Abstract
Kit ligand (Kitl), encoded by the Steel (Sl) locus, plays an essential role in hematopoiesis, gametogenesis, and melanogenesis during both embryonic and adult life. We have characterized a new spontaneous mutant of the Sl locus in mice designated KitlSl-20J that arose in the breeding colony at Jackson Laboratories. Heterozygous KitlSl-20J mice display a white belly spot and intercrossing results in an embryonic lethal phenotype in the homozygous state. Analysis of homozygous embryos demonstrated a significant reduction in fetal liver cellularity, colony forming unit-erythroid (CFU-E) progenitors, and a total absence of germ cells. Although expressed in vivo, recombinant mutant protein demonstrated loss of bioactivity that was correlated with lack of receptor binding. Analysis of the Sl gene transcripts in heterozygous KitlSl-20J mice revealed an in-frame tandem duplication of exon 3. A long-range polymerase chain reaction (PCR) strategy using overlapping primers in exon 3 amplified an approximately 7-kilobase (kb) product from DNA isolated from heterozygous KitlSl-20J mice but not from wild-type DNA that contained sequences from both introns 2 and 3 and an inverted intron 2 sequence, suggesting a complex rearrangement as the mechanism of the mutation. “Complexity analysis” of the sequence of the amplified product strongly suggests that local DNA motifs may have contributed to the generation of this spontaneous KitlSl-20J allele, likely mediated by a 2-step process. The KitlSl-20J mutation is a unique KitlSl allele and represents an unusual mechanism of mutation. (Blood. 2003;102:3548-3555)
Introduction
Many growth factors have pleiotropic effects on multiple cell lineages and influence the growth and development of multiple organ systems. One such growth factor encoded by the murine Steel (Sl) locus is Kit ligand (Kitl)1 (also known as stem cell factor [SCF]2 and mast cell growth factor [MGF]3 ). Kitl is the ligand of the receptor tyrosine kinase c-kit that is encoded by the dominant white spotting (W) locus.4,5 Originally identified through readily observable coat color abnormalities in mice, the study of W and Sl loci has contributed enormously to our understanding of the biology of hematopoietic,6,7 pigment,8,9 and germ cells10,11 both during early development and adult life.
Since the first description of W mouse coat color mutants in 192712 and Sl mutants in 1956,13,14 a large number of other mutations at both loci have been described that are associated with phenotypic abnormalities in multiple cell lineages. In general, heterozygotes at these loci exhibit dilute coat color ventrally or a white belly spot and/or a forehead blaze.15 Some mutant alleles are associated with mild effects on hematopoiesis and fertility in the heterozygous state. Depending upon the specific mutant allele, homozygotes display lethality late in gestation and perinatally or can be viable with significant pigmentation defects, variable degrees of anemia, and reduced fertility.6
Alternately spliced mRNA results in the production of 2 isoforms of the mature Kitl protein.16 Transcripts that include exon 6 sequences encode a glycoprotein that contains a proteolytic cleavage site. Cleavage at this site results in the production of the major soluble form of the protein. Alternately spliced transcripts that lack exon 6 gives rise to a membrane-associated isoform due to the absence of the primary cleavage site.7,17 Although the biologic significance of the 2 isoforms is not completely understood, evidence from the study of specific KitlSl mutants (KitlSl-d, KitlSl-17H)18,19 as well as other experiments suggests a differential role for the 2 isoforms of the protein in the activation of c-kit and downstream signaling.20-24
Characterization of the KitlSl mutant alleles has identified an array of diverse alterations at the molecular level. The originally described mutant allele (KitlSl) and Steel-grizzly belly (KitlSl-gb) allele lack the entire coding sequence of the Sl gene, and homozygosity for these alleles is associated with embryonic lethality.25 The Steel-dickie (KitlSl-d) allele contains a 4-kilobase (kb) intragenic deletion that includes the sequences encoding the membrane-spanning region of the protein that leads to the production of an obligatory secreted protein.26 Another characterized mutant allele, KitlSl-17H, has a point mutation in the 3′ splice acceptor site of intron 7 that is responsible for the generation of transcripts lacking sequences encoded by exon 8. The alteration in the reading frame results in a cytoplasmic tail containing novel amino acids and reduced presentation of the membrane-associated isoform of Kitl.27,28 By contrast, KitlSl-panda and KitlSl-contrasted alleles do not show any structural changes in the genomic sequence of the Sl gene. However, DNA rearrangements located over 100 kb 5′ to the coding region in these alleles are associated with reduced expression of the Kitl protein and a mutant phenotype.29 Recently 7 new N-ethylnitrosourea (ENU)-induced KitlSl mutant alleles have been described with point mutations in the coding region.30,31
In this study we describe a newly identified spontaneous KitlSl mutant allele designated KitlSl-20J that arose in the C57BL/6J breeding colony at The Jackson Laboratory (Bar Harbor, ME). Although many spontaneous and ENU-induced KitlSl mutant alleles with diverse alterations have been described at the molecular level, several features make this allele unique. Here we show that the KitlSl-20J allele contains a tandem duplication of exon 3 that represents an important internal gene duplication event not previously reported for the Sl gene. Analysis of the mutation at the genomic level suggests a complex rearrangement requiring a mechanism involving 2 distinct mutational events. “Complexity analysis” demonstrates that sequences in introns 2 and 3 of the Sl genome may have contributed to the generation of the mutation. The mutant KitlSl-20J protein is expressed but in vitro studies demonstrate a lack of binding to the receptor c-kit. The resulting phenotype is an intraembryonic lethal in the homozygous state.
Materials and methods
Animals
Spontaneous mutant mice with a white belly spot were observed in the breeding colony of C57Bl/6J mice at The Jackson Laboratory (Bar Harbor, ME). Complementation analysis performed there suggested a mutation at the Sl locus and the mice were designated KitlSl-20J.
RT-PCR analysis
Total RNA was isolated from spleens of heterozygous KitlSl-20J mice with a white belly spot and control C57Bl/6J mouse using Tripure Isolation Reagent (Roche Biochemicals, Indianapolis, IN) per the manufacturer's instructions. The primer pair used to amplify the entire coding sequence of the Sl gene transcripts was forward primer 5′AGCTAAACG GAGTCGCCACA3′ and reverse primer 5′GCCACTGTGCGAAGGTAA CA3′ (Gibco BRL custom primers, Life Technologies, Carlsbad, CA). Reverse transcriptase-polymerase chain reaction (RT-PCR) was performed using the Titan One Tube RT-PCR kit (Roche Biochemicals) per the manufacturer's protocol. The RT-PCR product was separated on a 3% agarose gel and individual DNA bands were isolated from the gel and cloned separately into a pCNTR shuttle vector provided in the General Contractor DNA cloning system (5Prime-3Prime Inc, Boulder, CO). Plasmid DNA isolated from individual selected clones was sequenced using M13 universal primer pair at the automated sequencing facility, Department of Biochemistry, Indiana University School of Medicine (Indianapolis, IN).
Long-range genomic PCR analysis
Tails from wild-type and heterozygous KitlSl-20J mice (white belly spotted) were digested overnight with proteinase K at 55°C and DNA were isolated by single phenol-chloroform extraction and ethanol precipitation. Long-range PCR analysis was performed using the Expand Long Template PCR system (Roche Biochemicals). The following primers were employed: forward primer (binding to exon 3/intron 3 junction of the Sl gene) 5′GGGATGGATGTTTTGGTATGT ACTTCACACATTTC3′ and reverse primer (binding to exon 3) 5′AACATCCATCCCG GCGACATAGTTGAGGGTTAT 3′. Approximately 500 ng DNA was used for amplification per the protocol provided by the manufacturer, with minor modifications. Buffer 3 was used with 10 μL of deoxynucleoside triphosphate (dNTP) mix from the Titan One Step RT-PCR kit (Roche Biochemicals). PCR amplification was performed in a Perkin Elmer GeneAmp 9700 thermocycler (Shelton, CT) using the manufacturer's program for long-range PCR. The amplified product was cloned into the vector provided in the TOPO XL PCR Cloning kit (Invitrogen, Life Technologies, Carlsbad, CA).
Fetal liver hematopoiesis
Heterozygous KitlSl-20J mice were intercrossed and fetal livers were dissected from 14.5-dpc (days after coitus) littermate embryos. Single-cell suspension of fetal livers was prepared by gentle homogenization and filtering through a 40-μm filter and cell counts and viability were determined using 0.4% trypan blue staining. A standard progenitor assay was performed in triplicate by seeding 5 × 104 unfractionated fetal liver cells in 1 mL methylcellulose medium containing 1% methylcellulose (Stem Cell Technologies, Vancouver, BC, Canada), 30% fetal calf serum (FCS; Hyclone, Logan, UT), 1% bovine serum albumin (BSA), 10-4 M 2-mercaptoethanol, 4 U/mL erythropoietin, 100 ng/mL recombinant rat SCF (rrSCF), 100 ng/mL granulocyte colony stimulating factor (G-CSF), 100 ng/mL megakaryocyte growth and development factor (MGDF; all from Amgen, Thousand Oaks, CA), and 100 ng/mL murine interleukin-3 (IL-3; Peprotech, Rocky Hill, NJ). Colony forming unit-erythroid progenitors (CFU-Es) were counted after 2 days in culture, whereas myeloid colonies and burst forming unit-erythroid progenitors (BFU-Es) were counted after 7 days using an inverted tissue culture microscope.
Construction of mammalian expression vectors and expression of Kitl proteins
Sl gene transcript encoding the secreted isoform of Kitl from wild-type (WT) and heterozygous KitlSl-20J mice was amplified using the following primer pair: forward primer 5′CGGCTCGAGGCCGCCACCATGAAGAAGACACAAACT3′ (Kozak sequence underlined) and reverse primer 5′CGCTCTAGATTAGGCTGCAACAGGGGGT AACATA3′. RT-PCR was performed as described in “RT-PCR analysis” and the amplified wild-type and KitlSl-20J cDNAs were cloned into a mammalian expression vector pCR3.1 using the Eukaryotic TA cloning kit (Invitrogen). For purification of Kitl proteins, expression vectors were constructed as described above except that the reverse primer (5′ATTCAATGGTGATGATGGGCTGCAACAGG GGGTAA 3′) amplified cDNA that encodes a soluble product fused with a 6-residue histidine tag incorporated before the stop codon.
CHOK1 cells were transfected with the cloned wild-type and mutant KitlSl-20J expression vectors using Fugene 6 transfection reagent (Roche Biochemicals), and stable clones were selected in medium containing 1 mg/mL G418 (dry powder; Bio Whittaker, Walkersville, MD). Serum-free conditioned medium was collected from stable clones grown to 80% confluency in five 10-cm2 tissue culture dishes and filtered through 0.45-μm filters (Fisher Scientific, Pittsburgh, PA). Subsequently, the conditioned medium was concentrated 20- to 40-fold in a Centriplus Centrifugal Filter Device (YM-10; Amicon, Millipore, Bedford, MA). Secretion of Kitl protein into the medium was confirmed by immunoblot analysis using anti-murine SCF antibody (Peprotech) at a dilution of 1:2000.
For receptor binding assay, after transfection of expression vectors into CHOK1 cells as described in the previous paragraph, histidine-tagged WT and KitlSl-20J proteins were purified from the conditioned medium using His-Bind Quick 900 cartridge (Novagen, Madison, WI) per the manufacturer's recommendations. Four milliliters of the final elute was concentrated and desalted using Centricon Centrifugal Filter Device (YM-10). Secreted WT and KitlSl-20J proteins were detected by immunoblot analysis.
Proliferation assay
C-kit-expressing MO7E cells were counted and seeded in 6 replicates at a density of 4 × 104 cells/well in 96-well plates. After 48 hours culture, 1.0 μCi (0.037 MBq) [3H] thymidine (Amersham, Piscataway, NJ) was added to each well for 6 hours at 37°C. Cells were then harvested using an automated cell harvester (96-well harvester, Packard Bioscience, Perkin Elmer, Boston, MA) and thymidine incorporation was determined in a scintillation counter (Beckman Coulter, Hialeah, FL).
Flow cytometric analysis of Kitl binding to receptor c-kit
Bone marrow-derived mast cells (0.5 × 106) were collected in 5 mL polystyrene tubes, washed with cold phosphate-buffered saline (PBS), and incubated for 1 hour in 5% bovine serum albumin (BSA) on ice to block nonspecific binding. For the ligand binding step, cells were incubated for 1 hour on ice with recombinant rat SCF (Amgen) or purified histidine-tagged wild-type and KitlSl-20J proteins. The cells were subsequently washed with cold PBS and incubated for 1 hour on ice with 10 μL biotinylated anti-murine SCF antibody (5 μg/mL; Peprotech). After subsequent washing with cold PBS, cells were incubated with 10 μL streptavidin-phycoerythrin (PE; BD-Pharmingen, San Diego, CA) for 30 minutes on ice. The cells were then washed with cold PBS and receptor-bound Kitl was analyzed using a FACScalibur (BD-Biosciences, San Jose, CA). At least 10 000 live events were acquired by gating on viable mast cells and data files were analyzed using CellQuest software (BD-Biosciences).
Complexity analysis
Results
Embryonic lethality of KitlSl-20J/KitlSl-20J embryos
Heterozygous KitlSl-20J mice appear normal except for coat color abnormalities (diluted ventral pigmentation or white belly spot) and are viable and fertile. In preliminary intercrossing of heterozygous KitlSl-20J mice, no offspring with severe coat color abnormalities indicative of homozygous mice were generated. Subsequent genetic analysis of viable offspring from 18 litters showed a mendelian ratio predicted for an embryonic lethal phenotype (1:2:0, +/+ vs KitlSl-20J/+ vs KitlSl-20J/KitlSl-20J; Table 1). In addition, comparison of the average litter sizes derived from wild-type C57Bl/6J and heterozygous KitlSl-20J crosses suggested that the KitlSl-20J mutation leads to embryonic lethality in the homozygous state (6.89 vs 4.28, C57Bl/6J vs KitlSl-20J/+; n = 18 litters; P < .05). Genotyping by RT-PCR analysis of embryos harvested at 14.5 dpc resulted in a segregation pattern similar to the expected mendelian ratio of 1:2:1 (Table 1). These data suggests that as with other severe mutant alleles such as KitlSl and KitlSl-gb, the KitlSl-20J/KitlSl-20J mutants die in utero between 14.5 dpc and birth.
Analysis of genotypes of fetal and perinatal mice resulting from intercrossing of heterozygous mice
. | . | Genotype, n (%) . | . | . | ||
|---|---|---|---|---|---|---|
| Day of analysis . | Litters . | +/+ . | KitlSI-20J/+ . | KitlSI-20J/KitlSI-20J . | ||
| 14.5 dpc | 4 | 7 (21) | 17 (50) | 10 (29) | ||
| After birth | 18 | 27 (35) | 49 (64) | 0 (0) | ||
. | . | Genotype, n (%) . | . | . | ||
|---|---|---|---|---|---|---|
| Day of analysis . | Litters . | +/+ . | KitlSI-20J/+ . | KitlSI-20J/KitlSI-20J . | ||
| 14.5 dpc | 4 | 7 (21) | 17 (50) | 10 (29) | ||
| After birth | 18 | 27 (35) | 49 (64) | 0 (0) | ||
Phenotypic analysis of KitlSl-20J/KitlSl-20J embryos
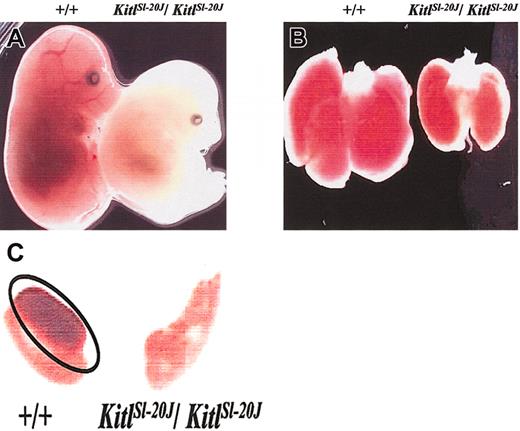
KitlSl-20J/KitlSl-20J embryos (genotype confirmed by RT-PCR) harvested at 14.5 dpc appeared pale and runted compared with their wild-type or heterozygote littermates (Figure 1A). The fetal livers in the homozygous embryos were also pale (Figure 1B) with more than 5-fold reduction in total fetal liver cellularity compared with wild-type littermates, suggesting impaired erythropoiesis. The cellular mechanism of fetal anemia in KitlSl-20J/KitlSl-20J embryos was evaluated using progenitor assay. The frequency of CFU-E progenitors per 50 000 unfractionated cells arising from KitlSl-20J/KitlSl-20J fetal liver was reduced 2.5-fold when compared with wild-type fetal livers (303 ± 23 vs 841 ± 180, KitlSl-20J/KitlSl-20J vs wild-type; mean ± SEM, n = 3-4; P < .05). By contrast there was no significant difference in the frequency of more primitive BFU-E progenitors between KitlSl-20J/KitlSl-20J and wild-type fetal livers (14 ± 0.7 vs 11.6 ± 2.5, KitlSl-20J/KitlSl-20J vs wild-type; mean ± SEM, n = 3-4; P > .05). This abnormal CFU-E/BFU-E ratio suggests a reduction in the net generation of erythroid progenitors during terminal differentiation as previously observed in other KitlSl mutant alleles.34
Phenotypic analysis of mutantKitlSl-20Jembryos. (A) Intact embryos harvested at 14.5 dpc from crosses of heterozygous mice. KitlSl-20J/KitlSl-20J embryo (right) appears more pale and runted compared with a wild-type littermate (left). (B) Fetal liver harvested from 14.5-dpc KitlSl-20J/KitlSl-20J embryo (right) appears paler and smaller than fetal liver harvested from a wild-type littermate (left). (C) Germ cell development in 13.5-dpc embryos. Genital ridges harvested at 13.5 dpc from KitlSl-20J/KitlSl-20J embryos were devoid of germ cells as demonstrated by lack of alkaline phosphatase staining when compared with genital ridges harvested from wild-type (+/+) littermates. Phosphatase-positive germ cells in wild-type genital ridges are outlined by an oval. Original magnifications: × 8 (A); × 12 (B, C).
Phenotypic analysis of mutantKitlSl-20Jembryos. (A) Intact embryos harvested at 14.5 dpc from crosses of heterozygous mice. KitlSl-20J/KitlSl-20J embryo (right) appears more pale and runted compared with a wild-type littermate (left). (B) Fetal liver harvested from 14.5-dpc KitlSl-20J/KitlSl-20J embryo (right) appears paler and smaller than fetal liver harvested from a wild-type littermate (left). (C) Germ cell development in 13.5-dpc embryos. Genital ridges harvested at 13.5 dpc from KitlSl-20J/KitlSl-20J embryos were devoid of germ cells as demonstrated by lack of alkaline phosphatase staining when compared with genital ridges harvested from wild-type (+/+) littermates. Phosphatase-positive germ cells in wild-type genital ridges are outlined by an oval. Original magnifications: × 8 (A); × 12 (B, C).
Since severe mutant Sl alleles can affect germ cell development, we also examined the genital ridges harvested from 13.5-dpc littermates by alkaline phosphatase staining. Primordial germ cells were not detectable in genital ridges harvested from KitlSl-20J/KitlSl-20J embryos as demonstrated by the lack of alkaline phosphatase staining when compared with wild-type embryos (Figure 1C).
In-frame tandem duplication of exon 3 in the mutant KitlSl-20J gene transcripts
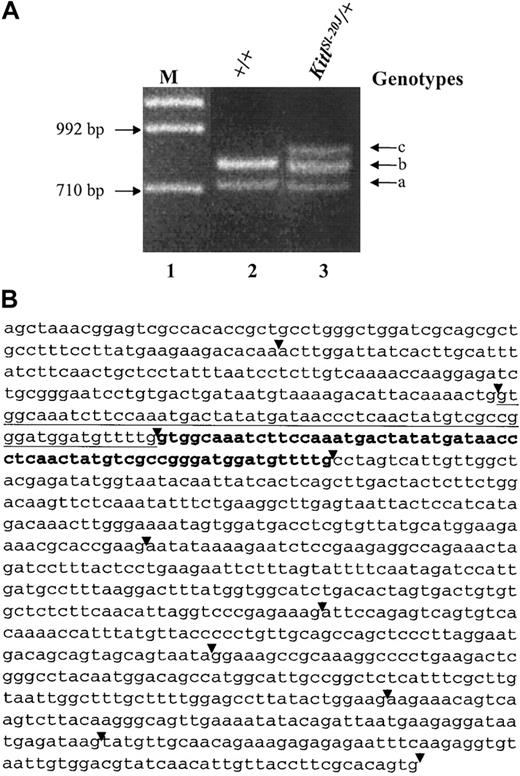
In an attempt to identify the mutation in the coding region of the KitlSl-20J allele, full-length Sl gene transcripts were amplified by RT-PCR using total RNA harvested from spleens of a heterozygous KitlSl-20J and wild-type C57BL/6J mouse. Analysis of the products yielded an additional band derived from mRNA of heterozygous KitlSl-20J mice (Figure 2A). The results were reproducible with RNA isolated from additional heterozygous KitlSl-20J mice. The presence of this additional band suggested an abnormal splice variant, use of an additional (second) promoter, or a mutation resulting in the insertion of alternative sequences in the coding region of the KitlSl-20J allele.
Analysis of RT-PCR-amplifiedSlgene transcripts. (A) Total RNA was isolated from the spleen of a C57BL/6J control mouse (+/+) and a heterozygous KitlSl-20J mouse (KitlSl-20J/+) with a white belly spot and RT-PCR was performed using primers that amplified the Sl gene transcripts. Two bands amplified with RNA isolated from +/+ mice corresponded to the membrane-associated (lane 2, band a) and soluble isoforms (lane 2, band b). An additional band (lane 3, band c) was detected with RNA isolated from heterozygous KitlSl-20J mice. M indicates marker lane. (B) Sequence analysis of the amplified soluble isoform of Sl gene transcript from heterozygous KitlSl-20J mouse. The primary copy of exon 3 is underlined and the tandem in-frame duplicated sequence is denoted in bold. The duplication of exon 3 was also present in the membrane-associated isoform of the mutant KitlSl-20J transcript (sequence not shown here). The exon boundaries are indicated by arrowheads.
Analysis of RT-PCR-amplifiedSlgene transcripts. (A) Total RNA was isolated from the spleen of a C57BL/6J control mouse (+/+) and a heterozygous KitlSl-20J mouse (KitlSl-20J/+) with a white belly spot and RT-PCR was performed using primers that amplified the Sl gene transcripts. Two bands amplified with RNA isolated from +/+ mice corresponded to the membrane-associated (lane 2, band a) and soluble isoforms (lane 2, band b). An additional band (lane 3, band c) was detected with RNA isolated from heterozygous KitlSl-20J mice. M indicates marker lane. (B) Sequence analysis of the amplified soluble isoform of Sl gene transcript from heterozygous KitlSl-20J mouse. The primary copy of exon 3 is underlined and the tandem in-frame duplicated sequence is denoted in bold. The duplication of exon 3 was also present in the membrane-associated isoform of the mutant KitlSl-20J transcript (sequence not shown here). The exon boundaries are indicated by arrowheads.
To further characterize the sequences amplified by RT-PCR analysis, individual bands generated from both wild-type and KitlSl-20J mice were cloned and sequenced. Sequence comparison of the amplified bands to the known sequence of the Sl gene (National Center for Biotechnology Information [NCBI] accession no. U44725) confirmed the wild-type identity of the 2 bands obtained from C57BL/6J mouse (Figure 2A lane 2, bands a-b) representing known soluble and membrane-associated isoforms. Sequence analysis of the top band obtained from heterozygous KitlSl-20J mouse (Figure 2A lane 3, band c) demonstrated an in-frame tandem duplication of exon 3 (Figure 2B sequence in bold) together with exon 6 sequence, corresponding to the soluble isoform of the mutant KitlSl-20J transcript. Sequence analysis of the middle band (Figure 2A lane 3, band b) revealed both wild-type sequences corresponding to the soluble isoform and sequences with a tandem duplication of exon 3 but lacking exon 6 that corresponded to the membrane-associated isoform of the mutant KitlSl-20J transcript. Sequence analysis of the lowest band (Figure 2A lane 3, band a) yielded the wild-type sequence of the membrane-associated isoform. Thus, the data suggested a genomic mutation in KitlSl-20J allele gave rise to duplicated exon 3 sequences in both soluble and membrane-associated isoforms of the mutant transcripts.
Long-range PCR product generated from DNA isolated from heterozygous KitlSl-20J mice confirms mutation at the genomic level
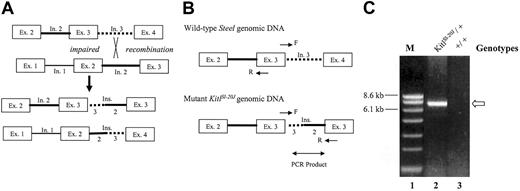
A possible mechanism giving rise to exon duplication is illegitimate recombination between repetitive elements present in introns (Figure 3A). To test for this mechanism we designed a long-range PCR strategy that used primers located in exon 3 such that a PCR product could be generated only in the presence of duplicated genomic exon 3 sequences (Figure 3B). Genomic DNA was isolated from heterozygous KitlSl-20J and wild-type C57BL/6J mice and subjected to long-range PCR analysis. The amplification of DNA from KitlSl-20J mice yielded a band of approximately 7 kb (Figure 3C lane 2), whereas no band was generated from genomic DNA isolated from the wild-type mouse (Figure 3C lane 3). A similar result was obtained from DNA isolated from a KitlSl-20J/KitlSl-20J embryo.
Proposed mechanism for exon duplication and long-range PCR strategy. (A) Impaired recombination between repetitive elements in intron 2 (solid line) and intron 3 (dotted line) of sister chromatins would generate one strand with the duplication of exon 3 and the other strand with the deletion of exon 3. (B) Schematic of the long-range PCR strategy. Primer pair in exon 3 (F indicates forward primer at the junction of exon 3 and intron 3; R, reverse primer at 5′ of exon 3) was designed to generate a PCR product only in the presence of duplicated exon 3 from the mutant KitlSl-20J genomic DNA. (C) Long-range PCR analysis of genomic DNA. An approximately 7-kb product (arrow) generated from DNA isolated from heterozygous KitlSl-20J mice (lane 2) not amplified from wild-type DNA (+/+, lane 3) supports impaired recombination as the mechanism of this mutation. M indicates marker lane.
Proposed mechanism for exon duplication and long-range PCR strategy. (A) Impaired recombination between repetitive elements in intron 2 (solid line) and intron 3 (dotted line) of sister chromatins would generate one strand with the duplication of exon 3 and the other strand with the deletion of exon 3. (B) Schematic of the long-range PCR strategy. Primer pair in exon 3 (F indicates forward primer at the junction of exon 3 and intron 3; R, reverse primer at 5′ of exon 3) was designed to generate a PCR product only in the presence of duplicated exon 3 from the mutant KitlSl-20J genomic DNA. (C) Long-range PCR analysis of genomic DNA. An approximately 7-kb product (arrow) generated from DNA isolated from heterozygous KitlSl-20J mice (lane 2) not amplified from wild-type DNA (+/+, lane 3) supports impaired recombination as the mechanism of this mutation. M indicates marker lane.
The proposed mechanism of illegitimate recombination predicted that the sequence between the duplicated exon 3 would include sequences derived from both introns 2 and 3 (Figure 3A). Analysis of the amplified long-range PCR product sequence yielded 5′ sequence from intron 3 at one end and 3′ sequence from intron 2 on the other end of the amplified product (data not shown). These data support illegitimate recombination as the mechanism of exon duplication.
Complexity analysis of intron sequences suggests a 2-step complex rearrangement as the likely mechanism mediating duplication of exon 3
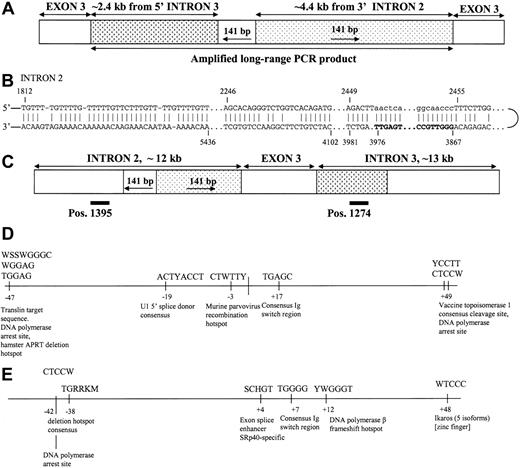
Sequence analysis of the amplified long-range PCR product also yielded a unique 141-base pair (bp) sequence at the junction of introns 3 and 2 that represents an inverted repeat of downstream sequence in intron 2 (Figure 4A). This strongly suggested a complex rearrangement in the genomic sequences of the KitlSl-20J mice resulting in the germ line-transmitted mutation.
Analysis of the long-range PCR product andSlgenomic sequences. (A) The amplified long-range PCR product comprises partial sequences of intron 3 and intron 2, as well as a 141-bp junction sequence that is an inversion of a downstream sequence in intron 2. (B) Possible mechanism of insertion of 141 bp mediated by hairpin loop formation in intron 2 (templating fragment shown in bold; inserted DNA sequence denoted by lowercase letters). (C) The position of the 2 copies of a direct repeat T(AC)14 in introns 2 and 3 are presented as black bars. Slipped mispairing between direct repeats would result in duplication of exon 3 and flanking intronic regions in the KitlSl-20J allele as shown in panel A. (D) Occurrence of known site-specific recombination motifs in intron 3 within 50 bp upstream and downstream of the breakpoint. (E) Occurrence of known site-specific recombination motifs in intron 2 within 50 bp upstream and downstream of the breakpoint (see “Discussion” for references).
Analysis of the long-range PCR product andSlgenomic sequences. (A) The amplified long-range PCR product comprises partial sequences of intron 3 and intron 2, as well as a 141-bp junction sequence that is an inversion of a downstream sequence in intron 2. (B) Possible mechanism of insertion of 141 bp mediated by hairpin loop formation in intron 2 (templating fragment shown in bold; inserted DNA sequence denoted by lowercase letters). (C) The position of the 2 copies of a direct repeat T(AC)14 in introns 2 and 3 are presented as black bars. Slipped mispairing between direct repeats would result in duplication of exon 3 and flanking intronic regions in the KitlSl-20J allele as shown in panel A. (D) Occurrence of known site-specific recombination motifs in intron 3 within 50 bp upstream and downstream of the breakpoint. (E) Occurrence of known site-specific recombination motifs in intron 2 within 50 bp upstream and downstream of the breakpoint (see “Discussion” for references).
The complexity or regularity of the DNA sequence in the vicinity of complex lesions has previously been found to be indicative of the possible mechanism of mutagenesis.33 Complexity analysis of DNA sequence, as devised by Gusev et al,32 was used to examine the potential contribution of the regularity of local DNA sequence structure to the mechanism of mutagenesis. This technique is based on the assessment of the occurrence of 4 different types of repetitive sequence element viz direct and inverted repeats and inversions thereof.32,33 Moreover, the type of repetitive element identified as making the most significant contribution to the change in local DNA sequence complexity is that which is most likely to be involved in mediating the mutation. The most probable path of lesion formation is that in which the change in complexity is maximized.
Complexity analysis suggested that the first step in the formation of the duplication/insertion rearrangement in the KitlSl-20J allele was likely to be the insertion of 141 bp mediated by an inverted repeat. This repeat appears to have promoted the formation of a hairpin loop (Figure 4B) that then templated the insertion of 141 nucleotides thereby repairing the loop and forming a more prominent structure. The second step, duplication, is readily explicable by a model of slipped mispairing. Two copies of a direct repeat, T(AC)14 (positions 1395 in intron 2 and 1274 in intron 3 as shown in Figure 4C), are thought to have been responsible for mediating this duplication; the nascent strand would have slipped backwards thereby templating the insertion of exon 3 and flanking intronic sequences. Inspection of the Sl gene sequence revealed a number of known site-specific recombination motifs35-38 in the vicinity of both the intron 3 and intron 2 breakpoints (Figure 4D-E). It is possible that one or more of these motifs were involved in the rearrangement process.
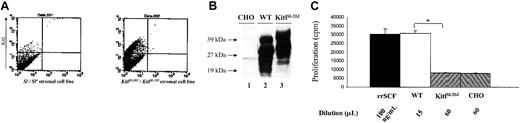
Biologic activity of expressed mutant KitlSl-20J protein
Flow cytometric analysis demonstrated that the mutant KitlSl-20J protein was expressed in fetal liver-derived stromal cells from homozygous KitlSl-20J embryos (Figure 5A). This suggested that the mutant protein lacked or had markedly reduced biologic activity. To determine the biologic activity of the mutant protein and compare it with wild-type Kitl, the wild-type cDNA encoding for secreted Kitl (1-165 amino acid [aa]) and the corresponding KitlSl-20J cDNA were cloned into mammalian expression plasmids and transfected into CHOK1 cells. As shown in Figure 5B, immunoblot of conditioned medium from stably transfected clones using an anti-murine SCF antibody demonstrated the expression and secretion of both wild-type and mutant KitlSl-20J protein into the medium. The observed pattern of multiple bands represents glycosylation of Kitl in mammalian expression systems and is consistent with published data.39 Since the biologic activity of Kitl appears to depend on homodimerization and exon 3 contains sequences implicated in the dimer interface,40 we tested the ability of the secreted Kitl proteins to form noncovalent dimers in solution by cross-linking assay using disuccinimide suberate (DSS) and its water-soluble analog bis-sulfosuccunimidyl suberate (BS3 ). These studies demonstrated that KitlSl-20J protein forms homodimers in solution similar to wild-type protein (data not shown).
Analysis of the biologic activity of mutant KitlSl-20Jprotein. (A) Flow cytometric analysis of fetal liver stromal cells derived from KitlSl-20J homozygous embryos (right) at 14.5 dpc demonstrates surface expression of Kitl protein. Fetal liver stromal cell line Sl/Sl4 derived from KitlSl/KitlSl embryos that lack expression of Kitl2 is used as the negative control (left). (B) Immunoblot analysis of conditioned medium from transfected CHOK1 cells. Conditioned medium was tested for expression of Kitl by immunoblotting with anti-murine SCF antibody. In lane 1, conditioned medium from nontransfected CHOK1 cells shows absence of Kitl. Lane 2 contains 15 μL conditioned medium from wild-type clone; lane 3, 30 μL conditioned medium from mutant (KitlSl-20J) clone. (C) Proliferation of MO7E cells in response to conditioned medium containing WT and KitlSl-20J proteins, measured by thymidine incorporation assay. Recombinant rat SCF and conditioned medium from nontransfected CHO cells (CHO) were used as controls (n = 6, mean ±SD; *P < .01, WT vs KitlSl-20J).
Analysis of the biologic activity of mutant KitlSl-20Jprotein. (A) Flow cytometric analysis of fetal liver stromal cells derived from KitlSl-20J homozygous embryos (right) at 14.5 dpc demonstrates surface expression of Kitl protein. Fetal liver stromal cell line Sl/Sl4 derived from KitlSl/KitlSl embryos that lack expression of Kitl2 is used as the negative control (left). (B) Immunoblot analysis of conditioned medium from transfected CHOK1 cells. Conditioned medium was tested for expression of Kitl by immunoblotting with anti-murine SCF antibody. In lane 1, conditioned medium from nontransfected CHOK1 cells shows absence of Kitl. Lane 2 contains 15 μL conditioned medium from wild-type clone; lane 3, 30 μL conditioned medium from mutant (KitlSl-20J) clone. (C) Proliferation of MO7E cells in response to conditioned medium containing WT and KitlSl-20J proteins, measured by thymidine incorporation assay. Recombinant rat SCF and conditioned medium from nontransfected CHO cells (CHO) were used as controls (n = 6, mean ±SD; *P < .01, WT vs KitlSl-20J).
We next determined the biologic activity of the expressed KitlSl-20J protein in stimulating proliferation of c-kit-positive MO7E cells using a thymidine incorporation assay. The proliferative response observed with 15 μL of conditioned medium containing wild-type Kitl (WT-CM) was similar to the response observed with 100 ng/mL of rrSCF confirming that WT-Kitl expressed in CHOK1 cells is biologically active (Figure 5C). By contrast, 60 μL of conditioned medium containing KitlSl-20J protein (KitlSl-20J -CM) stimulated significantly less proliferation of MO7E cells compared with WT-CM (8151 ± 379 vs 30 922 ± 1456, KitlSl-20J-CM vs WT-CM; mean ± SEM, n = 6; P < .01) and this proliferative response of KitlSl-20J-CM was not different from control conditioned medium (Figure 5C). These data demonstrated that the duplication of exon 3 results in loss of function of the expressed mutant KitlSl-20J protein when compared with wild-type Kitl.
Duplication of exon 3 results in defective binding of the expressed KitlSl-20J protein to the receptor c-kit
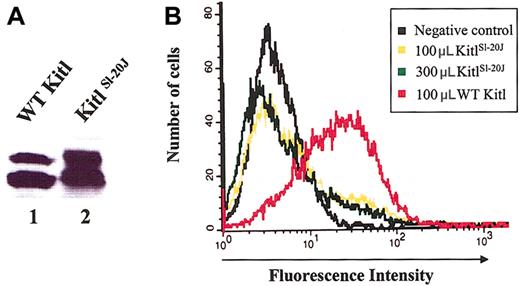
The lack of proliferative response of expressed mutant KitlSl-20J protein could be due either to a defect in binding of the mutant KitlSl-20J protein to c-kit or to an inability of the protein to activate the receptor after binding. To determine the effect of the mutation on receptor binding, we expressed KitlSl-20J and wild-type Kitl as histidine-tagged proteins. Conditioned medium was generated from transient transfection of CHOK1 cells and the expressed Kitl proteins were purified via binding of the histidine tag (Figure 6A). To analyze receptor binding, we used biotinylated anti-murine SCF antibody and streptavidin-PE to quantitate Kitl bound to the surface-expressed c-kit. Bone marrow-derived mast cells from WT mice, known to express high levels of c-kit, were used to test the binding of the Kitl protein, while mast cells derived from bone marrow of homozygous Wbanded mice41,42 that lack expression of c-kit (confirmed by flow cytometric analysis, data not shown) were used as a negative control. As expected, the binding of wild-type Kitl to c-kit could be readily detected on the surface of wild-type mast cells (Figure 6B). By contrast, no significant binding of wild-type Kitl could be detected on the surface of c-kit-deficient mast cells derived from homozygous Wbanded mice, demonstrating the specificity of this binding assay. Importantly, no significant binding of mutant KitlSl-20J protein to wild-type mast cells could be demonstrated even when 3-fold higher dilution of the mutant protein was compared with wild-type Kitl (Figure 6B). This result demonstrates that the lack of biologic activity of the mutant KitlSl-20J protein used is most likely due to defective c-kit binding.
Analysis of mutant KitlSl-20Jprotein binding to c-kit-positive cells by flow cytometry. (A) Immunoblot analysis of purified WT and KitlSl-20J proteins. (B) Receptor binding of 100 μL and 300 μL of KitlSl-20J protein compared with 100 μL WT protein. No significant binding was demonstrated with 3-fold higher dilution of KitlSl-20J protein.
Analysis of mutant KitlSl-20Jprotein binding to c-kit-positive cells by flow cytometry. (A) Immunoblot analysis of purified WT and KitlSl-20J proteins. (B) Receptor binding of 100 μL and 300 μL of KitlSl-20J protein compared with 100 μL WT protein. No significant binding was demonstrated with 3-fold higher dilution of KitlSl-20J protein.
Discussion
Mice harboring mutations in the Sl or W loci, encoding Kitl and its receptor c-kit, respectively, are easily recognized due to the associated abnormalities in coat color. The KitlSl-20J mice arose as a spontaneous mutation in the C57BL/6J breeding colony at The Jackson Laboratories and exhibit a white belly spot characteristic of a heterozygous mutation in the Sl or W locus. The results of complementation studies at Jackson Laboratories were consistent with a mutation in the Sl gene. Crossbreeding of the heterozygous KitlSl-20J mice did not generate any viable mice with severe hematologic or coat color abnormalities, and segregation patterns obtained both with viable newborn offspring and with embryos harvested at 14.5 dpc confirmed the embryonic lethality of the KitlSl-20J allele in the homozygous state.
Phenotypic analysis of homozygous KitlSl-20J embryos obtained from timed pregnancies at 14.5 dpc exhibited significantly reduced fetal liver cellularity and frequency of CFU-E progenitors. Since the frequency of BFU-E progenitors was not significantly different between the homozygous and mutant embryos, these data were suggestive of a defect in the generation of erythroid progenitors during terminal differentiation. These results are consistent with previous fetal liver hematopoietic studies using compound heterozygous mice (KitlSl/KitlSl-d), which demonstrated that mutant KitlSl alleles do not affect yolk sac-derived primitive hematopoiesis but significantly impair the terminal differentiation of the fetal liver-derived definitive erythroid cell lineage.34,43,44 Homozygous embryos harvested at 13.5 dpc also exhibited a significant lack of germ cells in the genital ridges as determined by alkaline phosphatase staining, a finding similar to the previously observed phenotype in other severe mutant KitlSl and W alleles.35,45
We then identified the molecular mutation in the KitlSl-20J allele by cloning and sequencing RT-PCR amplified products of Sl gene transcripts. Sequence analysis demonstrated that the mutation constituted an in-frame tandem duplication of exon 3 in both isoforms of the mutant Sl gene transcripts and confirmed that the KitlSl-20J mice were heterozygous for this mutation as predicted from the coat color phenotype.
Exon duplication is an important mechanism of mutation in evolution43 but is comparatively infrequent as a cause of pathology.36 Thus, although unambiguous intragenic multiexon duplications constitute approximately 0.1% of the inherited mutations logged in the Human Gene Mutation Database (www.hgmd.org), only 10 (0.02% of the total) involve the duplication of solitary exons. The murine mutation reported here may therefore be regarded as somewhat unusual. Although a number of mutations in the Sl gene have been identified, the KitlSl-20J mutation is the first example of exon duplication in this gene.
A model of impaired recombination resulting in duplication of exon 3 predicts a region comprising introns 3 and 2 between the duplicated exons. Sequence analysis of the amplified product of long-range PCR using overlapping exon 3 primers confirmed that the intronic region between the duplicated exons comprises introns 3 and 2. Analysis of the junction between introns 3 and 2 revealed a 141-bp sequence that represents an inversion of a downstream sequence of intron 2. These data thus suggest that the mutation in the KitlSl-20J allele represents a complex rearrangement involving a 2-step process of mutagenesis.
Meta-analytical studies have established that the local DNA sequence environment plays an important role in the generation of complex lesions.37 Inspection of the Sl gene sequence in the vicinity of the intron 2 and intron 3 breakpoints revealed a number of motifs known to be involved in site-specific recombination including a deletion hot-spot consensus sequence,38 a DNA polymerase frameshift hot spot,46 a consensus immunoglobulin switch region,47 and a translin target sequence48 (Figure 4D-E). One or more of these motifs could have been involved in the rearrangement process. Complexity analysis used to analyze DNA sequence flanking exon 3 of the Sl gene revealed that the KitlSl-20J mutation most likely originated via a 2-step process of insertion and duplication. We propose that the insertion was mediated by an inverted repeat and that this was followed by the duplication of exon 3, explicable by a model of slipped mispairing.
We next studied the consequence of the tandem duplication of exon 3 on the biologic properties of the mutant KitlSl-20J protein that results in an embryonic lethal phenotype. Exon 3 comprises 63 bp and starts with the first base of the in-frame triplet codon (G of GTG→valine). Thus, duplication of exon 3 would be predicted to result in the duplication of the 21 amino acids encoded in the mutant KitlSl-20J protein. Two groups have recently published the crystal structure of the soluble form of human Kitl.40,49 According to the published structure,49 human Kitl is a short-chain helical cytokine comprising 4 helices (αA, αB, αC, and αD) and 2β-strands (β1 located between αA and αB and β2 located between αC and αD). Since mouse Kitl is 88% identical in primary sequence to the human ortholog and murine Kitl effectively binds to human c-kit, it is reasonable to assume considerable similarities in the 3-dimensional structure between protein from these species. Exon 3 encodes for the residues 19 to 39 that form the end of the first helix (αA), encompasses the β1 strand, and ends 4 residues before the start of the second helix (αB). There is an additional 1-turn helix (αB′), between the β1 and αB, encoded by exon 3. Since flow analysis for surface expression of membrane-associated Kitl demonstrated that the protein was expressed in the homozygous KitlSl-20J fetal liver-derived stromal cells, we expressed the wild-type and mutant KitlSl-20J proteins in CHOK1 cells for comparative biologic studies. Expressed wild-type and mutant KitlSl-20J protein exhibited multiple bands on immunoblot analysis representing posttranslational glycosylation that is consistent with published data.39 Neither the glycosylation patterns nor the apparent quantity of mutant KitlSl-20J protein expressed was significantly different from the wild-type protein. The apparent slight increase in molecular weight seen in Figure 5B is due to the addition of exon 3-coded residues.
Proliferation assays performed with conditioned medium generated from transfected CHOK1 cells demonstrated a significant reduction in the ability of mutant KitlSl-20J protein to stimulate MO7E cells when compared with wild-type. Previous study of the biologic activity of various truncated mutant human Kitl forms has demonstrated that the truncation of the human soluble Kitl after N-terminal 127 amino acid residues resulted in a significant loss of cell proliferation activity but not significant effect on binding to the receptor c-kit.50 Since the binding of Kitl to c-kit can be separated from the activation of c-kit downstream signaling events, the lack of proliferative response demonstrated with mutant KitlSl-20J protein could in theory be due to a defect in binding of the mutant protein to the receptor or due to a defect in receptor activation. To test the consequence of the mutation on receptor binding we studied the binding of Kitl to receptor c-kit by flow cytometry. Receptor binding analysis demonstrated that the mutant KitlSl-20J protein was largely defective in binding to the receptor when compared with wild-type Kitl. These experimental data are supported by the study of human-murine Kitl chimeras that has implicated residues 1-35 in receptor binding to c-kit that include the residues 19-35 encoded by exon 3.51
The phenotype of KitlSl-20J/KitlSl-20J embryos as described here is similar to the null phenotype seen with lethal KitlSl mutant alleles (KitlSl, KitlSl-gb) that result in the deletion of the entire coding region. The loss of function of the KitlSl-20J protein when compared with wild-type Kitl suggests that the duplication of exon 3 has profound consequences on the biologic activity since the phenotype is very similar to these null mice and likely mediated by lack of receptor binding.
Taken together, the KitlSl-20J allele represents an unusual complex rearrangement of the Sl gene that results in the duplication of exon 3 and a severe biologic phenotype, suggesting an important role for exon 3 encoded sequences in receptor binding.
Prepublished online as Blood First Edition Paper, July 24, 2003; DOI 10.1182/blood-2003-05-1468.
Supported by National Institutes of Health grant R01 DK48605.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to acknowledge Dr David Ginsberg (Howard Hughes Medical Institute, University of Michigan, Ann Arbor, MI) and Dr Chris Wylie (Cincinnati Children's Research Foundation) for their help with long-range PCR design and germ cell staining, respectively. We thank Amgen for providing rat SCF. We thank members of the laboratory for helpful discussion and Matthew Hodgson and Keisha Steward for administrative assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal