Abstract
Cutaneous T-cell lymphoma (CTCL) is a malignancy of skin-homing T cells. A major feature of CTCL is profound immunosuppression, such that patients with advanced mycosis fungoides or Sézary syndrome have been compared with patients with advanced HIV disease and are susceptible to opportunistic infection. The etiology of this immunosuppression is unclear. We analyzed peripheral blood T cells of patients with CTCL with stage I to IV disease, using a sensitive beta-variable complementarity-determining region 3 spectratyping approach. Our data revealed a profound disruption of the complexity of the T-cell repertoire, which was universally observed in patients with advanced disease (stages III and IV), and present in up to 50% of patients with early-stage disease (stages I and II). In most patients, multiple monoclonal and oligoclonal complementarity-determining region 3 (CDR3) spectratype patterns in many different beta-variable families were seen. Equally striking was a reduction of normal T cells (as judged by absolute CD4 counts) across multiple beta-variable families. In general, CTCL spectratypes were reminiscent of advanced HIV spectratypes published elsewhere. Taken together, these data are most consistent with a global assault on the T-cell repertoire in patients with CTCL, a process that can be observed even in early-stage disease. (Blood. 2003;102:4059-4066)
Introduction
Primary cutaneous T-cell lymphomas (CTCLs) are a heterogeneous group of malignancies of memory T cells that home to skin.1,2 Mycosis fungoides (MF) and Sézary syndrome (SS) comprise most cases of CTCL. MF is characterized by erythematous patches, plaques, or tumors on the skin, with or without lymph node involvement. SS is characterized by erythroderma and the presence of malignant T cells in peripheral blood.3 Transformed T cells in CTCL are typically CD4+, produce T helper 2 (Th2) cytokines, and display skin-homing markers such as cutaneous lymphocyte antigen (CLA) and chemokine receptor CCR4.4-6 An important diagnostic correlate in SS is the identification of a unique T-cell receptor (TCR) rearrangement present in the circulating malignant T-cell population, indicative of an expanded (relatively or absolutely) transformed T-cell clone. Patients who present with early-stage MF often survive for decades, whereas those with advanced disease (stage III or higher) have survival times measured in months.7,8 Death is often the result of bacterial sepsis that has been attributed to an incompletely characterized immunodeficiency in the context of a compromised integument.9 A Th2 cytokine profile of the malignant cells, decreased T-cell responses to antigen, diminished T-cell cytotoxicity, as well as to a relative paucity of normal T lymphocytes, have all been suggested to contribute to this immunodeficiency.5,10,11
We have frequently observed a decreased number of normal T cells in patients with CTCL, particularly those with obvious peripheral blood involvement. This T lymphopenia is seen not only in SS patients in whom absolute T-cell counts are high because of an expanded population of leukemic malignant T cells11 but also in patients with normal total T-lymphocyte counts. In many of these latter patients, a large percentage of the T-cell population actually represents an expanded malignant clone, so that the absolute counts of normal nonmalignant T cells are greatly diminished. The cause of this relative lymphopenia is unknown but suggests a systemic dysregulation of normal T-cell production or survival. We designed the present study to examine the diversity of the T-cell repertoire in CTCL. In particular, we performed quantitative analysis of peripheral blood T-cell receptor beta-variable (BV) family expression by flow cytometry and used complementarity-determining region 3 (CDR3) spectratype analysis of the same peripheral blood T cells by using established BV primers. This latter technique is a measure of the complexity of the T-cell repertoire12,13 and is dramatically altered in diseases characterized by immunodeficiency, such as HIV infection,14 Omenn syndrome,15 and idiopathic CD4 lymphopenia.16
Our results indicate that there is a profound reduction in the complexity of the T-cell repertoire in CTCL that in advanced disease is comparable to that seen in HIV-infected patients. Although uniformly present in patients with stages III and IV disease, this loss of TCR complexity is also evident in some patients with very early disease (eg, stage IA). Moreover, our results indicate that a dramatic decrease of normal T cells occurs in a nonrandom fashion. Taken together, these results are consistent with a process that affects the entire T-cell population, leading to an oligoclonal dysplasia at the expense of the normal T-cell repertoire.
Patients, materials, and methods
Patients and control volunteers
Twenty patients with a histologically established diagnosis of CTCL were recruited from the Cutaneous Oncology Clinic at the Dana-Farber Cancer Institute after obtaining their informed consent. The characteristic of the patients are listed in Table 1. None of the patients were clinically infected at the time point of sampling. Six patients with psoriasis of moderate to severe severity (5 men, 1 woman; mean age, 63 years; range, 42-75 years), 1 patient with idiopathic erythroderma (male, 57 years), and 7 healthy control volunteers (3 men, 4 women; mean age, 58 years; range, 40-70 years) were also recruited for comparison. The patients with psoriasis were all being treated with phototherapy, and the patient with idiopathic erythrodermia had had 6 cycles of extracorporeal photochemotherapy (ECP) and methotrexate (MTX) 12.5 mg weekly. The study was approved by the institutional ethics committee.
Patient characteristics
Patient . | Sex/age, y . | Stage . | FACS analysis with anti-BV AB (% of BV + CD4 T cells) . | Antibodies, CD4 count/mm3 . | CD4/CD8 ratio . | Disease duration . | Current treatment . | Previous treatment . |
|---|---|---|---|---|---|---|---|---|
| Numerically expanded clone | ||||||||
| 1 | M/68 | IVB | BV 5.1 (73) | 538 | 2 | 1 y | PDN; topical steroids | MTX; UVB; etanercept (for concurrent arthritis) |
| 2 | F/50 | IVA | BV 14 (81) | 1293 | 14 | 3 y | Topical steroids; antihistamines | Topical steroids |
| 3 | M/50 | III | BV 18 (94) | 1727 | 13.5 | 4-5 y | 12 cycles ECP; MTX | PUVA; topical steroids |
| 4 | F/94 | III | BV 2 (96) | 8251 | 37.3 | 3 y | Topical steroids; antihistamines | PUVA; MTX, 25 mg |
| 5 | M/76 | III | BV 20 (93.6) | 838 | 13.3 | 5 y | 4 cycles ECP; IFN-α | Bexarotene; denileukin diftitox |
| 6 | F/72 | III | BV 1 (87) | 5965 | 7.9 | 3 y | 10 cycles ECP; bexarotene | Topical steroids; topical tacrolimus |
| Without an identifiable expanded population | ||||||||
| 7 | F/58 | IVA | No clone detectable | 223 | 4.4 | 5-6 y | 18 cycles ECP; IFN-α | Bexarotene; denileukin diftitox |
| 8 | M/61 | IV | No clone detectable | 1019 | 7.4 | 7 y | 4 cycles ECP; PDN | PUVA; IFN, total skin electron beam; bexarotene |
| 9 | M/74 | III | No clone detectable | 264 | 3.4 | 4 y | 6 cycles of ECP; topical steroids | Narrow UVB |
| 10 | M/59 | III | No clone detectable | 1005 | 2.1 | 6 mo | UVB (3 × wk) | None |
| 11 | M/72 | IB | No clone detectable | 701 | 7.2 | 4 y | 9 cycles ECP | PUVA |
| 12 | M/64 | IB | No clone detectable | 505 | 1.8 | 4 y | PUVA (2 × wk) | Topical steroids; nitrogen mustard |
| 13 | M/73 | IB | No clone detectable | 624 | 1.75 | 3 y | Denileukin diftitox | Denileukin diftitox; topical steroids; topical calcipotriol |
| 14 | M/70 | IIB | No clone detectable | 255 | 3.4 | 8 y | Topical steroids | PUVA; MTX; electron beam; ECP |
| 15 | F/53 | IB | No clone detectable | 811 | 1.2 | 5 y | Topical steroids and tacrolimus | PUVA; denileukin diftitox |
| 16 | M/77 | IB | No clone detectable | 443 | 1.1 | 3 y | Topical steroids | Topical steroids |
| 17 | F/30 | IA | No clone detectable | 460 | 0.5 | 3 y | Topical steroids | None |
| 18 | F/44 | IA | No clone detectable | 819 | 4.0 | 5 y | Topical steroids; MTX | Topical steroids |
| 19 | F/57 | IA | Not done | Not done | Not done | 10 y | No therapy (in spontaneous remission for 10 y) | None |
| 20 | F/60 | IA | Not done | Not done | Not done | 5 y | PUVA every 2 weeks | PUVA; topical steroids |
Patient . | Sex/age, y . | Stage . | FACS analysis with anti-BV AB (% of BV + CD4 T cells) . | Antibodies, CD4 count/mm3 . | CD4/CD8 ratio . | Disease duration . | Current treatment . | Previous treatment . |
|---|---|---|---|---|---|---|---|---|
| Numerically expanded clone | ||||||||
| 1 | M/68 | IVB | BV 5.1 (73) | 538 | 2 | 1 y | PDN; topical steroids | MTX; UVB; etanercept (for concurrent arthritis) |
| 2 | F/50 | IVA | BV 14 (81) | 1293 | 14 | 3 y | Topical steroids; antihistamines | Topical steroids |
| 3 | M/50 | III | BV 18 (94) | 1727 | 13.5 | 4-5 y | 12 cycles ECP; MTX | PUVA; topical steroids |
| 4 | F/94 | III | BV 2 (96) | 8251 | 37.3 | 3 y | Topical steroids; antihistamines | PUVA; MTX, 25 mg |
| 5 | M/76 | III | BV 20 (93.6) | 838 | 13.3 | 5 y | 4 cycles ECP; IFN-α | Bexarotene; denileukin diftitox |
| 6 | F/72 | III | BV 1 (87) | 5965 | 7.9 | 3 y | 10 cycles ECP; bexarotene | Topical steroids; topical tacrolimus |
| Without an identifiable expanded population | ||||||||
| 7 | F/58 | IVA | No clone detectable | 223 | 4.4 | 5-6 y | 18 cycles ECP; IFN-α | Bexarotene; denileukin diftitox |
| 8 | M/61 | IV | No clone detectable | 1019 | 7.4 | 7 y | 4 cycles ECP; PDN | PUVA; IFN, total skin electron beam; bexarotene |
| 9 | M/74 | III | No clone detectable | 264 | 3.4 | 4 y | 6 cycles of ECP; topical steroids | Narrow UVB |
| 10 | M/59 | III | No clone detectable | 1005 | 2.1 | 6 mo | UVB (3 × wk) | None |
| 11 | M/72 | IB | No clone detectable | 701 | 7.2 | 4 y | 9 cycles ECP | PUVA |
| 12 | M/64 | IB | No clone detectable | 505 | 1.8 | 4 y | PUVA (2 × wk) | Topical steroids; nitrogen mustard |
| 13 | M/73 | IB | No clone detectable | 624 | 1.75 | 3 y | Denileukin diftitox | Denileukin diftitox; topical steroids; topical calcipotriol |
| 14 | M/70 | IIB | No clone detectable | 255 | 3.4 | 8 y | Topical steroids | PUVA; MTX; electron beam; ECP |
| 15 | F/53 | IB | No clone detectable | 811 | 1.2 | 5 y | Topical steroids and tacrolimus | PUVA; denileukin diftitox |
| 16 | M/77 | IB | No clone detectable | 443 | 1.1 | 3 y | Topical steroids | Topical steroids |
| 17 | F/30 | IA | No clone detectable | 460 | 0.5 | 3 y | Topical steroids | None |
| 18 | F/44 | IA | No clone detectable | 819 | 4.0 | 5 y | Topical steroids; MTX | Topical steroids |
| 19 | F/57 | IA | Not done | Not done | Not done | 10 y | No therapy (in spontaneous remission for 10 y) | None |
| 20 | F/60 | IA | Not done | Not done | Not done | 5 y | PUVA every 2 weeks | PUVA; topical steroids |
PDN indicates prednisone; MTX, methotrexate; UVB, ultraviolet B; PUVA, Psoralens ultraviolet A; IFN-α, interferon α; and ECP, extracorporeal photochemotherapy.
Preparation of cells and isolation of T cells
Peripheral blood mononuclear cells (PBMCs) from patients and healthy donors were isolated from heparinized venous blood by density gradient centrifugation over Ficoll (Histopaque; Sigma, St Louis, MO). T cells and their subsets (CD3+, CD4+, CD8+) were selected with the use of immunomagnetic beads as described by the manufacturer (Miltenyi Biotec, Auburn, CA). The purity of the sorted population was analyzed by flow cytometry and ranged between 96% and 98%.
Dilution experiments
In dilution experiments 1 × 106 T cells from a healthy donor (representing a polyclonal population) were mixed with increasing numbers of monoclonal Jurkat cells (0% to 97%).
CDR3 size spectratyping
Total RNA was extracted from 3 to 5 × 106 cells with Trizol reagent (Life Technologies, Grand Island, NY). Total RNA (2-5 μg; A260/A280 = 1.9-2.1) was reverse transcribed by using oligo-dT primers and Powerscript reverse transcriptase (RT; Clontech, Palo Alto, CA). TCR BV segments were amplified with 1 of 24 BV subfamily-specific primers as well as constant beta (CB) primers that recognize both CB1 and CB2 regions as described previously.17 Sequences of BV 1 to 9, 11, 13 to 16, 18, and 20 primers were as in Choi et al18 ; of CB and BV 10 primers as in Genevee et al19 ; of BV 22, 24 as in Moss et al20 ; of BV 12 as in Hall and Finn21 ; of BV 17 and 19 as in Bragado et al22 ; and of BV 21 and 23 as in Hand et al.23 BV subfamilies are numbered as in Wei et al.24 Polymerase chain reaction (PCR) products were applied to a 5% polyacrylamide sequencing gel, and the size distribution of each fluorescent PCR product was determined by electrophoresis on an automated 377 DNA sequencer (Applied Biosystems, Foster City, CA). With this technique, an amplified TCR BV subfamily migrates as a series of bands, each one corresponding to a different CDR3 length, separated from one another by 3 nucleotides. Data were analyzed by using the GeneScan software (Applied Biosystems) that assigns a size and peak area to the different PCR products.
Direct sequencing of PCR products
Amplified PCR products of selected patients were purified by using the QIAquick PCR purification kit (Qiagen, Valencia, CA). The purified products were directly sequenced in both directions with BV- and CB-specific primers with the use of the ABI BigDye terminator sequencing kit (version 3) and a 3100 DNA sequencer (Applied Biosystems).
Flow cytometry
Three-color flow cytometric analysis was performed on PBMCs by using the following monoclonal antibodies against the TCR BV chain: phycoerythrin (PE)-conjugated antibodies to BV 1, BV 2, BV 5.1, BV 5.2, BV 5.3, BV 7, BV 9, BV 11, BV 12, BV 13.1, BV 13.6, BV 14, BV 16, BV 17, BV 18, BV 20, BV 21.3, BV 22 (Immunotech/Beckman Coulter, Brea, CA); PE-conjugated antibodies to BV 3, BV 8, BV 23 (BD Bioscience, San Diego, CA); CD3 peridinin chlorophyll (PerCp) and CD4 PerCp (BD Bioscience). Isotype controls used were immunoglobulin G1 (IgG1) PE, IgG2a PE, IgG2b PE, rat IgG1 PE (Immunotech/Beckman Coulter), and IgG1 PerCp (Becton Dickinson, San Jose, CA).
Integrated graphic representation by combining CDR3 spectratyping and the absolute numbers of BV+ CD4+ T cells
Qualitative alterations of the TCR repertoire obtained by CDR3 spectratyping were combined with the quantity of specific BV+ CD4- T cells (ie, absolute number of specific BV+ CD4+ T cells) for each BV subfamily and plotted in form of a “landscape.” A similar analysis has been described previously.25 The area under the entire CDR3 profile is estimated as 100%. If several peaks are present in a CDR3 profile, the absolute number of BV+ CD4+ T cells of this individual BV subfamily is segregated accordingly into several peaks. However, if only one peak occurs, it will represent the absolute number of CD4+ T cells of that particular BV subfamily. The overall TCR BV repertoire is then plotted as a function of the 24 BV subfamilies (x-axis), CDR3 length distribution (y-axis), and the absolute number of BV+ CD4+ T cells/mm3 (z-axis).
Scoring of CDR3 profiles and statistical analysis
Scoring of CDR3 profiles was performed by determining the number of contracted BV CDR3 size profiles in each subject. Contracted profiles were defined as follows: oligoclonal (1-4 peaks), monoclonal (1 peak), or absent (no peaks detectable). The analysis was performed by 2 different investigators in a blinded fashion. Statistical analysis was performed by using the Kruskal-Wallis nonparametric analysis of variance (ANOVA) test and subsequently by the Mann-Whitney U test (with the Bonferroni correction) for the unpaired samples. Correlation was tested by using the Spearman rank analysis. P < .05 was considered statistically significant.
Results
Patients with CTCL with a malignant clonal expansion have a highly contracted TCR repertoire
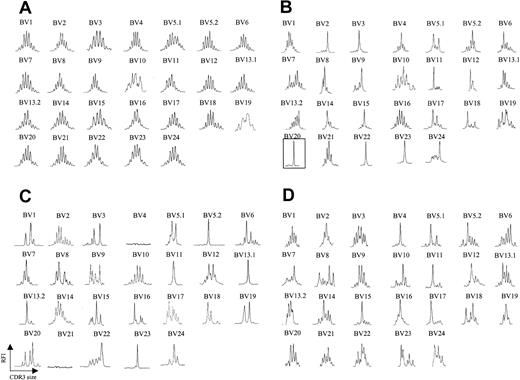
We analyzed peripheral blood from 20 patients with CTCL by fluorescence activated cell sorting (FACS) with a panel of antibodies specific for 21 different BV regions. Six patients were identified as having a clearly identifiable, numerically expanded population of a BV family, consistent with a circulating malignant CTCL clone. The abundance of the expanded clone ranged from 73% to 96% of total CD4+ T cells analyzed, and, in some (but not all) patients, the absolute CD4 count was elevated (Table 1). Although there are no definite phenotypic markers that identify the malignant clone, a high percentage (> 40%) of lymphocytes expressing a certain BV may represent a tentative criterion. Peripheral blood T cells were isolated from each of these patients, and CDR3 BV spectratyping was performed. Figure 1A shows a representative spectratype from peripheral blood T cells obtained from a healthy volunteer. A Gaussian distribution of CDR3 lengths is observed in all BV families examined, indicating a highly diverse T-cell repertoire across all BV families examined. In clear contrast, all 6 patients with identifiable circulating malignant T-cell populations had strikingly abnormal spectratypes, and, moreover, these abnormalities involved multiple BV families. A representative example is shown in Figure 1B. In each of these 6 patients, an apparent clonal spectratype signature of a single peak could be identified in the BV family that was expanded by FACS analysis, indicating close correlation of these techniques (eg, patient 5, Table 1; Figure 1B). Direct sequencing of the PCR products from these single spectratype peaks revealed a single sequence, consistent with an expanded clonal population of T cells (data not shown). Unexpectedly, however, apparent clonal populations could be also demonstrated in BV families that were not expanded by FACS analysis (Figure 1B). Additionally, spectratype patterns consisting of fewer than 5 peaks (which we will term oligoclonal), as well as apparent loss of BV families, were also noted. Absence of these BV families by spectratype analysis persisted even when an increased amount of cDNA was used and 40 cycles of PCR were performed. Taken together, these data suggested that in the disease process CTCL affects much of the T-cell population.
TCR BV CDR3 spectratyping profiles are strongly restricted in CTCL patients. (A) CDR3 spectratypes of CD3+ T cells from a representative healthy donor showing highly diverse profiles, reflecting a heterogeneous TCR repertoire. (B) A patient with stage III CTCL with a numerically expanded malignant clone (BV 20), demonstrating multiple monoclonal and oligoclonal profiles. (C-D) Similarly contracted profiles as well as some apparent deletions (BV 4, BV 21) are also found in patients with stage III CTCL without a numerically expanded clone (C) and even in some patients with stage IA CTCL (D). The y-axis represents relative fluorescence intensity (RFI), and the x-axis is CDR3 size (nucleotides).
TCR BV CDR3 spectratyping profiles are strongly restricted in CTCL patients. (A) CDR3 spectratypes of CD3+ T cells from a representative healthy donor showing highly diverse profiles, reflecting a heterogeneous TCR repertoire. (B) A patient with stage III CTCL with a numerically expanded malignant clone (BV 20), demonstrating multiple monoclonal and oligoclonal profiles. (C-D) Similarly contracted profiles as well as some apparent deletions (BV 4, BV 21) are also found in patients with stage III CTCL without a numerically expanded clone (C) and even in some patients with stage IA CTCL (D). The y-axis represents relative fluorescence intensity (RFI), and the x-axis is CDR3 size (nucleotides).
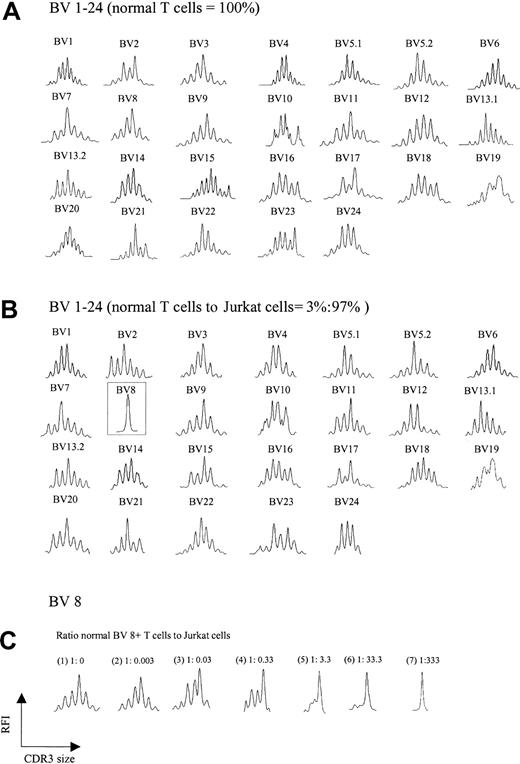
A trivial explanation for the aberrant spectratypes shown in Figure 1B was the possibility that abundant cDNA reverse transcribed from a single expanded clone somehow interferes with the detection of normal polyclonal CDR3 cDNA, creating a false impression of loss of complexity. To test this possibility, increasing numbers of Jurkat T-cell leukemia cells (BV8) were added to T cells from a healthy donor, and spectratyping was performed on the mixture. Figure 2A-B shows the comparison of the normal spectratype to the spectratype of a mixture containing 97% Jurkat/3% normal T cells. A normal background polyclonal spectratype appears in all BV families with the exception of BV 8, which shows a single peak, consistent with an abundance of Jurkat cells. Not until Jurkat cells exceeded 99% of the total mixture did subtle artifactual abnormalities begin to emerge in other BV families (not shown). Further experiments showed that for a given BV family, the appearance of a single peak on a spectratype profile required a ratio of about 1:300 normal to clonal T cells; lower ratios yielded more than one peak (Figure 2C). Thus, the ratio of an expanded clonal T cell to all other normal T cells in our patients would not be expected to interfere with the analysis of a normal spectratype. This expectation indicates that the abnormalities we observed in multiple BV families were an intrinsic feature of CTCL and not a technical artifact.
TCR BV repertoire in normal T cells and cell mixtures of these with Jurkat cells. (A) CDR3 spectratypes of T cells from a healthy donor showing highly diverse profiles reflecting a heterogeneous TCR repertoire. (B) Spectratype profiles of the cell mixtures containing 3% normal T cells and 97% Jurkat cells. As observed in panel A, polyclonal spectratype profiles were readily found across the other TCR BV subfamilies, even though 97% of the analyzed cells were composed of a clonal population (note the dominant peak in BV 8, representing Jurkat cells). (C) Spectratype profiles of BV 8 from pure normal T cells (1) and normal T cells spiked with increasing numbers of Jurkat cells (2-7). A single dominant peak was observed at a ratio of normal BV 8+ T cells to Jurkat cells of 1:333.
TCR BV repertoire in normal T cells and cell mixtures of these with Jurkat cells. (A) CDR3 spectratypes of T cells from a healthy donor showing highly diverse profiles reflecting a heterogeneous TCR repertoire. (B) Spectratype profiles of the cell mixtures containing 3% normal T cells and 97% Jurkat cells. As observed in panel A, polyclonal spectratype profiles were readily found across the other TCR BV subfamilies, even though 97% of the analyzed cells were composed of a clonal population (note the dominant peak in BV 8, representing Jurkat cells). (C) Spectratype profiles of BV 8 from pure normal T cells (1) and normal T cells spiked with increasing numbers of Jurkat cells (2-7). A single dominant peak was observed at a ratio of normal BV 8+ T cells to Jurkat cells of 1:333.
These data indicated that CTCL is not simply an expansion of a single malignant T-cell clone against the background of a normal T-cell repertoire, but rather a disease that affects the nonmalignant T-cell compartment as well. We next compared spectratypes of CD3+ T cells from a total of 20 patients with CTCL (including the 6 described earlier; Table 1) to those of 7 healthy donors, 6 patients with widespread psoriasis, and 1 patient with episodic idiopathic erythroderma who did not have CTCL. In a subset of these patients, CD3 T cells were separated into CD4 and CD8 subsets prior to analysis. In parallel, we examined peripheral blood T cells from these patients by flow cytometry. Absolute CD4 counts were reported for patients with CTCL by the clinical laboratories at Brigham and Women's Hospital.
Patients with all stages of CTCL show markedly abnormal CDR3 spectratypes, whereas control donors and patients with psoriasis have normal spectratypes
There was a significant correlation of degree of spectratype abnormality with stage of CTCL (rspearman = 0.69; P < .05), with 100% of patients with stage III or higher showing markedly abnormal spectratypes (eg, 5 or more contracted profiles). Although some patients with stage I disease had normal spectratype profiles (not shown), other patients with stage I disease had significant abnormalities of their T-cell repertoire, seemingly out of proportion to their limited disease involvement (Figures 1D and 3A). A similar significant (rspearman = 0.54; P < .05) correlation with stage could be seen when the number of single (monoclonal) peaks were enumerated in patients with different stages of CTCL (Figure 3B). In data described earlier, we noted that a clone must be present in a roughly 300:1 ratio to all other T cells of the same BV to display as a single peak. Although only 50% of patients with stages I and II disease showed monoclonal peaks, such single peaks could be detected in 100% of patients with stages III and IV disease. In contrast, no monoclonal peaks could be identified when spectratypes from healthy volunteers or patients with psoriasis were analyzed. In addition, a patient with idiopathic episodic erythroderma had an essentially normal spectratype (not shown). When the number of contracted profiles in spectratypes of CD3 (Figure 3C-D), CD4, and CD8 (not shown) positive T cells from healthy volunteers, patients with psoriasis, and patients with CTCL were compared, there was a significant difference between patients with CTCL and both patients with psoriasis and healthy volunteers. There was no significant difference between healthy volunteers and patients with psoriasis compared in this fashion. Thus, persistent antigenic stimulation of the immune system per se (eg, psoriasis) does not result in spectratype abnormalities in peripheral blood T cells. Analysis of both CD4 and CD8 T cells from patients with CTCL demonstrated spectratypes similar to those seen in CD3 T cells; therefore, the aberrant BV spectratypes were not a result of a single CD4 clone and multiple CD8 clones (not shown).
Correlation of degree of spectratype abnormality with stage of disease. A significant correlation was found between the number of contracted (A) and monoclonal (B) spectratypes and the clinical stage. (C) Quantification and statistical analysis of the number of contracted profiles in CTCL (▪), psoriasis (▴), and healthy donors (▾). A contracted profile was defined as having 0 to 4 distinct peaks. CDR3 spectratyping was performed from CD3+ T cells. Statistically significant perturbations of the TCR repertoire of patients with CTCL were found as compared with patients with psoriasis and healthy control donors. Horizontal bars show mean values. Statistical analysis is indicated. (D) Quantification and statistical analysis of the number of monoclonal profiles in patients with CTCL (▪) and psoriasis (▴) and healthy donors (▾). Markedly increased numbers of monoclonal profiles were found in patients with CTCL as compared with patients with psoriasis and healthy control donors. Horizontal bars show mean values. (E-F) BV families appear to be affected in a nonrandom fashion. The percentage of patients with CTCL with contracted (E) and monoclonal (F) profiles is shown.
Correlation of degree of spectratype abnormality with stage of disease. A significant correlation was found between the number of contracted (A) and monoclonal (B) spectratypes and the clinical stage. (C) Quantification and statistical analysis of the number of contracted profiles in CTCL (▪), psoriasis (▴), and healthy donors (▾). A contracted profile was defined as having 0 to 4 distinct peaks. CDR3 spectratyping was performed from CD3+ T cells. Statistically significant perturbations of the TCR repertoire of patients with CTCL were found as compared with patients with psoriasis and healthy control donors. Horizontal bars show mean values. Statistical analysis is indicated. (D) Quantification and statistical analysis of the number of monoclonal profiles in patients with CTCL (▪) and psoriasis (▴) and healthy donors (▾). Markedly increased numbers of monoclonal profiles were found in patients with CTCL as compared with patients with psoriasis and healthy control donors. Horizontal bars show mean values. (E-F) BV families appear to be affected in a nonrandom fashion. The percentage of patients with CTCL with contracted (E) and monoclonal (F) profiles is shown.
BV families appear to be affected in a nonrandom fashion
We did not observe a random distribution of BV oligoclonal and monoclonal profiles across all BV families (Figure 3E-F). For example, BV 23 was abnormal (oligo or monoclonal) in 45% of the CTCL patients examined. When only patients in stages III and IV were analyzed, abnormalities in BV 23 were seen in 60% of patients. In contrast, certain BV families (eg, BV 5.2, 6, 9, 20) were rarely affected. Some patients with CTCL showed apparent deletions of BV families (eg, BV 4 and 21 in Figure 1C), even when higher amounts of cDNA and 40 cycles of PCR were used. Most frequently absent was BV 21, which was undetectable in 30% of patients with CTCL (not shown), but was invariably present and normally complex in healthy control donors and patients with psoriasis.
Monoclonal BV spectratype peaks can be identified in patients who do not have expansion of the same BV family by FACS analysis, and such peaks correlate with a single clonal population of T cells by sequence analysis
We have described earlier the correlation of single spectratype peaks with clonal expansions identified by FACS analysis and single sequences confirmed by sequence analysis in 6 patients. In an additional 9 patients with CTCL, spectratypes that showed at least one monoclonal BV family were obtained. Although in 7 cases (in 5 different patients) this occurred in BV families for which we did not have a matching antibody, in 24 cases (in 10 different patients), FACS analysis of the BV family that showed a single peak by spectratype did not show relative expansion as compared with healthy control values of BV. Sequencing of PCR products from the spectratype analysis of these single peaks again showed only a single sequence, consistent with a clonal population of T cells (data not shown).
Analysis of clonal and oligoclonal populations in BV families that are not numerically expanded by FACS analysis suggests a widespread loss of normal T cells
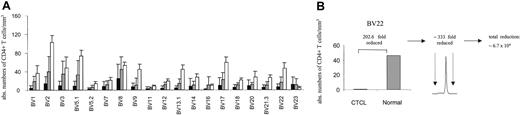
By combining spectratype data with absolute CD4 counts and BV FACS analysis, we attempted to assess the nature of the apparent loss of repertoire complexity in CTCL, both qualitatively and quantitatively. Figure 4A shows average absolute CD4 counts in each BV subfamily for patients with stage I/II CTCL (n = 7) and stage III/IV (n = 10, without the numerically expanded clones), compared with normal reference values.26 For patients with stage III/IV disease, T-cell numbers in all BV families were reduced with the exception of BV 23. An instructive example is shown in Figure 4B. In this patient, the absolute number of CD4+ BV 22+ cells is approximately 200-fold lower than normal values by FACS and cell count, yet a monoclonal pattern is shown in the BV 22 spectratype in this patient. If a monoclonal peak indicates that the ratio of clonal to normal T cells is 300, then one can conclude that the number of normal, nonclonal T cells in this BV family is at least 200 × 300, or 60 000-fold reduced as compared with normal values. Although imprecise, these data suggest that a significant loss of normal T cells is occurring across multiple BV families (at least in patients with stage III/IV) in a nonrandom fashion associated with a relative clonal expansion of BV-specific cells.
BV+ CD4+ T cells are markedly depleted in CTCL. (A) The absolute number of BV+ CD4+ T cells was calculated by determining the percentage of BV+ CD4+ T cells by flow cytometry and obtaining absolute CD4 cell counts. The means ± SDs of stage III/IV (▪; n = 10, without the numerically expanded clones) and stage I/II (▦) CTCL patients (n = 7) are shown in comparison to normal values (□) for each BV subfamily. Normal values represent the average number of CD4+ T cells (1100/mm3)26 and the normal mean percentage of BV+ CD4+ T cells in adult blood (mean ± SD are from a cohort of 85 normal specimens, according to Beckman Coulter). With the exception of BV 23 a markedly decreased number of BV+ CD4+ T cells were observed, particularly in patients with stage III/IV CTCL. (B) The combination of the reduction of absolute CD4+ values of specific BV subfamilies with a corresponding monoclonal spectratype profile reveals that a further depletion of BV 22+ CD4 T cells is actually present in this subfamily. An instructive example is shown.
BV+ CD4+ T cells are markedly depleted in CTCL. (A) The absolute number of BV+ CD4+ T cells was calculated by determining the percentage of BV+ CD4+ T cells by flow cytometry and obtaining absolute CD4 cell counts. The means ± SDs of stage III/IV (▪; n = 10, without the numerically expanded clones) and stage I/II (▦) CTCL patients (n = 7) are shown in comparison to normal values (□) for each BV subfamily. Normal values represent the average number of CD4+ T cells (1100/mm3)26 and the normal mean percentage of BV+ CD4+ T cells in adult blood (mean ± SD are from a cohort of 85 normal specimens, according to Beckman Coulter). With the exception of BV 23 a markedly decreased number of BV+ CD4+ T cells were observed, particularly in patients with stage III/IV CTCL. (B) The combination of the reduction of absolute CD4+ values of specific BV subfamilies with a corresponding monoclonal spectratype profile reveals that a further depletion of BV 22+ CD4 T cells is actually present in this subfamily. An instructive example is shown.
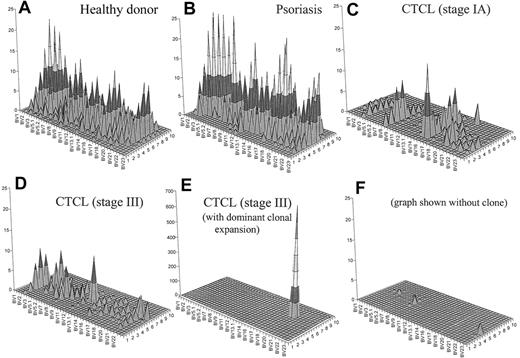
To generate an image that combines both CD4+ T-cell depletion and reduced T-cell repertoire complexity, we have generated topographic TCR “landscapes” that graphically demonstrate the loss of TCR repertoire complexity (Figure 5). A normal polyclonal diverse T-cell repertoire is shown in Figure 5A. A similarly complex landscape can be observed in psoriasis (Figure 5B). In contrast, a patient with CTCL with early-stage disease is shown in Figure 5C. The loss of complexity is evident even at this stage of disease in the absence of an expanded clone by FACS. Even more striking is the landscape of a patient with stage III disease without an identifiable expanded clone by FACS analysis (Figure 5D). In a patient with stage III with a numerically expanded clone identified by FACS, a single large peak appears to obscure the background (Figure 5E). However, even when this clone is subtracted from the landscape, there is still very little background complexity (Figure 5F).
Combination of a qualitative (CDR3 spectratyping) and quantitative (flow cytometry) analysis of the TCR repertoire. These graphs give a better visual assessment of the global reduction of the TCR diversity in CTCL. Panel A shows an overall diverse polyclonal CDR3 profile from a representative healthy donor. Panel B demonstrates a similarly diverse profile from a patient with psoriasis. Panel C shows a patient with CTCL with early-stage disease. The loss of complexity is evident even at this stage of disease in the absence of an expanded clone by FACS. Panel D shows a similarly restricted profile from a patient with stage III CTCL without a numerically expanded clone. (E) A highly restricted profile from a patient with CTCL with a malignant clone. (F) With this clone subtracted from the landscape, there is still very little background complexity. The x-axis displays the 24 BV subfamilies, the z-axis shows the CDR3 length distribution (in amino acids), and the z-axis indicates the absolute number of BV+ CD4+ T cells per cubic millimeter.
Combination of a qualitative (CDR3 spectratyping) and quantitative (flow cytometry) analysis of the TCR repertoire. These graphs give a better visual assessment of the global reduction of the TCR diversity in CTCL. Panel A shows an overall diverse polyclonal CDR3 profile from a representative healthy donor. Panel B demonstrates a similarly diverse profile from a patient with psoriasis. Panel C shows a patient with CTCL with early-stage disease. The loss of complexity is evident even at this stage of disease in the absence of an expanded clone by FACS. Panel D shows a similarly restricted profile from a patient with stage III CTCL without a numerically expanded clone. (E) A highly restricted profile from a patient with CTCL with a malignant clone. (F) With this clone subtracted from the landscape, there is still very little background complexity. The x-axis displays the 24 BV subfamilies, the z-axis shows the CDR3 length distribution (in amino acids), and the z-axis indicates the absolute number of BV+ CD4+ T cells per cubic millimeter.
Discussion
Cancers in general, and lymphomas in particular, are considered to be the result of an accumulation of multiple genetic lesions in a single cell.27,28 Occasionally, these genetic lesions are inherited but most are acquired somatically, often as the result of exposure to genotoxic agents. In most cases, a single transformed cell and its clonal progeny are responsible for the growth of a malignant population of cancer cells. Typically, nonmalignant cells of the same lineage do not appear altered. Certainly, the overwhelming majority of melanocytes are normal in patients with melanoma, and most lung epithelial cells appear normal (at least histologically) in patients with lung cancer. In late stages of leukemias, and lymphomas, normal hematopoietic populations are altered,29,30 but this alteration is typically ascribed to the mechanical and metabolic consequences of the overcrowding of anatomical compartments such as lymph nodes, peripheral blood, and bone marrow.
Our results with CTCL appear to be at odds with this paradigm. Rather than observing a single clonal population of malignant T cells against a background of normal T cells, we observed widespread and profound defects throughout the T-cell repertoire. In our study, we examined T-cell populations in patients with CTCL at the level of BV families by FACS profile and CDR3 spectratype. Of the 30 BV subfamilies, we were able to analyze approximately 70% by FACS and 80% by CDR3 spectratype, giving us a reasonable working overview of the T-cell repertoire. Our results show a global and profound reduction in the complexity of the T-cell repertoire, to a degree previously seen in patients with advanced HIV disease or after myeloablative bone marrow transplantation.14,17 In all patients examined with stage III and stage IV disease, a significant fraction of the BV repertoire is strikingly abnormal. This observation is also true in roughly half of all patients examined with stages I and II disease. Within each BV family, the expected Gaussian distribution of CDR3 lengths is not seen; in contrast, single peaks or a more limited number of lengths are observed in spectratype profiles. When sequenced, these single peaks represent single CDR3 sequences, indicating a clonal population of T cells.
Clonal T-cell populations are present in peripheral blood T cells in patients with CTCL with all stages of disease, and these populations often involve multiple different BV families. This finding of multiple clones in some patients is consistent with other reported results.31 Most often, these BV families containing clones are not numerically expanded relative to the total CD4 count by FACS analysis. However, they do appear to be expanded at the expense of normal T cells within their BV family. Possible explanations for this expansion are that a set of reactive clones is generated to an antigenic stimulus, or that there is a regulatory mechanism such that some T cells expand to fill the empty “space” created by the loss of other T-cell populations. In addition, in several patients certain BV families are undetectable. Although the numerically expanded clones in the 6 patients we analyzed did not favor any BV family, the BV spectratype abnormalities in our patients with CTCL overall were nonrandom. For example, BV 23 spectratypes were abnormal in 45% of all patients with CTCL, and in 60% of patients with stage III and IV. BV 21 spectratypes were undectable in 30% of all patients and in 40% of patients with stage III and IV. Certain BV families nearly always showed normal spectratypes, including BV 5.2, 6, 9, and 20.
Prior medical therapy of patients does not seem to be a contributing factor to the spectratype abnormalities we observed. Several of the patients with psoriasis had been on phototherapy or methotrexate, and their spectratypes were invariably normal. In addition, several of the patients with stage I had had no therapy more potent than topical steroids. Finally, our patient with idiopathic erythroderma had been on photopheresis and methotrexate for many months and had a completely normal spectratype. Although we cannot rule out the possibility that cytotoxic agents like denileukin diftitox (Ontak; Ligand Pharmaceuticals, NJ) depleted additional populations of normal T cells, we consider it unlikely, as we did not observe further drops in absolute CD4 counts in patients on this drug. Additionally, we would expect denileukin diftitox to deplete normal T cells in a BV- and clone-independent fashion. The fact that we saw similar abnormalities both in patients who were treated and not treated with denileukin diftitox make the possible contribution of this therapy unlikely. In summary, we could not attribute the significant changes in BV spectratypes observed to any prior therapy for this CTCL.
The reduction of TCR complexity in CTCL could be caused by a combination of different factors, including specific interactions with the tumor cells (eg, clonal expansion of T cells responding to CTCL-associated antigens) and inappropriate activation and/or suppression of T cells (ie, through activation-induced apoptosis by cytokines or by the liberation of free unprocessed antigen). Persistent antigenic stimulation has been invoked as a possible oncogenic stimulus for T cells in CTCL.32 Although antigenic stimuli may play some role in the abnormal spectratypes we see, our data with patients with psoriasis strongly suggest that persistent antigenic stimulation per se does not result in significant spectratype abnormalities in peripheral blood T cells. Psoriasis is widely believed to be an autoimmune T-cell-mediated disease that is chronic and incurable.33 Many of our patients with psoriasis had had their disease for many years and some for decades. Although occasional abnormal spectratypes were seen in some BV families, clonal patterns were never seen in these patients, and on balance the complexity of the T-cell repertoire in psoriasis was comparable to that in healthy individuals. In certain elderly patients with no evidence for disease, clonal expansions of single cells in a BV family yielding a single peak by spectratype have been noted.34-36 Invariably, however, the remainder of BV spectratypes in such patients is normal. Typically, these benign clonal expansions occur predominantly in CD8+ cells and may be related to prior Epstein-Barr virus (EBV) or cytomegalovirus (CMV) viral infection.
We have shown that the spectratype patterns that we observed are not a dilutional artifact—more than 97% of cells in a mixture of normal and leukemic cells can be clonal, and the patterns of unrelated BV families are normally complex. However, a significant amount of evidence has accumulated to suggest that the size of the T-cell compartment in peripheral blood is tightly controlled under normal conditions.37 Depletion of large numbers of normal T cells leads to antigen-independent proliferation and expansion of normal naive T cells. Similarly, there is a process by which large numbers of normal T cells that have clonally expanded in response to antigen ultimately undergo apoptosis, leaving only a fraction of the population to survive as long-lived memory cells. Our data in patients with CTCL are most consistent with a chronic process that results in widespread T-cell dyscrasia, accompanied by T-cell depletion. In this setting, surviving T cells may include those with a growth and survival advantage, and these favored cells may expand to occupy the putative depleted T-cell niches. Such cells include not only the dominant malignant clone but also other clones that may have accumulated genetic lesions that favor growth and/or survival without permitting unregulated autonomous expansion. Our spectratypes are in some ways similar to those of patients receiving T-cell-depleted bone marrow after myeloablative therapy for leukemia.17 In such patients, the small number of T cells transplanted along with hematopoietic progenitor cells appear to expand in an antigen-independent fashion. Because so few T cells are transferred, they do not represent all possible BV families, or CDR3 lengths within those families. The spectratypes on peripheral blood of such patients after several months (when T cells have presumably expanded to fill empty niches) are quite similar to our patients with stage III and IV CTCL.
The other clinical disorder in which comparable spectratypes are observed is in advanced HIV infection.14 In such patients, wholesale depletion of CD4+ cells is observed, but the spectratype patterns are similar to those of patients with advanced-stage CTCL. Losses in BV family complexity are seen across multiple families; however, whether this is due to compensatory expansion of certain clones of CD4+ cells with a growth advantage is unknown. In both patients with HIV-1 and patients with advanced-stage CTCL (data not shown), the appearance of T-cell receptor excision circles (TREC) DNA is very low.38 Because TRECs are only seen in naive T cells that have not clonally expanded, these data are consistent with compensatory proliferation of T cells attempting to fill the relatively empty T-cell microenvironment in peripheral blood.
Finally, it is intriguing that only certain BV families appear to be abnormal in CTCL. As noted earlier, BV 23 is frequently affected, and BV 21 is frequently absent, in these patients. In contrast, BV 5.2, 6, 9, and 20 are rarely abnormal. The only known stimuli that affect T cells at the level of BV sequences are superantigens.39 Viral and bacterial superantigens have been reported to bind to and activate T cells via the BV segment of the T-cell receptor, independent of the antigen recognition site. Superantigens from Staphylococcus aureus have been implicated in oligoclonal T-cell expansion in SS.40 We are unaware of any known bacterial superantigens that have affinity for BV 23 or 21, and the spectrum of viral superantigens is at present incomplete. Whether superantigens play a role in the compensatory T-cell proliferation that we propose occurs in these patients is at present unknown.
It has been appreciated for many years that patients with CTCL, particularly those with advanced disease, are profoundly immunosuppressed. The cause of death in these patients is typically infection9 and less frequently unrestrained tumor growth. In fact, patients with CTCL have been compared clinically with patients with advanced HIV-1 with regards to their immune status. In the present study, we report that the T-cell repertoire is dramatically abnormal in patients with CTCL. This is always the case in advanced disease but can also be observed in patients with stage IA disease, including some with single patches involving less than 5% of their surface area. We do not have a clear-cut explanation for this dramatic loss of complexity, but we feel that the presence of an agent that affects the T-cell population as a whole is the most logical interpretation. Whether this agent is an undiscovered T-cell tropic retrovirus, a different microbial pathogen, or a noninfectious process is at present unknown. Because of the striking similarities of CTCL spectratypes and HIV-1 spectratypes, we favor the first possibility. Previous reports implicating the lymphotropic retrovirus HTLV-1 (human T-cell lymphotrophic virus type 1) in CTCL have so far remained conflicting,41,42 but involvement of a virus related to HTLV-1 remains a possibility. The precise nature of such a putative agent is the subject of intensive study.
Prepublished online as Blood First Edition Paper, June 26, 2003; DOI 10.1182/blood-2003-04-1044.
Supported by a Specialized Program of Research Excellence (SPORE) grant in skin cancer from the National Cancer Institute, and a Dermatology Foundation Research Grant. N.Y. was supported by grants from the Novartis Foundation and Foundation Rene Touraine.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal