript first-strand synthesis system for reverse transcription-polymerase chain reaction (RT-PCR) protocol (Invitrogen). Aliquots of the RT reactions were used in PCR reactions containing 4 mM MgCl2, 1.3 μM each primer, and 1 × DNA Master SYBR Green 1 (Roche Molecular Biochemicals, Indianapolis, IN). PCR reactions were amplified on a LightCycler (Roche Molecular Biochemicals) under the following conditions: 45 cycles of 95°C for 1 second, 60°C for 5 seconds, and 72°C for 15 seconds, followed by a melting curve cycle. A standard curve was generated for each set of PCR primers in order to measure the concentration of PCR product. The levels of each PCR product were then normalized to those of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to determine the relative amount of gene expression. Oligonucleotides for PCR were as follows: PU.1 sense, 5′-GAGAACTTCCCTGAGAACCACTTC-3′; PU.1 antisense, 5′-CATGTAGGAAACCTGGTGACTGAG-3′; Ig-α (Mb-1) sense, 5′-CCTCTCCTCCTCTTCTTGTCATAC-3′; Ig-α (Mb-1) antisense, 5-GTTAGACTGAAGGCTGAACCACCA-3′; Ki67 sense, 5′-AGACTCTCCAAGACAGGTCTCAGT-3′; Ki67 antisense, 5′-GTGACCTTCTCTTACTCCCTGATG-3′; proliferating cell nuclear antigen (PCNA) sense, 5′-CCACTCCACTGTCTCCTACAGTAA-3′; PCNA antisense, 5′-GTGACAGAAAAGACCTCAGGACAC-3′; Bcl-xL sense, 5′-CGTATCAGAGCTTTGAGCAGGTAG-3′; Bcl-xL antisense, 5′-GGCTCTAGGTGGTCATTCAGATAG-3′; Survivin sense, 5′-CTTGAAACTGGACAGACAGAGAGC-3′; Survivin antisense, 5′-CCACTGTCTCCTTCTCTAAGATCC-3′; IL-13R sense, 5′-GGAGAGGAGGAGGGAAAGATAG-3; IL-13R antisense, 5′-CACAGGCAGTTATCACAAGACC-3′; EBV-induced gene 3 (EBI3) sense, 5′-TCTTCTCTCTCAAGTACCGACTCC-3′; and EBI3 antisense, 5′-GGGTCTGATATCAAGGATCCAGTC-3′. GAPDH oligonucleotide sequences have been published previously.25
Results
Overall changes in gene expression induced by LMP2A in murine cells in vivo
In order to identify the molecular basis for the B-cell abnormalities observed in LMP2A transgenic mice, we used DNA microarray analysis to directly compare gene transcription in B cells from LMP2A transgenic mice and nontransgenic littermates. To identify alterations in gene transcription that may be induced by LMP2A during B-cell development, CD19+ bone marrow B cells were selected on methylcellulose containing IL-7 and used for microarray analysis. Nonactivated, purified CD19+ splenic B cells from nontransgenic and LMP2A transgenic mice were also used to identify transcriptional alterations maintained by LMP2A in peripheral B cells. Identical, duplicate experiments were carried out for each tissue comparison.
Of more than 10 000 unique transcripts, expression of 227 genes was statistically different in bone marrow B cells from LMP2A transgenic mice compared with the same cells from nontransgenic littermates. Of the 227 genes, 148 (65%) were up-regulated and 79 (35%) were down-regulated (Figure 1A). Similarly, of the 298 genes affected by LMP2A in splenic B cells, 203 (68%) were up-regulated and 95 (32%) were down-regulated (Figure 1A), suggesting that LMP2A induces similar transcriptional changes in both developing and peripheral B cells.
Overall changes in gene expression induced by LMP2A. (A) The percentage of genes increased or decreased by LMP2A expression in various cell types is indicated. Gene expression in primary murine bone marrow and splenic B cells from LMP2A transgenic mice was compared with that in B cells from nontransgenic littermates. Gene expression in LMP2A-positive human B-lymphoma lines (BJAB) and EBV-transformed LCLs was compared with that in LMP2A-negative BJABs and LCLs, respectively. (B) Genes were annotated and placed in categories based on protein function. The percentages of genes in each category relative to the overall changes in gene expression are shown.
Overall changes in gene expression induced by LMP2A. (A) The percentage of genes increased or decreased by LMP2A expression in various cell types is indicated. Gene expression in primary murine bone marrow and splenic B cells from LMP2A transgenic mice was compared with that in B cells from nontransgenic littermates. Gene expression in LMP2A-positive human B-lymphoma lines (BJAB) and EBV-transformed LCLs was compared with that in LMP2A-negative BJABs and LCLs, respectively. (B) Genes were annotated and placed in categories based on protein function. The percentages of genes in each category relative to the overall changes in gene expression are shown.
Overall changes in gene expression induced by LMP2A in human cells in vitro
We also compared gene expression in 2 types of human cell lines expressing LMP2A in order to determine whether the LMP2A-mediated alterations in gene transcription that we observed in murine B cells in vivo are also induced in human B cells. We used an established EBV-negative B-lymphoma cell line, BJAB, that has been genetically engineered to express LMP2A as well as LCLs infected with LMP2A-positive and -negative EBV. By comparing gene expression in LMP2A-positive and -negative EBV-infected LCLs, we can separate the biologic effects of LMP2A from those of other viral latency genes. In LMP2A-expressing BJAB cells, we found that expression of approximately 159 genes was altered when compared with LMP2A-negative BJABs. Of the 159 transcripts, 75 (47%) were up-regulated and 84 (53%) were down-regulated (Figure 1A). Similarly, of the 139 genes whose transcription was altered by LMP2A expression in EBV-infected LCLs, 77 (55%) were up-regulated and 62 (45%) were down-regulated (Figure 1A). Together these results suggest that overall changes in gene expression in LMP2A-expressing murine bone marrow and splenic B cells freshly isolated in vivo are more similar to each other than to the human cell lines, whereas the human LMP2A-expressing BJABs and LCL lines are more similar to each other than to the murine primary cells.
Categorization of genes affected by LMP2A
In order to further define the effects of LMP2A in B cells, genes identified from each microarray were placed into categories based on protein function. Those genes with unknown function or whose category placement was unclear were designated as other/unknown. Figure 1B shows the relative percentage of genes in each category whose expression was altered by LMP2A in the different cell types. Overall, the patterns of gene expression were similar in each of the LMP2A-expressing B cells. For instance, roughly 10% of the genes whose expression was altered by LMP2A in bone marrow, spleen, BJAB, and LCLs are associated with protein transport and sorting (Figure 1B, pink bars). Similarly, LMP2A-mediated transcriptional changes in genes encoding ubiquitination components, transcription factors, signaling molecules, enzymes and protein metabolism factors, cytoskeletal elements, mediators of cell-cell interactions, and cell cycle and apoptosis proteins were comparable in all of the cell types tested (Figure 1B). In contrast, LMP2A-induced alterations in tumor-associated genes were observed only in bone marrow and splenic B cells and BJAB cells. When compared with BJAB and LCL cell lines, a larger percentage of genes involved in inflammation and immunity were affected by LMP2A in primary murine B cells, most likely due to the physiologic constraints of the improperly developed B cells in vivo. The category of genes most affected by LMP2A in human cell lines included genes involved in DNA/RNA metabolism, suggesting that LMP2A expression induces a general deregulation of DNA replication and gene transcription (Figure 1B). Taken together, these results indicate that many of the LMP2A-induced transcriptional changes in both primary murine B cells and human B-cell lines involve genes that encode functionally similar proteins.
Genes altered by LMP2A
Genes in which transcription was altered by LMP2A expression in murine and human B cells are listed in Tables 1 (increased) and 2 (decreased). These tables contain only the categories of genes that we believe may be associated with the development of EBV-associated malignancies. Tables S1 and S2, which list the genes contained in the remaining categories, can be viewed on the Blood website—see the Supplemental Tables link at the top of the online article. It is possible that genes listed in the supplemental tables may also contribute to EBV-associated pathologies; however, it was necessary to limit the amount of information presented to the focus of this article.
One category of genes in which expression was altered by LMP2A included those encoding proteins involved in cell cycle induction and inhibition of apoptosis. Expression of many cell cycle genes was increased by LMP2A, including Cyclin D1, Cdk5, Cdk9 (bone marrow); Cdc20, Cdc2a, Cdc6, Cks1, Cyclin A2, Cyclin B2 (spleen); and Cyclin F, Cyclin I (LCLs) (Table 1). Other indicators of an active cell proliferative state were increased by LMP2A, including Pcna, Prothymosin alpha (bone marrow); Ki67 (Spleen); Numa1 (BJAB); and Plk, Rcl (LCL). Genes encoding antiapoptotic proteins were also increased by LMP2A, including Survivin (bone marrow and spleen) and Api6, Bcl-xL (spleen). Expression of many negative regulators of cell cycle induction was inhibited by LMP2A, including Bin1 (bone marrow and spleen); Btg2 (spleen); Btg3 (BJAB); and Bcaa, Rb1 (LCL) (Table 2). In addition, expression of various proapoptotic genes was decreased, including Caspase-6, Ubcr (bone marrow); P53 (spleen); Pdcd4, Ucrq (BJAB); and Atm, Pten (LCL). These results indicate that LMP2A alters gene transcription to favor cell cycle induction and inhibition of apoptosis, either of which may contribute to the development of B-cell lymphoma.
As mentioned previously, transcription of many genes involved in DNA and RNA metabolism was altered by LMP2A expression. In both the murine primary B cells and the human B-cell lines, LMP2A expression resulted in increased expression of genes involved in chromatin modification, DNA replication, and RNA transcription/translation. These genes encode various types of proteins, including histones, zinc finger proteins, helicases, polymerases, eukaryotic translation factors, ribosomal proteins, and tRNA synthetases (Table 1). Additionally, murine bone marrow and/or splenic B cells from LMP2A transgenic mice demonstrated increased expression of genes encoding histone methyltransferases, base excision repair proteins, high mobility group (Hmg) proteins, mini chromosome maintenance-deficient (Mcmd) proteins, ribonucleotide reductases, and topoisomerases (Table 1). In addition to splicing factors, the LMP2A-expressing BJABs and LCLs expressed increased levels of genes encoding the Swi-Snf and Baf53 chromatin remodeling components, respectively. LMP2A expression in primary murine B cells and/or human B-cell lines resulted in decreased expression of genes encoding histones, polymerases, zinc finger proteins, Rad family proteins, heterogeneous nuclear ribonucleoproteins, and ribosomal proteins (Table 2). In addition, LMP2A-expressing human cell lines exhibited decreased expression of eukaryotic translation factors and BJABs expressed decreased levels of Hmg proteins. These results indicate that LMP2A alters chromatin remodeling and general transcription and translation regulation in cells, which could significantly alter normal cellular functions.
Expression of many genes encoding proteins involved in immunity was altered in bone marrow and splenic B cells from LMP2A transgenic mice, including immunoglobulin transcripts, complement components, Ly family members, tumor necrosis factor (TNF) members, and interferon (IFN)-induced proteins (Table 1). Specific genes in which expression was induced by LMP2A included Cd84, Il-5, Il-13r, Imap (bone marrow) and Cd7, Cd9, Cd68, Ebi3 (spleen). In contrast, there were few immunity-associated factors increased in LMP2A-expressing BJABs and none induced in LCLs. As we have previously reported, LMP2A globally down-regulates the expression of B-cell-specific factors in bone marrow B cells from transgenic mice, including Ig-α (Mb-1), Rag2, VpreB, λ5, and Tdt.25 VpreB, Ig-α (Mb-1), and Ig genes, in addition to Cd19 and Cd22, were down-regulated in splenic B cells from LMP2A transgenic mice (Table 2). Interestingly, several genes encoding proteins involved in immune regulation were also down-regulated in LMP2A-expressing primary murine B cells and human cell lines, including immunoglobulins, interleukins, complement components, TNF members, and IFN-induced proteins. In addition, bone marrow and splenic B cells from LMP2A transgenic mice expressed decreased levels of genes encoding MHC class I and II proteins (Table 2). LMP2A-expressing human cell lines demonstrated decreased levels of immunity-associated genes, including Lfa-1, Tcr alpha, VpreB (BJAB) and Cd69, Il-7, Cd20 (LCL). These results indicate that LMP2A interferes with the expression of a variety of factors involved in inflammation and immunity, both positively and negatively, when expressed in B cells.
Many genes encoding signaling proteins were unregulated by LMP2A in murine primary B cells and human cell lines, including G-protein signaling components, Ras-associated factors, and various phosphatases and kinases (Table 1). Additional factors increased by LMP2A in murine bone marrow and/or splenic B cells included Notch and Wnt signaling components and Socs-1, Enc1, C-kit, and Zap-70. Many genes encoding signaling proteins were decreased in both murine primary B cells and human cell lines, including Ras-associated factors, various kinases and phosphatases, and G-protein signaling components (Table 2). Specific genes involved in B-cell signaling and decreased by LMP2A expression included Syk (bone marrow); Blk, Btk, Shp, Blnk, Swap70 (spleen); CkII, Grb2, Vav3 (BJAB); and Calmodulin 1, Itpr1, Pip3 (LCL). These results further suggest that LMP2A inhibits normal B-cell signaling events, most likely by interfering with numerous signaling pathways.
Expression of genes encoding transcription factors was also increased by LMP2A in all cell types tested, including various homeobox transcription factors, Gli1, Tcf-1, Ap-2, Pax-3 (bone marrow); C/ebp, Creg, Dp1, Etv6, Ews, Id2, Tal1, Tcf-7, Xbp (spleen); Nfkbia (BJAB); and Atf5, Creg, Cutl1, Fkhrl1 (LCL) (Table 1). As we have previously reported, many transcription factors induced during B-cell development and critical for proper B-cell function were repressed in bone marrow and/or splenic B cells from LMP2A transgenic mice, including Pu.1, E2a, Ebf, Pax-5, Lef-1, and Spi-B (Table 2).25 Other transcription factors repressed by LMP2A included E4f, Ets2, Nf-y (bone marrow); Bcl-6, Lmo2, Nfkb2 (spleen); and Egr4, Nur77 (BJAB). These results indicate that LMP2A interferes with a variety of transcription factors necessary for proper B-cell development, activation, and Ig rearrangement, which are all necessary for proper B-cell function.
Several tumor-associated genes were up-regulated by LMP2A in murine primary B cells and BJABs, including Lag, Npn2, Pim1, Tcl1β1 (bone marrow); Evi2, Lag, Pim2 (spleen); and Ptov1 (BJAB) (Table 1). Although the exact function of many of these genes is unknown, expression of each has been shown to exhibit a strong correlation with malignant development. Interestingly, expression of genes encoding components of the proteosome complex was up-regulated by LMP2A in all cell types tested (Table 1). We have previously shown that LMP2A associates with Nedd4 family ubiquitin ligases and that this association results in the internalization and degradation of LMP2A and its associated proteins; therefore, it is intriguing that LMP2A appears to also induce transcription of the ubiquitination components as well.26,28,29
Similar patterns of gene expression in LMP2A-expressing cells and HRS cells
A comparison of the microarray data from this study and results from microarray analyses published recently has indicated that expression of many genes is similarly altered in LMP2A-expressing cells and HRS cells of Hodgkin lymphoma. Surprisingly, many of the similarities exist only in primary bone marrow and splenic B cells from LMP2A transgenic mice and not in the LMP2A-expressing human cell lines. As we have shown previously, LMP2A induces a global down-regulation of B-cell-specific transcription factors and signaling molecules, many of which are also significantly down-regulated in HRS cells.25 These include E2a, Ebf, Pax-5, Pu.1, Ig-α (Mb-1), Rag2, Cd19, Cd22, Blk, Swap70, Syk, Blnk, Spi-B, Cd20, Lfa-1, and Igk (Table 3). Such transcriptional alterations induced by LMP2A may explain why the identity of HRS cells as germinal center B cells has been unclear until recently.
LMP2A-expressing cells and Hodgkin lymphoma
Gene* . | Expression . | Tissue† . | Reference no. . |
|---|---|---|---|
| B-cell factors/signaling | |||
| E2A (E47) | Decreased | BM, spleen | 30 |
| Early B-cell factor (EBF) | Decreased | BM, spleen | 30 |
| Pax-5 (BSAP) | Decreased | BM, spleen | 30,31 |
| PU.1 | Decreased | BM | 30,32,33 |
| Ig-α (Mb-1) | Decreased | BM, spleen | 33,34 |
| Rag2 | Decreased | BM | 35 |
| CD19 | Decreased | Spleen | 33,34 |
| CD22 | Decreased | Spleen | 33,34 |
| Blk | Decreased | Spleen | 33,34 |
| SWAP70 | Decreased | Spleen | 33,34 |
| Syk | Decreased | BM | 34 |
| Blnk | Decreased | Spleen | 33 |
| Spi-B | Decreased | Spleen | 33 |
| CD20 | Decreased | LCL | 33 |
| LFA-1 | Decreased | BJAB | 33 |
| Igk | Decreased | BJAB | 33 |
| Cell cycle/apoptosis | |||
| Ki67 | Increased | Spleen | 36 |
| Cyclin A | Increased | Spleen | 37 |
| PCNA | Increased | BM | 38 |
| Bcl-xL | Increased | Spleen | 37 |
| Survivin | Increased | BM, spleen | 37 |
| Bcl-6 | Decreased | Spleen | 34 |
| Immunity | |||
| IL-5 | Increased | BM | 39 |
| IL-13R | Increased | Spleen | 40 |
| EB13 | Increased | Spleen | 41 |
Gene* . | Expression . | Tissue† . | Reference no. . |
|---|---|---|---|
| B-cell factors/signaling | |||
| E2A (E47) | Decreased | BM, spleen | 30 |
| Early B-cell factor (EBF) | Decreased | BM, spleen | 30 |
| Pax-5 (BSAP) | Decreased | BM, spleen | 30,31 |
| PU.1 | Decreased | BM | 30,32,33 |
| Ig-α (Mb-1) | Decreased | BM, spleen | 33,34 |
| Rag2 | Decreased | BM | 35 |
| CD19 | Decreased | Spleen | 33,34 |
| CD22 | Decreased | Spleen | 33,34 |
| Blk | Decreased | Spleen | 33,34 |
| SWAP70 | Decreased | Spleen | 33,34 |
| Syk | Decreased | BM | 34 |
| Blnk | Decreased | Spleen | 33 |
| Spi-B | Decreased | Spleen | 33 |
| CD20 | Decreased | LCL | 33 |
| LFA-1 | Decreased | BJAB | 33 |
| Igk | Decreased | BJAB | 33 |
| Cell cycle/apoptosis | |||
| Ki67 | Increased | Spleen | 36 |
| Cyclin A | Increased | Spleen | 37 |
| PCNA | Increased | BM | 38 |
| Bcl-xL | Increased | Spleen | 37 |
| Survivin | Increased | BM, spleen | 37 |
| Bcl-6 | Decreased | Spleen | 34 |
| Immunity | |||
| IL-5 | Increased | BM | 39 |
| IL-13R | Increased | Spleen | 40 |
| EB13 | Increased | Spleen | 41 |
Genes listed are similarly expressed in LMP2A-expressing cells and Hodgkin lymphoma cell lines or HRS cells.
Lists the tissue types in which gene expression was affected in LMP2A transgenic mice.
Increased expression of cell cycle and antiapoptotic proteins is thought to play an important role in the development of Hodgkin lymphoma. Expression of various cell cycle and antiapoptotic genes was increased in both LMP2A-expressing primary murine B cells and HRS cells, including Ki67, Cyclin A, Pcna, Bcl-xL, and Survivin (Table 3). Interestingly, decreased expression of Bcl-6, a transcription factor that regulates B-cell differentiation, was observed in both LMP2A-expressing splenic B cells and HRS cells. Transcription of 3 immunologic factors that suppress cell-mediated immunity, Il-5, Il-13r, and Ebi3, was up-regulated in both LMP2A-expressing primary B cells and HRS cells. IL-13 has been shown to be an autocrine growth factor for HRS cells, so expression of the IL-13 receptor would likely contribute to proliferation of HRS cells.42,43 EBI3 is an IL-12 p40 homolog that has been suggested to negatively interfere with IL-12 signaling.44
Expression levels of several key genes involved in B-cell development, cell cycle induction, apoptosis inhibition, and immunologic suppression were confirmed by real-time RT-PCR. We have previously shown by semiquantitative RT-PCR a 2- to 3-fold reduction in expression of the transcription factors E2A, EBF, and Pax-5 in B cells from LMP2A transgenic mice.25 Figure 2 demonstrates that, relative to the levels of expression in B cells from nontransgenic littermates, transcription of the B-cell factors Pu.1 and Ig-α (Mb-1) was down-regulated 2- to 3-fold in bone marrow and splenic B cells, respectively, from LMP2A transgenic mice. Additionally, expression of genes encoding the cell cycle inducers Pcna and Ki67 was increased in bone marrow and splenic B cells, respectively, from LMP2A transgenic mice. Importantly, expression levels of the antiapoptotic genes Bcl-xL and Survivin as well as the immunologic factors IL-13R and Ebi3 were each significantly increased in B cells from LMP2A transgenic mice. These results, taken together, strongly suggest that many of the transcriptional alterations characteristic of HRS cells are induced by LMP2A.
Quantitative real-time RT-PCR to measure changes in gene expression induced by LMP2A. Total RNA was isolated from CD19+ B cells from LMP2A transgenic mice and nontransgenic littermates. RNA was reverse transcribed and used in real-time PCR reactions using SYBR green to label double-stranded DNA products. Normalized levels of gene expression in B cells from LMP2A transgenic mice are shown relative to the levels in nontransgenic mice (set to a value of 1). Error bars represent the standard deviations of 4 separate experiments. WT indicates wild type. *Transcript levels were compared in splenic B cells, except for PU.1 and PCNA, in which expression was altered only in bone marrow B cells.
Quantitative real-time RT-PCR to measure changes in gene expression induced by LMP2A. Total RNA was isolated from CD19+ B cells from LMP2A transgenic mice and nontransgenic littermates. RNA was reverse transcribed and used in real-time PCR reactions using SYBR green to label double-stranded DNA products. Normalized levels of gene expression in B cells from LMP2A transgenic mice are shown relative to the levels in nontransgenic mice (set to a value of 1). Error bars represent the standard deviations of 4 separate experiments. WT indicates wild type. *Transcript levels were compared in splenic B cells, except for PU.1 and PCNA, in which expression was altered only in bone marrow B cells.
Similar patterns of gene expression in LMP2A-expressing cells and germinal center centroblasts
HRS cells are presumed derived from germinal center B cells that should have undergone apoptosis due to a lack of Ig expression.7,9 In a recent study, DNA microarray analysis was used to identify transcriptional changes that occur in germinal center B cells during the transition stages from naive B cells to activated, proliferating centroblasts, to noncycling centrocytes, and finally to memory B cells. Microarray analysis demonstrated that transition to the centroblast/centrocyte stage involves up-regulation of proliferation-associated genes and down-regulation of cytokine, chemokine, and adhesion receptors. Furthermore, the final transition to memory B cells is characterized by the return to a gene profile similar to naive B cells.45 A comparison of gene transcription in LMP2A-expressing cells has identified LMP2A-induced changes in gene expression that resemble those in germinal center centroblasts. In particular, increased expression of genes encoding cell cycle and proliferative proteins (Cyclin A2, Cyclin B2, Cyclin F, Cdc2, Cdc6, Cdc20, Cdk5, Pcna, Ki67, Mad2, Plk, Centromere protein A, Rpa3, Dp1, Tal-1, Kinesin-like 6, Rab6) as well as proteins involved in DNA metabolism (Hmg2, Ogg1, TopIIα) was identified in LMP2A-expressing cells and germinal center centroblasts (Table 4). Similarly, decreased expression of genes encoding immunologic factors (Ifn-αr2, IgD, Shp, Ccr1, Ccr6, Cd69) was observed in LMP2A-expressing cells and centroblasts (Table 4). These results suggest that LMP2A expression during EBV infection of B cells may induce and maintain a gene program that resembles an activated, proliferating germinal center centroblast, which may result in the development of Hodgkin lymphoma if left unchecked by the host's immune system.
LMP2A-expressing cells and germinal center centroblasts/centrocytes
Gene* . | Tissue† . |
|---|---|
| Increased | |
| Cyclin A2 | Spleen |
| Cyclin B2 | Spleen |
| Cyclin F | LCL |
| Cdc2 | Spleen |
| Cdc6 | Spleen |
| Cdc20 | Spleen |
| CDK5 | BM |
| PCNA | BM |
| Ki67 | Spleen |
| Mad2 | Spleen |
| PLK | LCL |
| Centromere protein A | LCL |
| RPA3 | LCL |
| Dp1 | Spleen |
| Tal-1 | Spleen |
| Kinesin like-6 | LCL |
| Rab6 | Spleen |
| HMG2 | Spleen |
| Ogg1 | LCL |
| Toplla | Spleen, LCL |
| CD9 | Spleen |
| Decreased | |
| Bin1 | BM, spleen |
| IFNAR2 | BM |
| IgD | Spleen |
| SHP | Spleen |
| CCR1 | BM |
| CCR6 | Spleen |
| CD69 | LCL |
Gene* . | Tissue† . |
|---|---|
| Increased | |
| Cyclin A2 | Spleen |
| Cyclin B2 | Spleen |
| Cyclin F | LCL |
| Cdc2 | Spleen |
| Cdc6 | Spleen |
| Cdc20 | Spleen |
| CDK5 | BM |
| PCNA | BM |
| Ki67 | Spleen |
| Mad2 | Spleen |
| PLK | LCL |
| Centromere protein A | LCL |
| RPA3 | LCL |
| Dp1 | Spleen |
| Tal-1 | Spleen |
| Kinesin like-6 | LCL |
| Rab6 | Spleen |
| HMG2 | Spleen |
| Ogg1 | LCL |
| Toplla | Spleen, LCL |
| CD9 | Spleen |
| Decreased | |
| Bin1 | BM, spleen |
| IFNAR2 | BM |
| IgD | Spleen |
| SHP | Spleen |
| CCR1 | BM |
| CCR6 | Spleen |
| CD69 | LCL |
Genes listed are similarly expressed in LMP2A-expressing B cells and germinal center B cells upon transition to centroblasts/centrocytes.33
Lists the cell types in which gene expression was affected by LMP2A in LMP2A transgenic mice and LMP2A-positive EBV-infected LCLs.
Discussion
In this study, we have analyzed gene transcription in LMP2A-expressing cells by DNA microarray analysis. We compared gene transcription in primary bone marrow and splenic B cells from LMP2A transgenic mice, human BJAB lines expressing LMP2A, and LMP2A-positive and -negative LCLs. We have found that bone marrow and splenic B cells from LMP2A transgenic mice exhibit similar patterns of gene transcription induced by LMP2A. Additionally, we have observed that transcriptional alterations induced by LMP2A in EBV-infected LCLs (versus LMP2A-negative EBV-infected LCLs) more closely resemble patterns observed in primary murine B cells than those in LMP2A-expressing BJAB cells. For instance, in contrast to primary murine B cells and LCLs, many genes involved in cell cycle induction were down-regulated by LMP2A expression in BJABs, including Cyclin A2, Cdc2, and Cks2 (Table 2). One reason for these differences may be that the LCLs were generated by immortalization of primary B cells in vitro by EBV infection. In contrast, the BJAB cell line is an established, immortalized line that contains pre-existing alterations in genes important for cell survival. It is likely that LMP2A-mediated transcriptional alterations would more closely resemble those in primary murine B cells and LCLs following BCR ligation in BJABs since alterations in downstream BCR signaling events in LMP2A-expressing BJABs have been clearly documented.14,16 Although the specific genes affected by LMP2A were different in the various cell types analyzed, they were often functionally similar, suggesting that LMP2A targets specific cellular pathways.
It is intriguing that transcriptional changes similar to those demonstrated in LMP2A-expressing cells have been described in HRS cells of Hodgkin lymphoma, particularly genes involved in cell cycle induction, apoptosis inhibition, and suppression of cell-mediated immunity (Table 3; Figure 2). These correlations not only implicate a role for LMP2A in the development of Hodgkin lymphoma, but provide further insight into the mechanisms used by LMP2A to establish viral latency in peripheral B cells. By allowing EBV-infected cells to avoid immune recognition and apoptosis induction, LMP2A is able to successfully establish and maintain viral latency. In addition, it has been well documented that Hodgkin lymphoma cells use various strategies to mediate immune evasion.46 The results of this study also indicate that LMP2A may be involved in the down-regulation of many B-cell-specific factors in HRS cells, which has made it difficult to determine the cellular origin of these cells until recently.7,9 It is important to note that many of the transcriptional abnormalities discussed here have been documented in EBV-negative as well as EBV-positive HRS cells. In fact, no clear correlation has been found between the expression of many of these genes and the EBV status of the HRS cells analyzed. This would suggest that a factor other than LMP2A may be responsible for altered gene regulation in EBV-negative HRS cells. However, it is possible that these cells were initially infected with EBV, but have lost viral gene expression during the late stages of malignant development. Additionally, it has been suggested that EBV-negative HRS cells may contain an as yet unidentified infectious agent, which may express a functionally homologous protein to LMP2A.47 Finally, altered gene expression in EBV-negative HRS cells may be due to a spontaneous genetic event that duplicates the effects of LMP2A.
An additional, striking correlation was made between transcriptional alterations in LMP2A-expressing cells and activated, proliferating germinal center centroblasts, particularly increased expression of genes involved in cell cycle induction and DNA metabolism and decreased expression of immunologic factors (Table 4). We propose that LMP2A expression in EBV-infected naive B cells induces transcriptional changes indicative of an activated, proliferating centroblast; however, constitutive signaling by LMP2A maintains the cells in a metabolically activated state, expressing cell cycle proteins, antiapoptotic proteins, and proteins involved in immune evasion. Since HRS cells are presumed to be derived from germinal center B cells that have lost Ig expression and should have undergone apoptosis,7,9 the results of this study suggest that, similar to the peripheral Ig-negative B cells in LMP2A transgenic mice, LMP2A allows HRS cells to survive and remain unrecognized by the immune system.
EBI3, which likely functions as an IL-12 antagonist, has recently been shown to be expressed in both HRS cells and EBV-immortalized LCLs.41,44 It has previously been shown that EBV LMP1 induces EBI3 expression through NF-κB activation; however, our studies suggest that LMP2A can also induce transcription of EBI3 in peripheral B cells in vivo. Additionally, although alterations in NF-κB transcription were not observed in LMP2A-expressing cells, a comparison of our results with a study identifying gene targets of NF-κB activation48 has demonstrated that expression of several NF-κB-induced genes was altered by LMP2A. These included increased expression of Wnt10, Pim-2, Pip5k, Mapk, Ras, Cyclin D, Igk, Oxyr, IgH, Ebi3, Caspase-11, and Pla2 (Tables 1,S1) and decreased expression of Lef-1, Rgs, VpreB, λ5, Gst, and Villin (Tables 2,S2). NF-κB activation has been shown to be required for proliferation and survival of not only peripheral B cells,49 but more specifically for proliferation and survival of HRS cells.50,51 Therefore, LMP2A may be able to up-regulate the expression of cell cycle and survival genes through activation of the NF-κB pathway, similar to LMP1. LMP2A also appears to affect the transcription of genes that are normally regulated by the Ras signaling pathway, which may provide another mechanism for increased cell survival.
Microarray data from LMP2A-expressing cells were also compared with analyses involving B cells transformed with v-abl, which blocks B-cell development.52 Many similarities were identified, including increased expression of Siat, Slc, Aif, Prdx5, Fkbp4, Cytochrome c, Eif-1, Csda, Gcn5, and C/ebp (Tables 1,S1) and decreased expression of Adam, Blnk, Ets, Tubby-like, Ig-α (Mb-1), Mad, Spi-B, Presinilin, Bcl-6, Ebf, Il-16, Btg, Tdt, Syk, Rag2, and Lef-1 (Tables 2,S2). Many of the B-cell developmental transcription factors whose expression was altered by LMP2A are normally regulated by the Wnt and Notch pathways, both of which play an important role in cell fate decisions. Interestingly, increased Notch signaling has been correlated with cell proliferation and survival in Hodgkin lymphoma and is proposed to be a contributing factor to the lack of B-cell-specific gene transcription in HRS cells.53 The effects of LMP2A on each of these cellular pathways should be further studied to elucidate the molecular mechanisms used by LMP2A to maintain viral latency and contribute to the development of human malignancies.
In summary, we have identified changes in gene transcription induced by LMP2A in both primary murine B cells and human B-cell lines. These results have identified many genes that are similarly altered in HRS cells of Hodgkin lymphoma and suggest that LMP2A may allow for the survival and persistence of HRS cells that lack a functional BCR. Future studies that address the mechanisms by which specific LMP2A-induced changes in gene expression promote the survival of B cells may provide better insight into the ways in which LMP2A maintains latency in EBV-infected B lymphocytes and how this contributes to the development of malignancies.
Prepublished online as Blood First Edition Paper, August 7, 2003; DOI 10.1182/blood-2003-04-1018.
R.L. is supported by Public Health Service grants CA62234, CA73507, and CA93444 from the National Cancer Institute and DE13127 from the National Institute of Dental and Craniofacial Research. R.L. is a Stohlman Scholar of the Leukemia and Lymphoma Society of America. T.P. is a Special Fellow of the Leukemia and Lymphoma Society of America. Microarray experiments were supported by a Howard Hughes microarray analysis support grant from the
Center for Genetic Medicine at Northwestern University.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Yonghong Zhang and Dr Eric Bremer from the CMIER facility for helping with the microarray hybridizations and data analysis. We would like to thank members of the Longnecker laboratory for help with these studies. In addition, we thank Dr Michelle Swanson-Mungerson for helpful discussions and kindly reading the manuscript prior to submission.

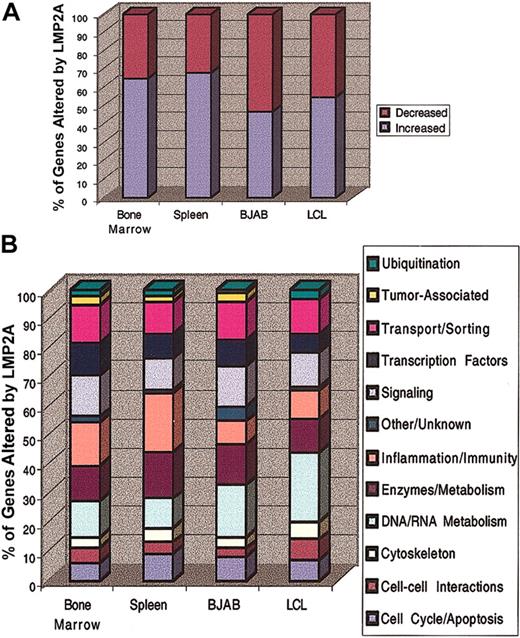

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal