Abstract
Mediastinal large B-cell lymphoma (MLBCL) is a recently identified subtype of diffuse large B-cell lymphoma (DLBCL) that characteristically presents as localized tumors in young female patients. Although MLBCL has distinctive pathologic features, it clinically resembles the nodular sclerosis subtype of classical Hodgkin lymphoma (cHL). To elucidate the molecular features of MLBCL, we compared the gene expression profiles of newly diagnosed MLBCL and DLBCL and developed a classifier of these diseases. MLBCLs had low levels of expression of multiple components of the B-cell receptor signaling cascade, a profile resembling that of Reed-Sternberg cells of cHL. Like cHLs, MLBCLs also had high levels of expression of the interleukin-13 (IL-13) receptor and downstream effectors of IL-13 signaling (Janus kinase-2 [JAK2] and signal transducer and activator of transcription-1 [STAT1]), tumor necrosis factor (TNF) family members, and TNF receptor-associated factor-1 (TRAF1). Increased expression of STAT1 and TRAF1 in MLBCL was confirmed by immunohisto-chemistry. Given the TRAF1 expression and known link to nuclear factor–κB (NF- κB), MLBCLs were also evaluated for nuclear translocation of c-REL protein. In almost all cases, c-REL was localized to the nucleus, consistent with activation of the NF-κB pathway. These studies identify a molecular link between MLBCL and cHL and a shared survival pathway.
Introduction
Diffuse large B-cell lymphomas (DLBCLs) are heterogeneous tumors with variable clinical presentations and responses to therapy.1 At diagnosis, these aggressive B-cell malignancies involve a wide variety of nodal and extranodal disease sites; however, it is unclear whether tumors presenting in specific anatomic locations exhibit unique molecular features. One likely exception is the recently identified entity, primary mediastinal (thymic) large B-cell lymphoma (MLBCL), a subtype of DLBCL defined by a combination of clinical and pathologic features.2,3
Unlike DLBCL, which commonly arises in elderly patients of both sexes, MLBCL typically presents in younger women. These patients have bulky mediastinal masses with frequent invasion of adjacent structures.2 Although patients with MLBCL rarely have extrathoracic disease at diagnosis, extranodal sites including the central nervous system (CNS), liver, adrenals, and kidneys are often involved at relapse.4 MLBCLs typically consist of tumor cells with a pale cytoplasm and a diffuse growth pattern associated with variable degrees of sclerosis.3,5 However, there are no histologic features that reliably distinguish these tumors from DLBCLs that often involve mediastinal regional lymph nodes.
Although MLBCLs have rearranged immunoglobulin (Ig) genes and express B-cell lineage markers such as CD19, CD20, CD22, and CD79a (Igα), these tumors do not express Ig.2 In fact, the discordant expression of specific components of the B-cell receptor (BCR) (Ig- and CD79a+) is a unique feature of MLBCLs.6 The anatomic location, morphologic appearance, and immunophenotypic signature of MLBCLs suggest that these tumors arise from thymic medullary B cells.2,7 MLBCLs and a subpopulation of normal thymic medullary B cells express MAL, an integral membrane protein and lipid raft component that is rarely found in DLBCL.8
MLBCLs have mutated, class-switched Ig genes without evidence of ongoing somatic mutation.9 These tumors also exhibit several characteristic genetic abnormalities, including gains of chromosomes 9p and 2p, and the associated JAK2 (9p24) and REL (2p16) loci.10,11 In contrast to DLBCLs, MLBCLs do not exhibit BCL2 rearrangements and rarely manifest translocations involving BCL6.12
Because MLBCL is currently considered a subtype of DLBCL (as opposed to a distinct disease), these tumors are treated similarly with empiric adriamycin-containing combination chemotherapy regimens. Involved-field radiation therapy is often added to the site of bulky, localized disease.13 However, combined modality therapy is only partially effective, because more than 40% of MLBCL patients die of their disease.13 Further, involved-field radiation therapy has been associated with long-term side effects including secondary malignancies and cardiac dysfunction.14 For these reasons, additional insights into the molecular signature of MLBCL and potential rational treatment targets are critically needed.
To delineate unique features of MLBCL compared with DLBCL, we performed gene expression profiling of these 2 tumors and found them to be markedly different. Importantly, we also identified a molecular link between MLBCL and another lymphoid neoplasm with shared clinical features, classical Hodgkin lymphoma (cHL).
Patients, materials, and methods
Case selection and histologic classification
Frozen tumor specimens from newly diagnosed, previously untreated MLBCL patients (34) and DLBCL patients (176) were analyzed according to protocol approved by the institutional review board of the Dana Farber/Harvard Cancer Center. MLBCL tumor specimens were derived from mediastinal masses or contiguous nodal biopsies, and DLBCLs were all nodal tumor specimens. Primary MLBCLs were identified using clinical criteria (predominant mediastinal mass with or without local extension and no extrathoracic disease) and pathologic features. The histopathology and immunophenotype of each primary MBLCL were reviewed by expert hematopathologists to confirm the diagnosis. The diagnosis of each DLBCL was originally made by expert hematopathologists at the participating institutions.
Target cRNAs of oligonucleotide microarrays
Total RNA was extracted from each frozen tumor specimen, and biotinylated cRNAs were generated as previously described.15,16 Samples were hybridized overnight to Affymetrix U133A and U133B oligonucleotide microarrays (Affymetrix, Santa Clara, CA), which include probe sets from more than 44 000 genes. Arrays were subsequently developed with phycoerythrin-conjugated streptavidin (SAPE) and biotinylated antibody against streptavidin and scanned to obtain quantitative gene expression levels.15 The raw gene expression values were then scaled to account for differences in global chip intensity.
Gene expression analysis
The top 15 000 genes from the U133A and U133B Affymetrix chips were selected based on their ranking as measured by a median absolute deviation (MAD) variation filter across all samples. From within this 15 000-gene pool, genes correlating with the class distinction of interest (mediastinal = 1 versus nonmediastinal = 2) were identified by ranking them according to their signal-to-noise ratio (SNR). For a given gene g, SNR (g) = (x̄1–x̄2)/(s1 + s2), where x̄1 and si denote, respectively, gene g's sample mean and sample standard deviation within class i = 1,2. Similar rankings were obtained by using the median in place of the mean or by using the t statistic.AMonte Carlo simulation of the permutation distribution of the SNRs was performed by permuting the sample labels indicating class membership (n = 1000); thereafter, the observed values in the data were compared with the 99th percentile of the permutation distribution (Supplemental Material: see the Supplemental Document link at the top of the online article on the Blood website).
MLBCL versus DLBCL classifier
The discriminatory power of the gene expression signature was evaluated by building classifiers for the MLBCL versus DLBCL distinction. Naive Bayes (NB) and weighted voting (WV) classifiers that included 10 to 1000 genes were built and evaluated with regard to prediction errors using leave-one-out cross-validation (LOO-CV) (see Supplemental Document). The classifier with the lowest balanced error rate (proportion of samples wrongly classified, averaged within classes) was chosen for further analysis (see Supplemental Document). A more rigorous estimate of the error rate was also computed based on a 2-level CV procedure in which the choice of how many genes to use is automatically made within the CV loop (see Supplemental Document).
Analysis of coregulated genes
Genes that were coregulated (ie, genes whose expression values follow a similar pattern) with the top-ranked genes in MLBCL were identified using a 1 minus the Pearson correlation coefficient as the distance metric (see Supplemental Document).
Enrichment test for cHL genes in MLBCL
An enrichment test was used to evaluate the significance of the observed similarity between the MLBCL and cHL signatures. The cHL signature was defined using a set of genes independently identified by Schwering et al as differentially expressed in Hodgkin Reed-Sternberg (HRS) cell lines and normal purified B cells (centroblasts, centrocytes, and naive and memory B cells) using Affymetrix U95 oligonucleotide arrays.17 The similarity between the MLBCL and HRS signatures was assessed using the following recently described procedure.18 A total of 15 000 genes were selected from the U133A/B chips according to a MAD-based variation filter and ranked according to their SNR with respect to the “MLBCL versus DLBCL”class membership. The 294 genes with reduced expression in HRS cell lines17 were then located within our ranked list of 15 000 genes, and their proximity to the genes with lower levels of expression in MLBCL was measured by a Kolmogorov-Smirnoff (KS) score (with a higher score corresponding to a higher proximity). A similar procedure was used to place the 195 genes with increased expression in HRS cell lines17,19 within the 15 000 genes ranked in the opposite direction. Thereafter, permutation of the “MLBCL versus DLBCL” sample labels, associated reranking of the 15 000 genes, and computation of the corresponding KS scores were performed multiple times (n = 1000) so as to compare the observed KS scores with those that could be expected by chance under a random class labeling. Empirical P values were then computed to quantify the signifi-cance of the observed similarities between the genes with increased and decreased expression in the HRS cell lines and MLBCLs (pmax). An alternative, less stringent, method was also used to compute an empirical P value (pmin). This method was based on the computation of the KS score for multiple (n = 1000) random sets of 294 or 195 genes. This approach determines how likely it would be to obtain the observed KS score if a random set of genes were selected (see Supplemental Document).
Immunohistochemistry
Immunohistochemistry with anti–signal transducer and activator of transcription-1 (anti-STAT1) (9H2, Cell Signaling Technology, Beverly, MA) or anti–tumor necrosis factor (TNF) receptor-associated factor-1 (anti-TRAF1) (H3; Santa Cruz Biotechnology, Santa Cruz, CA) murine monoclonal antibodies was performed using 3 to 5 μ-thick formalin-fixed, paraffin-embedded specimens by standard immunohistochemical methods. Slides were deparaffinized and pretreated with 10 mM citrate, pH 6.0, (Zymed, South San Francisco, CA) in a steam pressure cooker (Decloaking Chamber, BioCare Medical, Walnut Creek, CA). Slides were then treated with Peroxidase Block (DAKO USA, Carpinteria, CA) for 5 minutes to quench endogenous peroxidase activity and incubated with a 1:5 dilution of goat serum in 50 mM Tris (tris(hydroxymethyl)aminomethane)–HCl, pH 7.4, for 20 minutes to block nonspecific binding sites. The anti-STAT1 antibody and anti-TRAF1 antibodies were diluted in 50 mM Tris-HCl, pH 7.4, with 3% goat serum (anti-STAT1 1:1000, anti-TRAF1 1:250) and applied to slides at room temperature for 1 hour. Goat antimouse horseradish peroxidase–conjugated antibody (Envision detection kit, DAKO USA) was applied for 30 minutes and developed using a diaminobenzidine (DAB) chromogen kit (DAKO USA).
c-REL immunohistochemistry was performed as outlined above with the following modifications. Formalin-fixed, paraffin-embedded sections were dewaxed and antigen retrieved in 1 mM EDTA (ethylenediaminetetraacetic acid) as published.20 Following blocking of nonspecific binding with 5% nonfat dried milk in Tris-buffered saline, pH 7.5, slides were incubated overnight with either affinity-purified rabbit anti–c-REL antibody at 1 μg/mL (PC139, Oncogene Research Products, Darmstadt, Germany) or pooled nonspecific rabbit Ig (Sigma, St Louis, MO). The slides were then washed and incubated with biotin-conjugated goat antirabbit secondary antibody (Vector, Burlingame, CA), followed by fluorescein isothiocyanate (FITC)–avidin (Molecular Probes, Eugene, OR). The sections were mounted and counterstained with propidium iodide (Molecular Probes). In separately reported studies, the association between nuclear c-REL immunostaining and nuclear factor–κB (NF-κB) activation was confirmed using molecular techniques and similarly prepared untreated and CD40 ligand–activated B cells (G.C. et al, manuscript in preparation).
Fluorescence in situ hybridization (FISH)
Air-dried touch preparations were prepared on Superfrost Plus slides from fresh frozen tumor specimens and stored in a desiccator at room temperature until use. Nuclei were hybridized to commercially available probes flanking or spanning the IgH, BCL2, and BCL6 loci (Vysis, Downer's Grove, IL) using conditions recommended by the manufacturer. After counterstaining with 4,6 diamidino-2-phenylindole (DAPI), interphase nuclei were scored for various chromosomal aberrations by fluorescence microscopy.
Results
Clinical and pathologic features of MLBCL
The clinical characteristics of the MLBCL and DLBCL patients included in this analysis are outlined in Table 1. The 34 MLBCL patients were predominantly young females (median age, 32 years) with localized and bulky disease (78% stage I/II). Those MLBCL patients with advanced-stage disease had contiguous involvement of the pleura and/or pericardium. In contrast, DLBCL patients were older (median age, 64 years) with an equal sex distribution and a higher incidence of advanced-stage disease (65% stage III/IV, Table 1). Pathologic review of the diagnostic MLBCL specimens confirmed the presence of sclerosis in 94% of cases; 77% of cases with available Ig immunohistochemistry had undetectable Ig, and 12% had equivocal Ig expression (Table 1). In the subset of MLBCLs with available genetic data, there was only one tumor with a BCL6 rearrangement (n = 19) and none with BCL2 translocations (n = 17) (Table 1), consistent with previous reports.12
Clinical and pathological features of MLBCL and DLBCL patients and tumors
Clinical feature . | MLBCL, n = 34 . | DLBCL, n = 176 . |
|---|---|---|
| Median age, y (range) | 32 (14-59) | 64 (20-92) |
| Sex (%) | ||
| Female | 24 (71) | 84 (48) |
| Male | 10 (29) | 92 (52) |
| Stage (%) | ||
| I/II | 25 (74) | 53 (30) |
| III/IV | 9 (26) | 115 (65) |
| Unknown | 0 (0) | 8 (5) |
| Pathology (%) | N/A | |
| Sclerosis | ||
| Present | 32 (94) | |
| Absent | 2 (6) | |
| Immunoglobulin by IHC* | ||
| Present | 3 (12) | |
| Absent | 20 (77) | |
| Equivocal | 3 (12) | |
| Not available | 8 | |
| FISH† | N/A‡ | |
| BCL2 rearrangements | 0 | |
| BCL6 rearrangements | 1 |
Clinical feature . | MLBCL, n = 34 . | DLBCL, n = 176 . |
|---|---|---|
| Median age, y (range) | 32 (14-59) | 64 (20-92) |
| Sex (%) | ||
| Female | 24 (71) | 84 (48) |
| Male | 10 (29) | 92 (52) |
| Stage (%) | ||
| I/II | 25 (74) | 53 (30) |
| III/IV | 9 (26) | 115 (65) |
| Unknown | 0 (0) | 8 (5) |
| Pathology (%) | N/A | |
| Sclerosis | ||
| Present | 32 (94) | |
| Absent | 2 (6) | |
| Immunoglobulin by IHC* | ||
| Present | 3 (12) | |
| Absent | 20 (77) | |
| Equivocal | 3 (12) | |
| Not available | 8 | |
| FISH† | N/A‡ | |
| BCL2 rearrangements | 0 | |
| BCL6 rearrangements | 1 |
N/A indicates not available.
MLBCL immunoglobulin status by immunohistochemistry (IHC) available for 26 patients (% positive calculated on available cases)
MLBCL interphase FISH available: BCL2, 17 patients; BCL6, 19 patients
To be described in a subsequent analysis
MLBCLs have a unique transcriptional profile
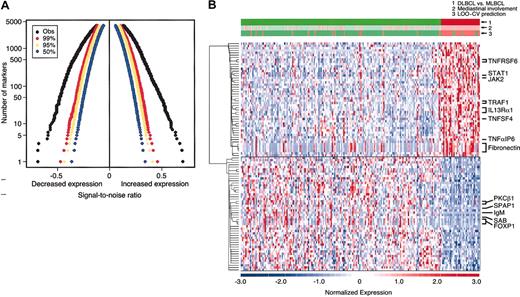
The distinctive clinical and pathologic characteristics of MLBCL—predominantly young women with Ig-negative, localized, and sclerotic tumors—suggested that this entity might also have a unique molecular signature. To address this possibility, diagnostic tumor specimens from the MBLCL and DLBCL cohorts were subjected to transcriptional profiling. The genes were sorted by their degree of correlation with the MLBCL versus DLBCL distinction according to the signal-to-noise metric (see “Patients, materials, and methods”). Permutation of the sample labels indicated that MLBCL had significantly lower expression of more than 1000 genes and significantly higher expression of more than 1000 additional genes, compared with DLBCLs (P < .01) (Figure 1A).
DLBCL and MLBCL genes. (A) Permutation analyses of differentially expressed genes in MLBCL and DLBCL. The observed signal-to-noise ratios (SNRs, x-axis) of ranked genes with lower levels of expression (left) and higher levels of expression (right) (black) in MLBCL versus DLBCL are compared with those expected by chance in 99% (red), 95% (yellow), and 50% (blue) of permutations. The y-axis indicates the number of genes that are differentially expressed in each direction. (B) Comparative gene expression profiles of DLBCL and MLBCL. At the top, the actual clinical/pathologic diagnosis of DLBCL versus MLBCL (green versus red), presence or absence of mediastinal disease (pink versus light green), and molecular prediction of DLBCL versus MLBCL (green versus red) are compared. The top 50 genes that were expressed at higher levels in MLBCL are shown in the upper half of the figure and the top 50 that were more abundant in DLBCL are shown in the lower half of the figure. Red indicates high relative expression; blue, low expression. Color scale at the bottom indicates relative expression in standard deviations from the mean. Each column is a sample and each row is a gene. Genes are clustered using hierarchical clustering. Expression profiles of 176 DLBCLs are on the left; profiles of the 34 MLBCLs are on the right.
DLBCL and MLBCL genes. (A) Permutation analyses of differentially expressed genes in MLBCL and DLBCL. The observed signal-to-noise ratios (SNRs, x-axis) of ranked genes with lower levels of expression (left) and higher levels of expression (right) (black) in MLBCL versus DLBCL are compared with those expected by chance in 99% (red), 95% (yellow), and 50% (blue) of permutations. The y-axis indicates the number of genes that are differentially expressed in each direction. (B) Comparative gene expression profiles of DLBCL and MLBCL. At the top, the actual clinical/pathologic diagnosis of DLBCL versus MLBCL (green versus red), presence or absence of mediastinal disease (pink versus light green), and molecular prediction of DLBCL versus MLBCL (green versus red) are compared. The top 50 genes that were expressed at higher levels in MLBCL are shown in the upper half of the figure and the top 50 that were more abundant in DLBCL are shown in the lower half of the figure. Red indicates high relative expression; blue, low expression. Color scale at the bottom indicates relative expression in standard deviations from the mean. Each column is a sample and each row is a gene. Genes are clustered using hierarchical clustering. Expression profiles of 176 DLBCLs are on the left; profiles of the 34 MLBCLs are on the right.
The top 50 genes with significantly higher or lower expression in MLBCL (as compared with DLBCL) are visually displayed in Figure 1B. The MLBCL transcriptional signature also included 3 genes previously reported to be expressed at high levels in this disease: the cell surface protein and lipid raft component, MAL; the recently described interleukin-4 (IL-4)–induced gene, FIG1; and the adhesion molecule, CD58/LFA3 (Table 2).8,21,22 These tumors also had lower levels of IgM, consistent with the previously noted absence of Ig in most cases of MLBCL (Figure 1B and Table 3.)40
Genes with higher levels of expression in MLBCL*
Gene class and specific genes . | Fold change† . |
|---|---|
| Cytokines and cytokine pathways | |
| IL-13Rα1‡ | 1.9 |
| JAK2‡ | 2.0 |
| STAT1‡ | 1.7 |
| Interleukin-4—induced gene 1 (FIG1)§ | 1.7 |
| NF—IL-3‡ | 2.0 |
| γIFN-inducible protein 10 (IP10/CXCR3 ligand)‡ | 2.0 |
| γIFN-inducible protein 30 (IFI30/GILT) | 1.6 |
| IFN-induced protein 35 | 1.5 |
| RANTES (CCL5)‡ | 1.9 |
| Chemokine receptor 1 (CCR1) | 1.5 |
| RON (macrophage-stimulating 1 receptor/c-met—related TK) | 1.7 |
| Interleukin-15 | 1.7 |
| TNF family members and induced proteins | |
| OX40 ligand (TNFSF4)‡ | 2.0 |
| FAS ligand receptor/CD95 (TNFRSF6)ठ| 1.7 |
| Herpes virus entry mediator (HVEM, TNFRSF14) | 1.5 |
| TRAF1‡ | 1.8 |
| TNF-α—induced protein 3 (A20)‡ | 1.6 |
| TNF-α—induced protein 6 | 2.3 |
| Costimulatory molecules and inflammatory mediators | |
| SLAM | 1.7 |
| CD80 (B7.1)‡ | 1.4 |
| CD86 (B7.2)‡ | 1.4 |
| Cyclooxygenase 1 | 1.9 |
| Prostaglandin receptor (IP) | 2.6 |
| Cell adhesion molecules/extracellular matrix components | |
| Fibroblast growth factor receptor-1 | 1.4 |
| CD58 (lymphocyte-function associated antigen 3)ठ| 1.5 |
| Integrin α M (CD11b) | 1.8 |
| Fibronectin | 3.0 |
| Fibroblast activation protein α (seprase) | 2.0 |
| Decorin | 2.0 |
| Collagen type III α 1 | 2.1 |
| Collagen type IV α 1 | 2.3 |
| Collagen type IV α 2 | 1.6 |
| Matrix metalloproteinase 14 | 2.0 |
| Tissue inhibitor of metallo-proteinase 1‡ | 1.9 |
| Other | |
| MAL§ | 2.8 |
| SNFT (Jun dimerization protein) | 2.8 |
| Adenosine deaminase | 1.9 |
Gene class and specific genes . | Fold change† . |
|---|---|
| Cytokines and cytokine pathways | |
| IL-13Rα1‡ | 1.9 |
| JAK2‡ | 2.0 |
| STAT1‡ | 1.7 |
| Interleukin-4—induced gene 1 (FIG1)§ | 1.7 |
| NF—IL-3‡ | 2.0 |
| γIFN-inducible protein 10 (IP10/CXCR3 ligand)‡ | 2.0 |
| γIFN-inducible protein 30 (IFI30/GILT) | 1.6 |
| IFN-induced protein 35 | 1.5 |
| RANTES (CCL5)‡ | 1.9 |
| Chemokine receptor 1 (CCR1) | 1.5 |
| RON (macrophage-stimulating 1 receptor/c-met—related TK) | 1.7 |
| Interleukin-15 | 1.7 |
| TNF family members and induced proteins | |
| OX40 ligand (TNFSF4)‡ | 2.0 |
| FAS ligand receptor/CD95 (TNFRSF6)ठ| 1.7 |
| Herpes virus entry mediator (HVEM, TNFRSF14) | 1.5 |
| TRAF1‡ | 1.8 |
| TNF-α—induced protein 3 (A20)‡ | 1.6 |
| TNF-α—induced protein 6 | 2.3 |
| Costimulatory molecules and inflammatory mediators | |
| SLAM | 1.7 |
| CD80 (B7.1)‡ | 1.4 |
| CD86 (B7.2)‡ | 1.4 |
| Cyclooxygenase 1 | 1.9 |
| Prostaglandin receptor (IP) | 2.6 |
| Cell adhesion molecules/extracellular matrix components | |
| Fibroblast growth factor receptor-1 | 1.4 |
| CD58 (lymphocyte-function associated antigen 3)ठ| 1.5 |
| Integrin α M (CD11b) | 1.8 |
| Fibronectin | 3.0 |
| Fibroblast activation protein α (seprase) | 2.0 |
| Decorin | 2.0 |
| Collagen type III α 1 | 2.1 |
| Collagen type IV α 1 | 2.3 |
| Collagen type IV α 2 | 1.6 |
| Matrix metalloproteinase 14 | 2.0 |
| Tissue inhibitor of metallo-proteinase 1‡ | 1.9 |
| Other | |
| MAL§ | 2.8 |
| SNFT (Jun dimerization protein) | 2.8 |
| Adenosine deaminase | 1.9 |
Gene list derived from the top 400 genes with higher levels of expression in MLBCL. All genes significant at P < .01
Fold change compared with DLBCL
Genes with lower levels of expression in MLBCL*
Gene class and specific genes . | Fold change† . |
|---|---|
| B-cell receptor components and signaling cascade | |
| IgMठ| 5.1 |
| B-cell lymphocyte kinase (BLK)‡ | 2.7 |
| SH3 domain-binding protein (SAB) (BTK-associated binding protein) | 2.0 |
| B-cell linker protein (BLNK)‡ | 1.5 |
| PKCβI‡ | 2.0 |
| NFATc‡ | 2.0 |
| AKT-1 | 1.5 |
| CD22‡ | 2.6 |
| Transcription factors | |
| Spi-B‡ | 1.6 |
| Ikaros | 1.5 |
| FOXP1 | 2.0 |
| Class II MHC componentsठ| |
| DO⇠| 1.9 |
| DQβ | 2.0 |
| DMα | 1.5 |
| Germinal center and other B-cell markers | |
| CD10 | 3.1 |
| SPAP1 (SH2 domain-containing phosphatase anchor protein, IRTA 4) | 2.4 |
| Protein tyrosine phosphatase receptor type C (PTPRCAP) | 1.4 |
| TNF family members | |
| Transmembrane activator and CAML interactor (TAC1) (TNFRSF13B) | 1.5 |
| CD27 (TNFRSF7) | 1.7 |
| TRAF5‡ | 1.5 |
| Cytokines | |
| Interleukin-16‡ | 1.4 |
Gene class and specific genes . | Fold change† . |
|---|---|
| B-cell receptor components and signaling cascade | |
| IgMठ| 5.1 |
| B-cell lymphocyte kinase (BLK)‡ | 2.7 |
| SH3 domain-binding protein (SAB) (BTK-associated binding protein) | 2.0 |
| B-cell linker protein (BLNK)‡ | 1.5 |
| PKCβI‡ | 2.0 |
| NFATc‡ | 2.0 |
| AKT-1 | 1.5 |
| CD22‡ | 2.6 |
| Transcription factors | |
| Spi-B‡ | 1.6 |
| Ikaros | 1.5 |
| FOXP1 | 2.0 |
| Class II MHC componentsठ| |
| DO⇠| 1.9 |
| DQβ | 2.0 |
| DMα | 1.5 |
| Germinal center and other B-cell markers | |
| CD10 | 3.1 |
| SPAP1 (SH2 domain-containing phosphatase anchor protein, IRTA 4) | 2.4 |
| Protein tyrosine phosphatase receptor type C (PTPRCAP) | 1.4 |
| TNF family members | |
| Transmembrane activator and CAML interactor (TAC1) (TNFRSF13B) | 1.5 |
| CD27 (TNFRSF7) | 1.7 |
| TRAF5‡ | 1.5 |
| Cytokines | |
| Interleukin-16‡ | 1.4 |
Gene list derived from the top 400 genes with lower levels of expression in MLBCL. All genes significant at P < .01
Fold change compared with DLBCL
Previously identified as having reduced expression in Hodgkin Reed-Sternberg cells: IgM, CD22, BLK, PKCβI/II, BLNK, NFATc, Spi-B, class II DOβ, TRAF5, and IL-16.17
Previously identified as having reduced expression in MLBCL: Ig2 and MHC class II.41
Class prediction
The discriminatory power of the gene expression signature was evaluated by building naive Bayes (NB) and weighted voting classifiers for the MLBCL versus DLBCL distinction. These classifiers were tested using a leave-one-out cross-validation strategy. A 100-gene NB model achieved the lowest balanced error rate (11%) (Figure 1B and Supplemental Document). Of interest, 6 patients whose DLBCL was classified by the NB model as MLBCL had predominant, although not exclusive, mediastinal disease (Figure 1B), and 2 patients whose tumors did not involve the mediastinum exhibited c-REL amplification (data not shown). In addition, 45 patients whose disease involved regional mediastinal lymph nodes as well as other nodal and extranodal sites were identified by the NB model as having DLBCL rather than primary MLBCL (Figure 1B). These data highlight the potential value of a molecular classifier in entities that are currently defined with only clinical and pathologic criteria.
MLBCL transcriptional profile resembles that of cHL
Inspection of the MLBCL transcriptional profile revealed striking similarities to that of cHL (Figure 1B; Tables 2, 3). Like Hodgkin Reed-Sternberg (HRS) cells, MLBCLs had low levels of expression of multiple B-cell signaling components and coreceptors (Table 3 and Figure 2).17,42-44 MLBCLs also had high levels of expression of cytokine pathway components, TNF family members, and extracellular matrix elements previously identified in cHL (Table 2).45 These observations are of particular interest because MLBCL and the most common subtype of cHL (nodular sclerosis) have similar clinical presentations—in younger patients with local/mediastinal tumors characterized by reactive fibrosis.
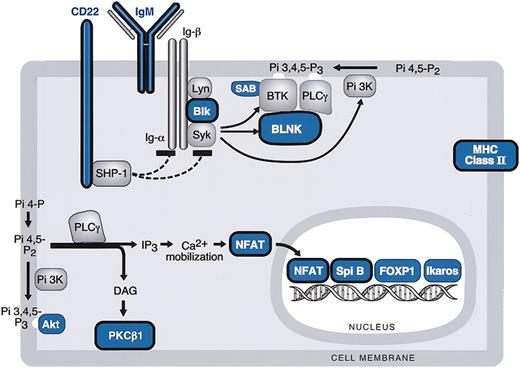
BCR signaling cascade components in MLBCL. Components of the BCR signaling pathway that are expressed at significantly lower levels in MLBCL are indicated in blue, and components that are reported to be expressed at reduced levels in HRS cells are circled in black. Genes are derived from the top 400 with low levels of expression in MLBCL (Table 3).
BCR signaling cascade components in MLBCL. Components of the BCR signaling pathway that are expressed at significantly lower levels in MLBCL are indicated in blue, and components that are reported to be expressed at reduced levels in HRS cells are circled in black. Genes are derived from the top 400 with low levels of expression in MLBCL (Table 3).
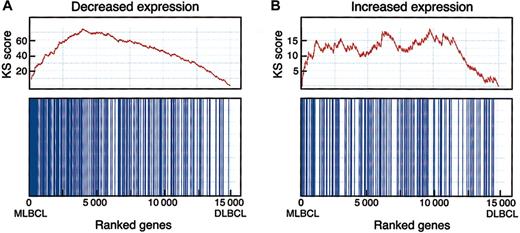
To determine the statistical significance of a potential MLBCL-cHL connection, we performed an enrichment test using an independently identified series of differentially expressed genes in HRS cell lines (Figure 3).17,19 When compared with random class labelings, the Kolmogorov-Smirnov (KS) statistic indicated that the observed MLBCL-cHL similarity was highly significant. Interestingly, this similarity was primarily attributable to genes with low levels of expression in HRS cell lines (versus normal B cells) and primary MLBCL (versus DLBCL) (pmax = .012, pmin < .001) (Figure 3). In contrast, the similarity between genes with high levels of expression in HRS cell lines and primary MLBCL was less significant (pmax = .213, pmin = .007). This may reflect the contribution of the tumor microenvironment to the gene signatures of primary MLBCL and/or the absence of this contribution from profiles of HRS cell lines maintained in vitro.19,45
Enrichment test for differentially expressed HRS genes in MBLCL. A graphic rendition of the computation of the KS scores for HRS genes that are expressed at low (A) and at high (B) levels in MLBCL is shown. The bottom panel for each signature indicates with vertical bars the location of the HRS genes within the ranked set of 15 000 differentially expressed genes in MLBCL and DLBCL (with the MLBCL genes to the left and the DLBCL genes to the right). The top panel shows the assignment of “rewards” and “penalties” to the overall KS score as the list of 15 000 ranked genes is scanned from the MLBCL end (left) to the DLBCL end (right). Every “hit” (ie, the encounter of an HRS gene during the scan) increases the KS score, and every “miss” (the encounter of a non-HRS gene) decreases the score, resulting in the indicated stepwise curve. The final score corresponds to the highest value (on the y-axis) in the plot. High enrichment would correspond to a steep climb upward to the left, whereas lack of enrichment would correspond to a lack of clear upward trend in the curve. The KS statistic indicates that genes reported to have low levels of expression in HRS cell lines are similarly decreased in MLBCL (pmax = .012, pmin < .001). In contrast, the similarity between genes with higher levels of expression in HRS cell lines and primary MLBCL was less significant (pmax = .213, pmin = .007), likely reflecting the importance of tumor microenvironment to primary MLBCL and cHL signatures45 and the absence of these features in the signatures of isolated HRS cell lines maintained in vitro.19
Enrichment test for differentially expressed HRS genes in MBLCL. A graphic rendition of the computation of the KS scores for HRS genes that are expressed at low (A) and at high (B) levels in MLBCL is shown. The bottom panel for each signature indicates with vertical bars the location of the HRS genes within the ranked set of 15 000 differentially expressed genes in MLBCL and DLBCL (with the MLBCL genes to the left and the DLBCL genes to the right). The top panel shows the assignment of “rewards” and “penalties” to the overall KS score as the list of 15 000 ranked genes is scanned from the MLBCL end (left) to the DLBCL end (right). Every “hit” (ie, the encounter of an HRS gene during the scan) increases the KS score, and every “miss” (the encounter of a non-HRS gene) decreases the score, resulting in the indicated stepwise curve. The final score corresponds to the highest value (on the y-axis) in the plot. High enrichment would correspond to a steep climb upward to the left, whereas lack of enrichment would correspond to a lack of clear upward trend in the curve. The KS statistic indicates that genes reported to have low levels of expression in HRS cell lines are similarly decreased in MLBCL (pmax = .012, pmin < .001). In contrast, the similarity between genes with higher levels of expression in HRS cell lines and primary MLBCL was less significant (pmax = .213, pmin = .007), likely reflecting the importance of tumor microenvironment to primary MLBCL and cHL signatures45 and the absence of these features in the signatures of isolated HRS cell lines maintained in vitro.19
Decreased expression of BCR signaling cascade components in MLBCL
Like cHL, MLBCLs had low levels of expression of multiple components of the BCR signaling cascade including the cell surface immunoglobulin receptor, IgM; the Igα/β-associated SRC-family protein tyrosine kinase (PTK), BLK; the SYK PTK-regulated scaffolding protein, BLNK; and the Bruton tyrosine kinase (BTK) binding protein, SAB (Table 3 and Figure 2).46-48 MLBCLs also had low levels of phospholipase Cγ2 (PLCγ2)–regulated molecules including the critical lymphoid transcription factor, nuclear factor of activated T cells (NFATc), and protein kinase C β1 (PKCβ), also reported to be decreased in HRS cells (Table 3 and Figure 2).46
The phosphatidylinositol-3 kinase (PI-3K)–regulated downstream kinase, AKT, and a major AKT target, FORKHEADP1 (FOXP1), were also expressed at low levels in MLBCL46,49 (Table 3 and Figure 2). Lymphoid transcription factors including Spi-B and Ikaros were also less abundant in MLBCL (Table 3 and Figure 2).50
In addition to the above-mentioned B-cell transcription factors and components of the BCR signaling cascade, MLBCLs, like HRS cells, had low levels of expression of the CD22 coreceptor and several major histocompatibility complex (MHC) class II molecules (Table 3 and Figure 2).46,51 MBLCLs also expressed low levels of the germinal center (GC) metalloendopeptidase, CD10 (Table 3).
Increased expression of cytokine pathway components, TNF family members, and extracellular matrix elements in MLBCL
Like primary cHL, MLBCLs had increased expression of specific cytokine pathway members. For example, MLBCLs expressed high levels of the interleukin-13 receptor α1 (IL-13Rα1) and additional downstream effectors of IL-13 signaling previously identified in primary cHL (Table 2).23,24,45 In normal B cells, IL-13R signaling stimulates cellular proliferation and triggers Ig class switching.45 IL-13Rα1 associates with IL-4Rα to form a high-affinity IL-13R complex; signaling through this receptor results in phosphorylation of Janus kinase-2 (JAK2) and STAT1.52 Of note, MLBCLs expressed high levels of JAK2 and STAT1 and 2 IL-4–induced genes, FIG1 and NF–IL-3 (Table 2).21,53 Given the composition of the IL-13Rα1/IL-4α high-affinity receptor, the identification of 2 IL-4–induced genes (FIG1 and NF–IL-3) in MLBCL suggests that the pathway may be active in this disease (Table 2). Consistent with this possibility, NF–IL-3 was the gene most closely coregulated with the IL-13R in MLBCL (data not shown).
MLBCLs also had high levels of expression of γ interferon (IFN)–induced proteins including the chemokine and CXCR3 ligand, IP1054 ; IFI30/GILT55 ; and IFI35 (Table 2).56 HRS cell lines and primary cHLs also express γ IFN–induced proteins such as IP10, a chemoattractant for T and natural killer (NK) cells29 (Table 2). Like cHL, MLBCLs also had high levels of expression of the T-helper cell chemokine, RANTES (regulated on activation normal T cells expressed and secreted) (Table 2)29 and the costimulatory molecules CD80 (B7.1) and CD86 (B7.2).34,35 MLBCLs also had increased expression of signaling lymphocytic activation molecule (SLAM), a B- and T-cell surface molecule that potentiates lymphocyte expansion in a CD28-independent manner and serves as a measles virus receptor.57 In addition, components of the prostaglandin pathway, cyclooxygenase 1 and prostaglandin IP receptor, were also expressed at high levels in MLBCLs.58
Equally striking in MLBCL was the increased expression of TNF superfamily (SF) members previously implicated in the pathogenesis of cHL, including the OX40 ligand (TNFSF4) and the FAS ligand receptor (TNFRSF6) previously identified in MLBCL38 (Table 2). The herpes virus entry mediator (HVEM, TNFRSF14) and TRAF1 were also expressed at high levels in MLBCL. These results are of interest because HVEM associates with TRAF1 to stimulate NF-κB activation59 and TRAF-1 is expressed in HRS cells (Table 2) and up-regulated following NF-κB activation (Table 2).31,32,60
Signaling through TNF receptors triggers distinct signaling pathways leading to either activation of NF-κB transcription factors or apoptosis.61 In this regard, TRAF1 plays a unique regulatory role in lymphoid cells, interacting with cellular inhibitor-of-apoptosis proteins (cIAPs) to suppress TNF-induced apoptosis.62 In addition to TRAF1, MLBCLs also expressed higher levels of A20, another TNF-α–induced protein (Table 2) that protects against TNF-induced apoptosis.63 Of note, HRS cell lines with high basal expression of A20 and TRAF1 are reported to be resistant to apoptosis.33 Taken together, these data suggest that in MLBCL, like cHL, signaling through TNF receptors and associated factors favors NF-κB activation and resistance to apoptosis.
Primary MLBCL and cHL often exhibit sclerosis, underscoring the potential importance of extracellular matrix components in both diseases. Consistent with these observations, MLBCLs had higher levels of expression of a TNF-α–induced hyaluronan-binding protein (TNFAIP6)64 ; adhesion molecules including CD58/LFA3, integrinαM/CD11b, and fibronectin; fibroblast growth factor receptors and activation proteins; and additional extracellular matrix components including several types of collagen and decorin (Table 2).65,66
Expression of STAT1 and TRAF1 proteins in MLBCL
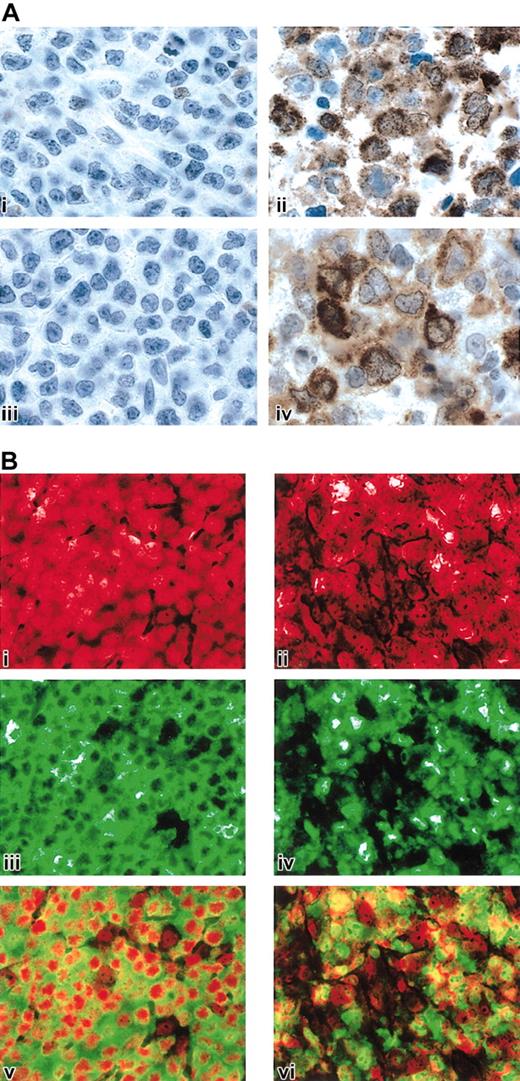
The critical roles of IL-13 signaling and NF-κB activation in cHL and the observed similarities between cHL and MLBCL prompted us to obtain protein confirmation for specific pathway members in MLBCL. Given the known functions of STAT1 in IL-13 signaling and TRAF1 in downstream activation of NF-κB,52,67 we performed STAT1 and TRAF1 immunohistochemistry in a series of MLBCLs and DLBCLs from our cohort with available formalin-fixed paraffin-embedded tissue as well as cases of nodular sclerosis cHL. As summarized in Table 4 and illustrated in Figure 4A, all MLBCLs exhibited tumor cell expression of STAT1, whereas none of the DLBCLs had definite STAT1 staining. The cHLs also expressed STAT1 in the HRS cells and surrounding inflammatory cells (likely macrophages) (Table 4).
Immunohistochemical analysis of STAT1 and TRAF1 expression in DLBCL, MLBCL, and nodular sclerosis HL
Diagnosis (no.) . | STAT1 (no.) . | TRAF1 (no.) . |
|---|---|---|
| DLBCL (5) | Negative* (5) | Negative (5) |
| MLBCL (6) | Positive† (6) | Positive (5) |
| Weakly positive‡ (1) | ||
| Nodular sclerosis HL (12) | Positive (6) | Positive (12) |
| Weakly positive (5) | ||
| Negative (1) |
Diagnosis (no.) . | STAT1 (no.) . | TRAF1 (no.) . |
|---|---|---|
| DLBCL (5) | Negative* (5) | Negative (5) |
| MLBCL (6) | Positive† (6) | Positive (5) |
| Weakly positive‡ (1) | ||
| Nodular sclerosis HL (12) | Positive (6) | Positive (12) |
| Weakly positive (5) | ||
| Negative (1) |
Staining observed in less than 5% of tumor cells
Strong staining observed in more than 90% of tumor cells
Weak staining observed in more than 90% of tumor cells
STAT1, TRAF1, and c-REL immunohistochemistry in MLBCL. (A) STAT1 and TRAF1 proteins in MLBCL. A control DLBCL (i, iii) and representative MLBCL (ii, iv) were stained for STAT1 (i, ii) and TRAF1 (iii, iv). Original magnification, × 1000. (B) c-REL immunohistochemistry in MLBCL. A control DLBCL (i,iii,v) and representative MLBCL (ii,iv,vi) are doubly stained for c-REL (iii,iv; green) and propidium iodide (i,ii; nuclear, red). The 2 channels of each case are superimposed in subpanels v and vi. Note predominant nuclear c-REL staining of MLBCL at right (vi) compared with the predominantly cytoplasmic c-REL staining of the control DLBCL on the left (v). Original magnification, × 400.
STAT1, TRAF1, and c-REL immunohistochemistry in MLBCL. (A) STAT1 and TRAF1 proteins in MLBCL. A control DLBCL (i, iii) and representative MLBCL (ii, iv) were stained for STAT1 (i, ii) and TRAF1 (iii, iv). Original magnification, × 1000. (B) c-REL immunohistochemistry in MLBCL. A control DLBCL (i,iii,v) and representative MLBCL (ii,iv,vi) are doubly stained for c-REL (iii,iv; green) and propidium iodide (i,ii; nuclear, red). The 2 channels of each case are superimposed in subpanels v and vi. Note predominant nuclear c-REL staining of MLBCL at right (vi) compared with the predominantly cytoplasmic c-REL staining of the control DLBCL on the left (v). Original magnification, × 400.
MLBCLs have immunohistochemical evidence of NF-κB activation
MLBCLs that lack Ig and associated BCR survival signals have likely developed alternative mechanisms for escaping cell death, similar to those described in cHL.45 In cHL, activation of the NF-κB pathway enhances the survival of surface immunoglobulin–negative (sIg-) Reed-Sternberg cells.44 Given the TRAF1 expression in MLBCL and components of the MLBCL transcriptional profiles suggestive of NF-κB activation (Tables 2 and 4; Figure 4A), we determined the subcellular location of the c-REL subunit of the NF-κB heterodimer in primary MLBCLs.68 In 5 of 6 MLBCLs assessed by c-REL immunohistochemistry, the protein was localized to the nucleus, consistent with activation of the NF-κB pathway (Figure 4B). These observations highlight a shared survival pathway in MLBCL and cHL.
Discussion
The unique profile of primary MLBCL—low levels of expression of BCR signaling pathway components, a distinctive cytokine pathway signature, and activation of NF-κB—is strikingly similar to that of a clinically related disorder, cHL. Both MLBCL and cHL of the nodular sclerosis subtype commonly present in young patients as mediastinal tumors with absent surface Ig and prominent fibrosis. MLBCL and classical HRS cells also exhibit common genetic abnormalities including gains of chromosome 2p and 9p. Found in about 20% of MLBCL and up to 50% of cHL, gains in chromosome 2p are associated with amplification of the REL locus, one potential mechanism for increased NF-κB activity and tumor cell resistance to apoptosis.11,25,68,69 Gains in chromosome 9p and the JAK2 locus are observed in about 75% of MLBCLs and 25% of cHLs and are unique to these diseases.10,11,26 In addition to these clinical, immunologic, and molecular similarities, there are also rare reported cases of composite cHL and MLBCL, further supporting a pathogenetic relationship between these tumors.70 The clonal relationship between HRS cells and non-Hodgkin lymphoma (NHL) cells in composite lymphomas was one of the strongest initial pieces of evidence that cHL was a B-cell lymphoma.71 In our own series, one patient with MLBCL subsequently relapsed with cHL (nodular sclerosis subtype).
The extent of similarity between the nodular sclerosis subtype of cHL and MLBCL is perhaps surprising given the degree to which the histopathologies and outcome of these 2 entities differ. Cells resembling HRS cells are usually not seen in MLBCL, and MLBCL cells retain the expression of multiple B-cell markers, including CD20, CD79a, BOB1, and OCT2,6,72 that are lost in HRS cells.17,73 In addition, nodular sclerosis cHL is characteristically associated with a polymorphous inflammatory infiltrate rich in plasma cells, neutrophils, and eosinophils, reactive cell types that are usually absent in MLBCL.45 The reactive infiltrate in cHL is recruited and maintained by additional chemokines and cytokines, such as IL-5, IL-6, and IL-10,45 that are not part of the MLBCL signature. Based on the existence of unusual patients with both nodular sclerosis cHL and MLBCL, we suggest these 2 entities arise from a common precursor cell with a growth and survival advantage stemming from genetic lesions that result in constitutive activation of NF-κB. Acquisition of distinct sets of secondary genetic lesions then results in either MLBCL or nodular sclerosis cHL, potentially explaining the differences in the clinical course of these diseases.
The reason for the lower levels of expression of Ig and BCR signaling components in MLBCL is not yet known. In a subset of cHLs, the lack of a functional BCR has been attributed to “crippling” somatic mutations of the rearranged immunoglobulin gene.74 In cHLs with functional gene rearrangements, the absence of Ig has been ascribed to deficiencies in transcription factors (OCT2, BOB1, PU1) necessary for Ig synthesis.73,75 In a recent study, 12 of 13 MLBCLs exhibited functional IgVH gene rearrangements, suggesting that destructive somatic mutations were not the primary cause of reduced surface Ig in this disease.9 More recently, abundant OCT2 and BOB1 transcription factors and Ig transcripts with a switched isotype were detected in MLBCL, suggesting that the transcriptional machinery for Ig synthesis was intact in these tumors.72 However, our MLBCL transcriptional profile reveals reduced levels of other critical B-cell transcription factors such as NFATc and Spi-B, which have known roles in regulating the expression of specific Ig isotypes.76,77
MLBCL has a prominent cytokine pathway signature, which likely reflects dynamic interactions between the tumor cells, infiltrating inflammatory cells, and the surrounding tumor stroma. Like cHLs, MLBCLs have increased abundance of high-affinity IL-13 receptor subunits and downstream effectors (JAK2 and STAT1, NF–IL-3), implicating this cytokine pathway in disease pathogenesis.45 Our studies also indicate that MLBCLs overexpress specific TNF family members that are known to interact with TRAF1 and subsequently activate NF-κB. TRAF1 transcripts and protein were also more abundant in MLBCL. Consistent with a likely role for NF-κB activation in MLBCL, MLBCLs exhibited c-REL nuclear staining.
The nuclear localization of c-REL and likely activation of NF-κB in MLBCL provide new clues regarding more rational targeted therapy in this disease. Given the limitations of current combined modality therapy, including treatment failures and longterm radiation toxicity, the NF-κB pathway and supporting cytokine network may represent more specific MLBCL treatment targets. In this regard, the current study both defines molecular differences in morphologically similar tumors (MLBCL versus DLBCL) and identifies shared features of morphologically distinct neoplasms (MLBCL and cHL).
Prepublished online as Blood First Edition Paper, August 21, 2003; DOI 10.1182/blood-2003-06-1841.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
K.J.S. and S.M. contributed equally to this study. T.R.G. and M.A.S. contributed equally to this study.
The online version of the article contains a data supplement.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal