Our previous study demonstrated that 2-methoxyestradiol (2ME2), an estrogen derivative, induces apoptosis in multiple myeloma (MM) cells; however, the related transcriptional events are unclear. In the present study, we used oligonucleotide microarrays to identify genes altered during 2ME2-induced apoptosis in MM cells. 2ME2 triggers an early transient induction of genes known to trigger cell death and repression of growth/survival-related genes. Many genes regulating cell defense/repair machinery also were transiently induced. Since 2ME2 also induces apoptosis in MM cells resistant to conventional therapies such as dexamethasone (Dex), we compared the gene profiles of 2ME2-treated and Dex-resistant MM cells. Our results suggest that 2ME2 overcomes Dex resistance by modulating genes that confer chemoresistance in MM cells. Microarray results were confirmed by Northern and Western blot analyses. A comparative analysis of selected genes from freshly isolated MM patient cells and 2ME2-treated MM.1S MM cells further provides an in vivo relevance of our in vitro studies. Collectively, these findings suggest genetic events mediating anti-MM activity of 2ME2, as well as mechanisms whereby 2ME2 overcomes Dex resistance in MM cells. These studies may therefore allow improved therapeutic use of 2ME2, based upon targeting genes that regulate MM cell growth and survival.
Introduction
2-Methoxyestradiol (2ME2), a natural metabolite of estradiol, is a potent antitumor and antiangiogenic agent.1 Studies using xenografts and metastatic disease models in mice have suggested that 2ME2 targets not only the tumor cell but also the tumor vasculature.2,3 Our recent studies showed that 2ME2 induces apoptosis in refractory multiple myeloma (MM) cells and evades the protective effects of growth factors present in the bone marrow millieu.4 These findings provide a potential rationale for evaluating 2ME2 as a novel therapeutic in MM.
Despite being a natural derivative of estradiol (E2), 2ME2 binds poorly to the estrogen receptors (ERs), and therefore the antiproliferative effects of 2ME2 are not mediated by ERs.5 Various other mechanisms of 2ME2 actions have been suggested, including inhibition of tubulin polymerization,6,7 sulfonation of 2ME2,8 and inhibition of superoxide dismutase.9 Recent studies have implicated a role for p53 and mitochondria-initiated signals in response to 2ME2.4 9-11 To date, however, 2ME2-triggered genetic events are unclear.
Gene expression profiling has provided framework to improve diagnosis and classification of cancer and to define novel mechanisms of drug actions.12 13 In the present study, we therefore used oligonucleotide microarrays to (1) identify gene expression changes in 2ME2-treated MM cells, (2) derive functional significance of selected altered genes, and (3) obtain a subset of genes that are significantly altered by 2ME2 treatment and are either overexpressed in MM cells or may confer Dex resistance. These profiling studies both elucidate the gene alteration patterns during 2ME2 activity in MM and provide the framework for clinical application of 2ME2, either alone or in combination with other conventional therapies, to improve patient outcome in refractory MM.
Materials and methods
Cell culture and reagents
Dex-sensitive MM.1S and Dex-resistant MM.1R human MM cell lines were kindly provided by Dr Steven Rosen (Northwestern University, Chicago, IL).14 Cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine. MM.1R cell line was cultured with Dex to confirm lack of drug sensitivity. Mononuclear cells were prepared from MM patient bone marrow (BM) samples by Ficoll-Hypaque density gradient centrifugation. The MM cells were freshly isolated from patients relapsing after prior therapy with Dex. An informed consent was obtained from all patients in accordance with the Helsinki protocol. Tumor cells (CD138+, 97% ± 2.0%) were isolated by CD138 positive selection using CD138 (Syndecan-1) Micro Beads and Auto MACS magnetic cell sorter, according to manufacturer's instructions (Miltenyi Biotec, Auburn, CA). CD138+ myeloma cells were viable (94%-97%) for 2-3 weeks in vitro.
MM.1S cells were treated with 3 μM 2ME2 (Sigma Chemical, St Louis, MO) for 6 hours, 12 hours, 24 hours, 48 hours, or 72 hours, and harvested MM.1S cells also were treated with 0.05 μM Dex (Sigma Chemical) or interleukin-6 (10 ng) for 24 hours, 48 hours, or 72 hours, as previously described.15 Cell viability was performed using trypan blue exclusion (> 95% at 6 hours and 12 hours; > 80% at 24 hours; < 45% at 48 hours; and < 80% at 72 hours of 2ME2 exposure to MM.1S MM cells).
Preparation of biotinylated probes and hybridization on microarrays
Total RNA was isolated from 1 × 107 lysed cells using TRIzol Reagent (Life Technologies, Rockville, MD) and double-stranded cDNA prepared from 5 μg of total RNA using Life Technologies Superscript choice system and an oligo (dT)–anchored T7 primer. Affymetrix huGene FL arrays (Santa Clara, CA) containing 12 000 genes were used for mRNA expression profiling. Biotinylated RNA was synthesized using the BioArray RNA transcript labeling kit (Enzo, Farmingdale, NY) with biotin-11-CTP and biotin-16-UTP for 5 hours at 37°C. In vitro transcription products were purified using RNeasy columns (Quiagen, Valencia, CA). Biotinylated RNA was then treated for 35 minutes at 94°C in a buffer containing 200 mM Tris [tris(hydroxymethyl)aminomethane] acetate, pH 8.1, 500 mM potassium acetate, and 150 mM magnesium acetate.
Affymetrix U95A arrays were hybridized with biotinylated in vitro transcription products (10 μg/chip) for 16 hours at 45°C using the manufacturer's hybridization buffer. Fluidic station 400 (Affymetrix) was used for washing and staining the arrays. The DNA chips were then analyzed using a gene array scanner (Affymetrix) with argon ion laser as an excitation source; emission was detected by a photomultiplier tube through a 570-nm long pass filter.
Data processing.
The .CEL (cell intensity) files are obtained using Affymetrix Microarrays Suite 5.0 software. The DNA-Chip Analyzer (DChip) developed by us (www.dchip.org, version 1.0) is used to normalize all CEL files to a baseline array with overall median intensity, and the model-based expression (PM/MM difference model) is used to compute the expression values.
Selection of 170 genes.
Using the DChip “Compare samples” function, 170 noncontrol probe sets are selected. The selected genes represent (1) greater than 1.3 lower bound of fold change in either direction, (2) greater than 100 expression value difference in either direction, and (3) theP value for testing of the zero expression value difference less than 0.05, in at least 1 of the 5 comparisons: control versus 6 hours, control versus 12 hours, control versus 24 hours, control versus 48 hours, and control versus 72 hours. Note that the P value in criteria 3 is based on a t-like statistic and is used to assess the relative magnitude of the expression value difference when considering the model-based standard errors of the expression values; therefore, this P value is used only as a filtering statistic rather than as an assessment of the real statistical significance.
Definition of 61 induced genes and 109 suppressed genes.
From the 170 genes, genes were separated into 2 groups. One group contains the genes that were induced at any time point compared to the control sample (“induced group”), which satisfied the criteria in 1, 2, and 3 for up-regulation. The other group is the “suppressed group,” satisfying the criteria for down-regulation.
Western blot analyses
Total protein lysates and cytosolic extracts were prepared as previously described.16 Briefly, equal amounts of proteins were resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes. Proteins were then transferred to nitrocellulose filters, blocked by incubation in 5% dry milk in PBST (0.05% Tween-20 in phosphate-buffered saline), and probed with anti–XBP-1 (kindly provided by Dr Laurie Glimcher, Harvard Medical School, Boston, MA); antitubulin, antiactin (Sigma Chemicals); anti-SHP2, anti–c-myc, anti–cyclin B, anti-p27KIP, anti Hsp-27, -70, -90 (Santa Cruz Biotech, Santa Cruz, CA); or antiubiquitin (kindly provided by Dr Lan Bo Chen, Dana Farber Cancer Institute, Boston, MA) Abs. Blots were then developed by enhanced chemiluminescence (ECL; Amersham, Arlington Heights, IL).
Northern blot analyses
Northern blot analyses were performed as previously described.17 Briefly, total RNA (20 μg/lane) was subjected to electrophoresis through a 1% agarose/2.2 M formaldehyde gel, transferred to nitrocellulose filters, and hybridized to32P-labeled DNA probes for XBP-1 (kindly provided by Dr Laurie Glimcher), and glyceraldehyde phosphate dehydrogenase (GAPDH) probe. Hybridizations were performed at 42°C for 24 hours in 50% (vol/vol) formamide, 2 × SSC (SSC: 0.15 M sodium chloride, 0.015 M sodium citrate), 1 × Denhardt solution, 0.1% (wt/vol) sodium dodecyl sulfate (SDS), and 200 μg/mL salmon sperm DNA. The filters were washed twice in 2 × SSC-0.1% SDS at room temperature, and then in 0.1 × SSC-0.1% SDS at 60°C for 30 minutes before exposure to Kodak X-Omat XAR film (Eastman Kodak, Rochester, NY) using an intensifying screen. The autoradiograms were scanned using an LKB Produkter (Bromma, Sweden) Ultrascan XL laser densitometer and analyzed with the Gelscan software package. Signal intensity was determined in a linear range and normalized to GAPDH.
Transient transfections
MM.1S cells were transiently transfected using “Cell line Nucleofector Kit V,” according to manufacturer's instructions (Amaxa Biosystems, Koeln, Germany) with vector containing green fluorescence protein (GFP) alone or with dominant-negative XBP-1 (DN-XBP-1). Following transfections, GFP-positive cells were selected by flow cytometry and treated with Dex (5 μM), Dex + IL-6 (10 ng), or 2ME2 (3 μM) for 48 hours. Cell viability was assessed by 3-(4,5-dimethylthiozol-2,5-diphenyltetrazolium bromide (MTT; Chemicon International, Temecula, CA) assay according to manufacturer's instructions. Cell viability was also assessed by trypan blue exclusion.
Transwell migration assay
Cell migration was assayed using a modified Boyden chamber assay, as described previously.18 Briefly, starved cells were treated with 3 μM 2ME2 for 12 hours and then added to a 8-mm pore size polycarbonate membrane precoated with indicated fibronectin concentrations separating the 2 chambers of a 6.5-mm transwell (Costar, San Diego, CA). Recombinant vascular endothelial growth factor165 (VEGF165) (10 ng/mL) was added to the lower chamber. After 2-5 hours, cells that had migrated into the lower compartment were counted using a Coulter counter ZBII (Beckman Coulter, Fullerton, CA).
Results and discussion
To delineate specific alterations in gene expression induced by 2ME2, MM.1S MM cells were treated with 2ME2 (3 μM) and harvested at different time intervals. Total cellular RNA was prepared and subjected to oligonucleotide microarray profiling, followed by data analysis using DChip. These studies identify genes that are either transiently induced or repressed in response to 2ME2 treatment. To interpret and organize the data, differentially expressed cDNAs are grouped according to their temporal expression pattern using a hierarchical clustering algorithm.19 20
Genes regulated in response to 2ME2
A total of 184 differentially expressed probe sets, including 170 noncontrol probe sets, are selected (Figures1-2). The primary gene microarray data also are provided at the Web sitehttp://biosun1.harvard.edu/complab/2me2/. Our results demonstrate that treatment of MM.1S cells with 2ME2 alters expression of genes encoding apoptotic and antiapoptotic signaling proteins, structural/cytoskeleton proteins, transcription factors, cell surface proteins, as well as proteins involved in intracellular transport and signaling via both endoplasmic reticulum (ER) and mitochondria.
Hierarchical clustering of gene expression data.
MM.1S cells were treated with 3 μM 2ME2 for the indicated times and subjected to oligonucleotide array analysis. The data were then analyzed using DNA-Chip analyzer (DChip); 61 genes are induced in response to 2ME2 treatment at indicated time intervals. Each row represents a gene. The expression index is standardized genewise to have mean 0 and standard deviation 1 (−3.0, blue to +3.0, red).
Hierarchical clustering of gene expression data.
MM.1S cells were treated with 3 μM 2ME2 for the indicated times and subjected to oligonucleotide array analysis. The data were then analyzed using DNA-Chip analyzer (DChip); 61 genes are induced in response to 2ME2 treatment at indicated time intervals. Each row represents a gene. The expression index is standardized genewise to have mean 0 and standard deviation 1 (−3.0, blue to +3.0, red).
Hierarchical clustering of gene expression data.
MM.1S cells were treated with 3 μM 2ME2 for the indicated times and subjected to oligonucleotide array analysis. The data were then analyzed using DChip; 109 genes are suppressed in response to 2ME2 treatment at indicated time intervals. Each row represents a gene. The expression index is standardized genewise to have mean 0 and standard deviation 1 (−3.0, blue to +3.0, red).
Hierarchical clustering of gene expression data.
MM.1S cells were treated with 3 μM 2ME2 for the indicated times and subjected to oligonucleotide array analysis. The data were then analyzed using DChip; 109 genes are suppressed in response to 2ME2 treatment at indicated time intervals. Each row represents a gene. The expression index is standardized genewise to have mean 0 and standard deviation 1 (−3.0, blue to +3.0, red).
As shown in Figure 1, the expression pattern of multiple genes changed progressively with time of 2ME2 treatment. First, significant numbers of genes (29 of total 58 induced genes) are up-regulated soon (6-12 hours) after 2ME2 treatment, suggesting that the initial stress response is mediated through gene induction. Second, apoptosis-related genes contributing to generation of reactive oxygen species (ROS) such as proline oxidase,21 nM23,22 and creatine kinase23 are detectable early, at 6 hours and 12 hours of 2ME2 exposure (Figure 1). The induction of ROS-related genes is consistent with prior studies demonstrating a role for ROS in 2ME2-triggered responses in other cell systems.9Interestingly, 2ME2 also simultaneously induces genes known to regulate cell defense/survival machinery. For example, 2ME2 increases the transcripts of growth factor receptors (G-protein coupled receptor), cell adhesion molecules (integrin β-5, mucin1), or eukaryotic initiation factor family (eIF5A). Prior studies have shown that up-regulation of cell adhesion molecules provides a survival advantage for MM cells in the BM microenvironment.24,25 Third, growth-related genes, such as the BTG family interleukin 2γ-receptor gene, are significantly down-regulated in response to 2ME2 treatment (Figure 2). Fourth, 2ME2 alters structural/cytoskeleton and cell cycle regulatory genes (tubulin, actin, c-myc, cyclin B) (Figures 1-2; Table1). The changes in these genes correlate with the 2ME2-induced growth arrest and apoptosis in MM cells. Fifth, 2ME2 down-regulates mRNA levels of genes encoding proteins that are both highly expressed in MM26 and may confer drug resistance (XBP-1, adrenomedullin, heat shock family, ubiquitins).16 27-29
Genes modulated by 2ME2 in MM cells
| Gene name . | GenBank no. . | 2ME2 (fold change over control) . | ||||
|---|---|---|---|---|---|---|
| 6 hr . | 12 hr . | 24 hr . | 48 hr . | 72 hr . | ||
| Growth/proliferation and cell adhesion | ||||||
| Interleukin 2 receptor, γ | D11086 | −1.8 | −1.8 | −2.1 | −3.0 | −2.5 |
| BTG family, member 2 | U72649 | −1.8 | −1.9 | −2.4 | −2.9 | −1.9 |
| Small inducible cytokine A3 | D90144 | −1.5 | −1.4 | −1.7 | −2.4 | 1.5 |
| Rho GDP dissociation inhibitor (GDI) | X69549 | −2.0 | −1.9 | −1.5 | −1.6 | −2.5 |
| G-protein coupled receptor | AF091890 | 1.3 | 2.6 | 1.2 | 1.3 | 1.3 |
| Mucin 1, transmembrane | X52228 | 2.1 | 1.5 | 1.6 | 1.5 | 1.8 |
| Integrin, β 5 | M35011 | 1.5 | 2.5 | 1.5 | 1.6 | 1.3 |
| LPS-induced TNF-α factor | AF010312 | −3.1 | −3.6 | −2.8 | −2.9 | −2.3 |
| TNF-α–induced protein 3 | M59465 | −2.3 | −2.0 | −2.8 | −3.0 | −2.6 |
| Antigen identified by monoclonal antibody Ki-67 | X65550 | −2.6 | −2.3 | −1.2 | −1.7 | −2.1 |
| Farnesyl diphosphate synthase | D14697 | −2.3 | −2.2 | −1.8 | −1.7 | −1.7 |
| Apoptosis/survival/drug resistance | ||||||
| Proline dehydrogenase (oxidase) 1 | U82381 | 2.8 | 2.1 | 2.4 | 2.1 | 2.2 |
| Creatine kinase | X15334 | 2.9 | 1.8 | 1.5 | 1.3 | 1.4 |
| Poly (ADP-ribose) polymerase | J03473 | −1.4 | −1.7 | −1.7 | −1.6 | −1.9 |
| Crystalin, β B1 | U35340 | 2.5 | 2.1 | 1.3 | 1.7 | 1.5 |
| C-myc-P64 | M13929 | 2.5 | 2.4 | 1.8 | 1.6 | 1.1 |
| Casein kinase 1, epsilon | L37043 | −2.1 | −1.7 | −1.8 | −1.7 | −1.9 |
| NM23/nonmetastatic cells 4 | Y07604 | −2.1 | −1.9 | −2.2 | −2.3 | −2.8 |
| C-terminal binding protein 1 | U37408 | −2.2 | −2.5 | −2.4 | −3.4 | −2.8 |
| Pyruvate kinase | M26252 | −3.4 | −4.6 | −3.1 | −5.0 | −6.1 |
| CDC28 protein kinase 2 | X54942 | −1.6 | −1.6 | 1.5 | 2.5 | 1.4 |
| Protein tyrosine phosphatase, nonreceptor type 6 | X62055 | −3.5 | −3.3 | −2.5 | −2.8 | −2.3 |
| Lectin, galactoside-binding, soluble, 1 (galactin 1) | AI535946 | −2.5 | −2.8 | −3.1 | −1.9 | −2.3 |
| V-raf-1 murine leukemia viral oncogene homolog 1 | X06409 | −2.5 | −3.1 | −2.5 | −2.2 | −1.8 |
| Myeloid cell leukemia sequence 1 (BCL2 related) | L08246 | −2.2 | −2.1 | −2.2 | −2.4 | −1.9 |
| Adrenomedullin | D14874 | −2.4 | −2.6 | −3.2 | −4.8 | −6.4 |
| X-box binding protein 1 | Z93930 | −1.9 | −2.4 | −2.5 | −3.2 | −1.8 |
| Pim-2oncogene | U77735 | −1.5 | −1.3 | −1.4 | −2.3 | −2.0 |
| Endoplasmic reticulum and translation | ||||||
| Eukaryotic translation initiation factor 5A | AI074025 | 2.0 | 1.6 | 2.1 | 1.5 | 1.4 |
| Eukaryotic translation initiation factor 4B | X55733 | −2.8 | −2.7 | −2.8 | −3.1 | −2.6 |
| Eukaryotic translation elongation factor 1 | L41498 | −2.0 | −2.3 | −2.2 | −2.1 | −1.0 |
| Eukaryotic translation initiation factor 3 | U94855 | −2.3 | −2.3 | −1.6 | −1.5 | −1.7 |
| TAP binding protein (tapasin) | AF029750 | −2.2 | −2.8 | −2.6 | −3.0 | −1.9 |
| Tumor rejection antigen (gp96) | X15187 | −1.9 | −2.4 | −2.4 | −2.3 | −1.4 |
| Endoplasmic reticulum stress inducible | AF055001 | −1.5 | −2.2 | −2.4 | −2.0 | 1.2 |
| Cytoskeleton/structural | ||||||
| Suppression of tumorigenicity 5 | U15131 | 2.0 | 2.1 | 1.6 | 1.7 | 1.6 |
| Tubulin, β, 2 | X02344 | −1.9 | −2.4 | −1.3 | −1.5 | −2.4 |
| Actin-related protein 2/3 complex | AF006088 | −1.2 | −1.5 | −2.6 | −1.3 | −1.5 |
| Vimentin | Z19554 | −2.1 | −2.9 | −1.7 | −1.4 | −1.4 |
| Lymphocyte specific protein 1 | M33552 | −4.0 | −4.2 | −2.7 | −2.6 | −2.2 |
| Adducin 1 (α) | X58141 | −1.7 | −1.8 | −2.2 | −2.3 | −1.9 |
| Ubiquitins and metabolism | ||||||
| Nucleosome assembly protein 1-like 1 | W26056 | −1.9 | −2.5 | −2.3 | −2.3 | −2.0 |
| γ-glutamyltransferase 2 | M30474 | −2.0 | −2.5 | −1.4 | −1.8 | −1.3 |
| Phosphomannomutase 2 | U85773 | 2.4 | 1.9 | 1.4 | 1.7 | 1.5 |
| Ubiquitin-conjugating enzyme E2M | AF075599 | 2.2 | 2.1 | 2.6 | 2.4 | 2.0 |
| Karyopherin α 2 (RAG) | U28386 | −1.1 | −1.2 | 1.2 | 1.2 | −2.1 |
| Solute carrier family 25 | J03592 | −2.6 | −2.7 | −2.1 | −1.9 | −1.5 |
| Cytochrome c oxidase subunit VIa polypeptide 1 | AI540925 | −1.6 | −1.8 | −1.3 | −1.2 | −1.3 |
| Ubiquitin-conjugating enzyme E2G 2 | AF032456 | 1.7 | 1.7 | 1.1 | 1.8 | 1.1 |
| Ubiquitin-conjugating enzyme E2E 3 | AB017644 | −1.6 | −1.6 | −1.8 | −1.6 | −1.9 |
| Splicing factor, arginine/serine-rich 6 | AL031681 | 2.0 | 1.5 | −1.0 | −1.2 | −1.1 |
| Leukotriene A4 hydrolase | J03459 | −2.4 | −2.4 | −2.0 | −2.8 | −2.1 |
| Ubiquitin-activating enzyme E1 | M58028 | −2.2 | −2.8 | −2.2 | −2.6 | −2.0 |
| FK506 binding protein 1A (12 kDa) | V00599 | −2.1 | −1.9 | −1.5 | −2.9 | −4.5 |
| RNA binding motif protein 3 | U28686 | 1.8 | 2.1 | 1.6 | 1.5 | 1.4 |
| Gene name . | GenBank no. . | 2ME2 (fold change over control) . | ||||
|---|---|---|---|---|---|---|
| 6 hr . | 12 hr . | 24 hr . | 48 hr . | 72 hr . | ||
| Growth/proliferation and cell adhesion | ||||||
| Interleukin 2 receptor, γ | D11086 | −1.8 | −1.8 | −2.1 | −3.0 | −2.5 |
| BTG family, member 2 | U72649 | −1.8 | −1.9 | −2.4 | −2.9 | −1.9 |
| Small inducible cytokine A3 | D90144 | −1.5 | −1.4 | −1.7 | −2.4 | 1.5 |
| Rho GDP dissociation inhibitor (GDI) | X69549 | −2.0 | −1.9 | −1.5 | −1.6 | −2.5 |
| G-protein coupled receptor | AF091890 | 1.3 | 2.6 | 1.2 | 1.3 | 1.3 |
| Mucin 1, transmembrane | X52228 | 2.1 | 1.5 | 1.6 | 1.5 | 1.8 |
| Integrin, β 5 | M35011 | 1.5 | 2.5 | 1.5 | 1.6 | 1.3 |
| LPS-induced TNF-α factor | AF010312 | −3.1 | −3.6 | −2.8 | −2.9 | −2.3 |
| TNF-α–induced protein 3 | M59465 | −2.3 | −2.0 | −2.8 | −3.0 | −2.6 |
| Antigen identified by monoclonal antibody Ki-67 | X65550 | −2.6 | −2.3 | −1.2 | −1.7 | −2.1 |
| Farnesyl diphosphate synthase | D14697 | −2.3 | −2.2 | −1.8 | −1.7 | −1.7 |
| Apoptosis/survival/drug resistance | ||||||
| Proline dehydrogenase (oxidase) 1 | U82381 | 2.8 | 2.1 | 2.4 | 2.1 | 2.2 |
| Creatine kinase | X15334 | 2.9 | 1.8 | 1.5 | 1.3 | 1.4 |
| Poly (ADP-ribose) polymerase | J03473 | −1.4 | −1.7 | −1.7 | −1.6 | −1.9 |
| Crystalin, β B1 | U35340 | 2.5 | 2.1 | 1.3 | 1.7 | 1.5 |
| C-myc-P64 | M13929 | 2.5 | 2.4 | 1.8 | 1.6 | 1.1 |
| Casein kinase 1, epsilon | L37043 | −2.1 | −1.7 | −1.8 | −1.7 | −1.9 |
| NM23/nonmetastatic cells 4 | Y07604 | −2.1 | −1.9 | −2.2 | −2.3 | −2.8 |
| C-terminal binding protein 1 | U37408 | −2.2 | −2.5 | −2.4 | −3.4 | −2.8 |
| Pyruvate kinase | M26252 | −3.4 | −4.6 | −3.1 | −5.0 | −6.1 |
| CDC28 protein kinase 2 | X54942 | −1.6 | −1.6 | 1.5 | 2.5 | 1.4 |
| Protein tyrosine phosphatase, nonreceptor type 6 | X62055 | −3.5 | −3.3 | −2.5 | −2.8 | −2.3 |
| Lectin, galactoside-binding, soluble, 1 (galactin 1) | AI535946 | −2.5 | −2.8 | −3.1 | −1.9 | −2.3 |
| V-raf-1 murine leukemia viral oncogene homolog 1 | X06409 | −2.5 | −3.1 | −2.5 | −2.2 | −1.8 |
| Myeloid cell leukemia sequence 1 (BCL2 related) | L08246 | −2.2 | −2.1 | −2.2 | −2.4 | −1.9 |
| Adrenomedullin | D14874 | −2.4 | −2.6 | −3.2 | −4.8 | −6.4 |
| X-box binding protein 1 | Z93930 | −1.9 | −2.4 | −2.5 | −3.2 | −1.8 |
| Pim-2oncogene | U77735 | −1.5 | −1.3 | −1.4 | −2.3 | −2.0 |
| Endoplasmic reticulum and translation | ||||||
| Eukaryotic translation initiation factor 5A | AI074025 | 2.0 | 1.6 | 2.1 | 1.5 | 1.4 |
| Eukaryotic translation initiation factor 4B | X55733 | −2.8 | −2.7 | −2.8 | −3.1 | −2.6 |
| Eukaryotic translation elongation factor 1 | L41498 | −2.0 | −2.3 | −2.2 | −2.1 | −1.0 |
| Eukaryotic translation initiation factor 3 | U94855 | −2.3 | −2.3 | −1.6 | −1.5 | −1.7 |
| TAP binding protein (tapasin) | AF029750 | −2.2 | −2.8 | −2.6 | −3.0 | −1.9 |
| Tumor rejection antigen (gp96) | X15187 | −1.9 | −2.4 | −2.4 | −2.3 | −1.4 |
| Endoplasmic reticulum stress inducible | AF055001 | −1.5 | −2.2 | −2.4 | −2.0 | 1.2 |
| Cytoskeleton/structural | ||||||
| Suppression of tumorigenicity 5 | U15131 | 2.0 | 2.1 | 1.6 | 1.7 | 1.6 |
| Tubulin, β, 2 | X02344 | −1.9 | −2.4 | −1.3 | −1.5 | −2.4 |
| Actin-related protein 2/3 complex | AF006088 | −1.2 | −1.5 | −2.6 | −1.3 | −1.5 |
| Vimentin | Z19554 | −2.1 | −2.9 | −1.7 | −1.4 | −1.4 |
| Lymphocyte specific protein 1 | M33552 | −4.0 | −4.2 | −2.7 | −2.6 | −2.2 |
| Adducin 1 (α) | X58141 | −1.7 | −1.8 | −2.2 | −2.3 | −1.9 |
| Ubiquitins and metabolism | ||||||
| Nucleosome assembly protein 1-like 1 | W26056 | −1.9 | −2.5 | −2.3 | −2.3 | −2.0 |
| γ-glutamyltransferase 2 | M30474 | −2.0 | −2.5 | −1.4 | −1.8 | −1.3 |
| Phosphomannomutase 2 | U85773 | 2.4 | 1.9 | 1.4 | 1.7 | 1.5 |
| Ubiquitin-conjugating enzyme E2M | AF075599 | 2.2 | 2.1 | 2.6 | 2.4 | 2.0 |
| Karyopherin α 2 (RAG) | U28386 | −1.1 | −1.2 | 1.2 | 1.2 | −2.1 |
| Solute carrier family 25 | J03592 | −2.6 | −2.7 | −2.1 | −1.9 | −1.5 |
| Cytochrome c oxidase subunit VIa polypeptide 1 | AI540925 | −1.6 | −1.8 | −1.3 | −1.2 | −1.3 |
| Ubiquitin-conjugating enzyme E2G 2 | AF032456 | 1.7 | 1.7 | 1.1 | 1.8 | 1.1 |
| Ubiquitin-conjugating enzyme E2E 3 | AB017644 | −1.6 | −1.6 | −1.8 | −1.6 | −1.9 |
| Splicing factor, arginine/serine-rich 6 | AL031681 | 2.0 | 1.5 | −1.0 | −1.2 | −1.1 |
| Leukotriene A4 hydrolase | J03459 | −2.4 | −2.4 | −2.0 | −2.8 | −2.1 |
| Ubiquitin-activating enzyme E1 | M58028 | −2.2 | −2.8 | −2.2 | −2.6 | −2.0 |
| FK506 binding protein 1A (12 kDa) | V00599 | −2.1 | −1.9 | −1.5 | −2.9 | −4.5 |
| RNA binding motif protein 3 | U28686 | 1.8 | 2.1 | 1.6 | 1.5 | 1.4 |
Finally, oligonucleotide microarray analysis also demonstrates 2ME2-induced alterations in genes encoding for proteins mediating unfolded protein response (UPR) pathway (X-Box binding-1, tapasin, Grp78/Bip, Grp94, MHC-I, peptidylpropyl isomerase [PPIases] [cyclophilin]-like 2), FK506 binding proteins) (Figures 1-2). Taken together, the 2ME2-triggered gene profiles may be useful in providing a framework to predict 2ME2 efficacy in MM patient cells by examining the expression levels of particular genes. We next focused our study on validating and deriving functional significance of the oligonucleotide microarray data for selected genes known to contribute in MM pathogenesis.
Effects of 2ME2 on the UPR pathway
In eukaryotic cells, secreted and membrane-bound proteins undergo folding, posttranslational modification, and oligomerization in the ER before transport to the plasma membrane. Cells cope with misfolded ER proteins in part through a mechanism called the unfolded protein response (UPR).30 The current microarray analysis reveals that 2ME2 modulates various genes involved in the UPR pathway by regulating ER function. Specifically, 2ME2 inhibits expression of X-box binding protein-1 (XBP-1), MHC-I, tapasin (transporter associated with antigen processing [TAP]), beta-2-microglobulin, and gp96 (grp94), indicating a global attenuating effect of 2ME2 on ER-related signaling components in MM cells (Figure 2).
Regulation of XBP-1 expression by 2ME2 in MM.1S and MM patient cells
A recent study has demonstrated that XBP-1, a basic-region leucine zipper protein in the CREB/ATF family of transcription factors, mediates B-cell differentiation into plasma cells via UPR pathway.31 Functional knockout of XBP-1 in mice leads to a number of defects associated with a deficiency in the UPR pathway. Moreover, it has been shown that XBP-1 mutant B cells in immune-deficient (RAG-2−/−) chimeric mice cannot differentiate into antibody-secreting plasma cells.31 A functional UPR system is therefore necessary for both differentiation of B cells into plasma cells and to ensure that only properly folded antibodies are secreted.
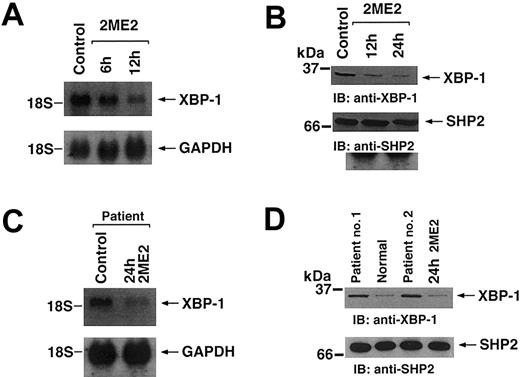
In the current study, oligonucleotide microarray analysis showed that 2ME2 represses the expression of XBP-1 gene (Figure 2). In order to validate oligonucleotide microarray results, we performed Northern blot analysis. MM.1S MM cells were treated with 2ME2, and total RNA was extracted for Northern blot hybridization using an XBP-1 cDNA probe. As shown in Figure 3A, treatment of MM.1S MM cells with 2ME2 significantly decreases (by 3- to 4-fold, as determined by densitometric analysis) XBP-1 mRNA levels.
Effect of 2ME2 on XBP-1 expression.
MM.1S cells were treated with 3 μM 2ME2 for the indicated times. (A) Total cellular RNA was subjected to Northern blot analysis. Data shown is representative of 3 independent experiments with similar results. (B) Total cell lysates were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti–XBP-1 (upper panel) or anti-SHP2 (lower panel) Abs. (C) Purified patient MM cells (CD138+) were treated with 2ME2 for the indicated times and analyzed for mRNA levels by Northern blotting. The blots were reprobed with GAPDH to confirm equal loading. (D) Total cell lysates from purified MM cells (CD138+) (patient 1 and patient 2), normal BM cells, and 2ME2-treated patient cells were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti–XBP-1 (upper panel) or anti-SHP2 (lower panel) Abs.
Effect of 2ME2 on XBP-1 expression.
MM.1S cells were treated with 3 μM 2ME2 for the indicated times. (A) Total cellular RNA was subjected to Northern blot analysis. Data shown is representative of 3 independent experiments with similar results. (B) Total cell lysates were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti–XBP-1 (upper panel) or anti-SHP2 (lower panel) Abs. (C) Purified patient MM cells (CD138+) were treated with 2ME2 for the indicated times and analyzed for mRNA levels by Northern blotting. The blots were reprobed with GAPDH to confirm equal loading. (D) Total cell lysates from purified MM cells (CD138+) (patient 1 and patient 2), normal BM cells, and 2ME2-treated patient cells were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti–XBP-1 (upper panel) or anti-SHP2 (lower panel) Abs.
To determine whether changes in the mRNA levels of XBP-1 correlate with alterations in protein levels, total protein lysates from 2ME2-treated MM.1S cells were subjected to SDS-PAGE analysis, transferred onto a filter, and immunoblotted with anti–XBP-1 Abs. 2ME2 also down-regulates XBP-1 protein expression in MM.1S MM cells (Figure 3B) without changes in the SHP2 protein levels (Figure 3B, lower panels).
We next examined whether XBP-1 expression is similarly modulated in response to 2ME2 in patient MM cells. CD138+ plasma cells from patient BM were obtained and treated with 2ME2 (3 μM), and total RNA was subjected to Northern blot analysis using an XBP-1 cDNA probe. As shown in Figure 3C, 2ME2 decreases (by 5-fold, as determined by densitometric analysis using GAPDH as control for normalization) XBP-1 transcripts in patient MM cells. Examination of XBP-1 protein expression also showed a similar decrease after exposure to 2ME2 in CD138+ plasma cells from patient BM (Figure 3D). Importantly, a comparative analysis of XBP-1 protein expression in CD138+ plasma cells from patient BM and normal BM-derived cells also demonstrate significantly (2- to 3-fold,P < .05) higher XBP-1 transcripts in MM cells than in normal cells (Figure 3D). A recent study using microarrays analysis on MM patient cells also showed higher XBP-1 transcripts in these cells.26 Our findings provide further evidence that novel anti-MM agent 2ME2 down-regulates XBP-1 expression in MM cells; however, whether XBP-1 mediates 2ME2 apoptotic effects remains to be examined.
Effect of IL-6 and Dex on XBP-1 expression
Interleukin-6 (IL-6) is a major growth and survival factor in MM cells, and we therefore next examined whether XBP-1 levels are affected during IL-6–induced proliferation in MM.1S MM cells. Treatment of MM.1S MM cells with rhIL-6 (10 ng/mL) induces significant (2.1 ± 0.2-fold; P < .05, n = 3) proliferation. As seen in Figure 4A-B (upper panels), both XBP-1 mRNA and protein levels were significantly induced (3.8 ± 0.4-fold for mRNA; 4.2 ± 0.3-fold for protein, as determined by densitometry) after exposure to IL-6, without any alterations in GAPDH and SHP2 levels, respectively (Figure 4A-B, lower panels). Our results are consistent with a prior study demonstrating XBP-1 as an IL-6 target gene.32 The clinical observation of elevated serum levels of IL-6 in MM patients with progressive disease,33 coupled with our present results, suggests that IL-6 may regulate XBP-1 expression in a paracrine manner and thereby contribute to growth/survival of MM cells.
Importantly, we and others have shown that IL-6 is not only a growth factor in MM cells but also an antiapoptotic factor protecting against Dex-induced apoptosis.15,34-37 We therefore next examined whether the protective effect of IL-6 against Dex involves XBP-1. As shown in Figure 4C (upper panel), treatment of MM.1S MM cells with Dex (10 μM) does not alter XBP-1 protein levels. This is in concert with our previous oligonucleotide microarray results showing no changes in XBP-1 transcripts in response to Dex treatment in MM.1S MM cells.16 IL-6–mediated protection against Dex may therefore be associated with an unopposed induction of XBP-1 by IL-6. This observation is further supported by our findings with 2ME2: unlike Dex-triggered killing, 2ME2-induced apoptosis is not blocked by IL-64 since 2ME2 down-regulates XBP-1 transcript and blocks the IL-6–induced antiapoptotic signal.
Effect of IL-6 on XBP-1 expression and its functional significance.
MM.1S cells were treated with IL-6 (10 ng) for the indicated times. (A) Total cellular RNA was subjected to Northern blot analysis. Data shown is representative of 3 independent experiments with similar results. (B) Total cell lysates were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti–XBP-1 (upper panel) or anti-SHP2 (lower panel) Abs. (C) Total cell lysates from Dex-, 2ME2-, Dex + IL-6, or 2ME2 + IL-6–treated MM.1S cells were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti–XBP-1 (upper panel) or anti-SHP2 (lower panel) Abs. (D) Effect of overexpression of dominant-negative XBP-1 (DN-XBP-1) in MM.1S cells. Cells were transiently transfected with cDNA expression construct containing green fluorescence protein (GFP) alone or with dominant-negative XBP-1 (DN-XBP-1). Following transfections, GFP-positive cells were selected by flow cytometry, treated with Dex (5 μM) or Dex + IL-6 (10 ng) for 48 hours, and analyzed for cell viability by MTT assay. Median viability was 53% for Dex in GFP-transfected cells and 32% for GFP + DN-XBP-1–transfected cells (P < .005, n = 3; mean ± SD of 3 independent experiments. Error bars indicate SD.) (E) Schematic representation of the role of XBP-1 in 2ME2-induced apoptosis and overcoming Dex resistance, as well as in IL-6–mediated protective effect against Dex. Arrows indicate the flow of signal; ×, no effects.
Effect of IL-6 on XBP-1 expression and its functional significance.
MM.1S cells were treated with IL-6 (10 ng) for the indicated times. (A) Total cellular RNA was subjected to Northern blot analysis. Data shown is representative of 3 independent experiments with similar results. (B) Total cell lysates were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti–XBP-1 (upper panel) or anti-SHP2 (lower panel) Abs. (C) Total cell lysates from Dex-, 2ME2-, Dex + IL-6, or 2ME2 + IL-6–treated MM.1S cells were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti–XBP-1 (upper panel) or anti-SHP2 (lower panel) Abs. (D) Effect of overexpression of dominant-negative XBP-1 (DN-XBP-1) in MM.1S cells. Cells were transiently transfected with cDNA expression construct containing green fluorescence protein (GFP) alone or with dominant-negative XBP-1 (DN-XBP-1). Following transfections, GFP-positive cells were selected by flow cytometry, treated with Dex (5 μM) or Dex + IL-6 (10 ng) for 48 hours, and analyzed for cell viability by MTT assay. Median viability was 53% for Dex in GFP-transfected cells and 32% for GFP + DN-XBP-1–transfected cells (P < .005, n = 3; mean ± SD of 3 independent experiments. Error bars indicate SD.) (E) Schematic representation of the role of XBP-1 in 2ME2-induced apoptosis and overcoming Dex resistance, as well as in IL-6–mediated protective effect against Dex. Arrows indicate the flow of signal; ×, no effects.
We next directly assayed for the functional significance of XBP-1 in MM cells. MM.1S cells were transiently transfected with cDNA expression construct containing GFP alone or cotransfected with dominant negative XBP-1 (DN-XBP-1). GFP-positive cells were selected and treated with Dex in the presence or absence of IL-6 and assayed for cell viability.
Dex treatment of cells overexpressing DN-XBP-1 had significantly decreased viability (32.5% ± 2.3%, P = .05, as determined by one-sided Wilcoxon rank-sum test) compared to GFP alone (53.3% ± 4.6%; P = .05, as determined by one-sided Wilcoxon rank-sum test) (Figure 4D). Moreover, a marked decrease in the ability of IL-6 to block Dex-induced cytotoxic effects was seen in cells overexpressing DN-XBP-1 versus cells with GFP alone (median viability for Dex + IL-6 was 79% in GFP-transfected cells compared to 43% viability in GFP + DN-XBP-1–transfected cells;P = .05, as determined by one-sided Wilcoxon rank-sum test) (Figure 4D). In contrast, overexpression of DN-XBP-1 did not increase sensitivity to 2ME2 treatment of MM cells (data not shown). Taken together, our data suggest the following: (1) XBP-1 is highly expressed in MM cells as compared to normal BM cells, (2) XBP-1 is not a common target of anti-MM agents, since 2ME2 (but not Dex) attenuates XBP-1 expression, (3) XBP-1 mediates the protective effect of IL-6 against Dex, and (4) blocking the function of XBP-1 sensitizes cells to Dex-, but not 2ME2-, induced cytotoxicity.
Effects of 2ME2 on structural/cytoskeleton-related genes
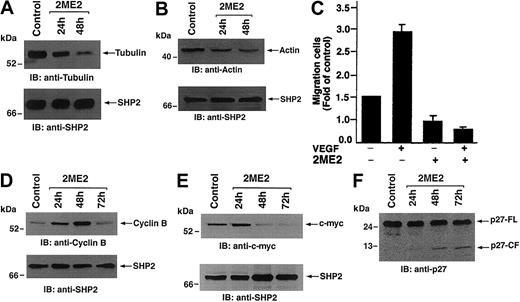
2ME2 is a known microtubule-interfering agent,38 and our oligonucleotide microarray analysis shows that 2ME2 triggers a significant decrease (6.3 ± 0.4-fold decrease,P < .05) in cytoskeletal/structural genes, including tubulin and actin (Figure 2; Table 1). To determine whether changes in the mRNA levels of tubulin and actin correlate with protein levels, total protein lysates from 2ME2-treated MM.1S cells were subjected to SDS-PAGE analysis, transferred onto a filter, and immunoblotted with antitubulin and antiactin Abs. As shown in Figure 5A-B (upper panels), 2ME2 decreases protein levels of both tubulin and actin in MM.1S MM cells, without any alteration in SHP2 levels (Figure 5A-B, lower panels).
To assess the functional significance of these changes, we next determined whether 2ME2-induced decreases in cytoskeleton proteins alter the migration of these cells. MM.1S MM cells were treated with rhVEGF (10 ng) for 24 hours and migration in transwell inserts examined, as in prior studies,39 in the presence or absence of 2ME2. As shown in Figure 5C, 2ME2 significantly (P < .05) decreases VEGF-induced migration of MM.1S MM cells. These findings confirm the biologic significance of our oligonucleotide microarray data.
Effect of 2ME2 on cytoskeleton and cell cycle regulatory proteins.
MM.1S cells were treated with 3 μM 2ME2 for the indicated times. (A-B) Total cell lysates were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with antitubulin or antiactin Abs (upper panels) or anti-SHP2 (lower panels) Abs. (C) 2ME2 affects VEGF-induced migration of MM.1S cells. MM.1S MM cells were treated with rVEGF (10 ng) for 24 hours, followed by exposure to 3 μM 2ME2 and analysis in a transwell migration assay. (D-F) Total cell lysates were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti–cyclin B or anti–c-myc, or anti-p27 Abs (upper panels) or anti-SHP2 (lower panels) Abs. FL indicates full length; CF, cleaved fragment.
Effect of 2ME2 on cytoskeleton and cell cycle regulatory proteins.
MM.1S cells were treated with 3 μM 2ME2 for the indicated times. (A-B) Total cell lysates were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with antitubulin or antiactin Abs (upper panels) or anti-SHP2 (lower panels) Abs. (C) 2ME2 affects VEGF-induced migration of MM.1S cells. MM.1S MM cells were treated with rVEGF (10 ng) for 24 hours, followed by exposure to 3 μM 2ME2 and analysis in a transwell migration assay. (D-F) Total cell lysates were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti–cyclin B or anti–c-myc, or anti-p27 Abs (upper panels) or anti-SHP2 (lower panels) Abs. FL indicates full length; CF, cleaved fragment.
Effect of 2ME2 on cell cycle regulatory proteins
Previous studies have reported that 2ME2 induces changes the expression of cell cycle regulatory genes.40 In the current study, cyclin B1, cdc28 protein kinase-2, and c-myc were transiently induced (Figure 1). Immunoblot analysis of 2ME2-treated MM.1S MM cell lysates with anti–cyclin B1 Abs demonstrated similar changes in cyclin B1 protein expression (Figure 5). These results suggest a role of cyclin B1 during 2ME2-induced growth arrest and apoptosis in these cells and are consistent with its function in other cell systems.41
Modulation of c-myc expression is a key event during plasma cell differentiation, and a recent study using cDNA microarray analysis showed that c-myc is highly expressed in MM cells.26 In the current study, 2ME2 induces a modest transient induction of c-myc early (6-12 hours) after 2ME2 exposure, followed by a decrease in c-myc transcript levels at later periods (Figure 1; Table 1). Examination of c-myc protein expression in 2ME2-treated MM.1S MM cell lysates demonstrated similar changes in c-myc protein expression in response to 2ME2 (Figure 5E). In order to further confirm whether 2ME2-induced alteration in c-myc affect the downstream target genes such as p27Kip, we analyzed the changes in p27Kip in response to 2ME2. 2ME2 does not alter the p27Kip mRNA levels but triggers degradation and proteolytic cleavage of p27Kip, suggesting an activation of c-myc–related signaling.
2ME2 overcomes Dex resistance by altering genes conferring Dex resistance in MM
One of our major current efforts is delineation of mechanisms mediating chemoresistance in MM cells. Initial treatment of MM patients with Dex achieves responses; however, relapse of MM ultimately occurs due to development of Dex resistance.42 To date, the molecular and genetic alterations conferring Dex resistance in MM cells are not well defined. Our recent study showed that 2ME2 overcomes Dex resistance, evidenced by triggering apoptosis in Dex-resistant (MM.1R) MM cells. We therefore next examined whether 2ME2 alters the expression of genes that are overexpressed in Dex-resistant cells. For these studies, we compared the gene expression profiles of Dex-resistant (MM.1R) cells and 2ME2-treated cells. These comparative analyses demonstrate that 2ME2 down-regulates various genes, which are overexpressed in MM.1R cells, including genes encoding heat shock family and ubiquitin-related proteins, suggesting mechanisms whereby 2ME2 overcomes Dex resistance.
Heat shock proteins
Hsps are a set of highly conserved proteins with various subfamilies, based on their molecular weight, including Hsp90, Hsp70, Hsp40, or Hsp27.43 Overexpression of Hsp70 protects cells from apoptosis, both upstream and downstream of effector caspase activation.43 Hsp27 is constitutively expressed in tumor cells at specific stages of development and differentiation.43 In the present study, treatment of MM.1S MM cells with 2ME2 (3 μM) for 72 hours decreased mRNA levels of Hsp70 and Hsp90 (Figure 1). A transient increase (24-48 hours) in Hsp70 early after 2ME2 treatment is suggestive of a stress response to 2ME2.
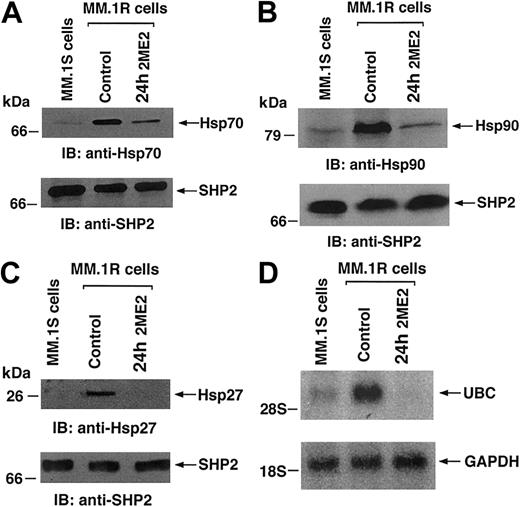
Our previous study showed that Dex-resistant (MM.1R) MM cells have significantly higher levels of Hsps as compared to Dex-sensitive (MM.1S) MM cells and may therefore confer Dex-resistance.16 Importantly, 2ME2 induces apoptosis in MM.1R cells, and we next examined the effect of 2ME2 on Hsp protein levels in MM.1R MM cells. Total protein lysates from MM.1S, MM.1R, and 2ME2-treated MM.1R cells were subjected to SDS-PAGE analysis, transferred onto a filter, and immunoblotted with anti-Hsp27, anti-Hsp70, and anti-Hsp90 Abs. As shown in Figure 6A-C (upper panels), MM.1R cells show significantly (3- to 4-fold, as determined by densitometric analysis) higher expression of all 3 Hsps as compared to MM.1S cells; moreover, treatment of MM.1R cells with 2ME2 decreases the levels of all 3 Hsps. Reprobing the immunoblot with anti–SHP-2 Ab confirms equal protein loading, indicating that the differences in the protein levels of hsp27 and hsp72 are specific (Figure 6A-C, lower panels). These findings suggest that neutralizing the antiapoptotic effects of Hsps, for example, by inhibiting their expression using antisense oligonucleotides, may sensitize MM cells to 2ME2 or overcome drug resistance. Conversely, other pharmacological stimuli or cytokines that trigger Hsp expression may protect normal cells from stress agents.
Ubiquitin-related genes
Our oligonucleotide microarray data demonstrate that 2ME2 alters genes encoding proteins involved in ubiquitin (ub)–proteasome pathway mediating cell growth and apoptosis. Increased expression of ubiquitin-related genes has been shown to be an early event in tumorigenesis and is associated with the drug-resistant phenotype.44,45 In this context, our prior gene profiling analysis demonstrated significantly higher ubiquitin and ubiquitin conjugating enzyme mRNA levels in Dex-resistant (MM.1R) cells than in Dex-sensitive (MM.1S) MM cells.16 In the present study, 2ME2 down-regulates ubiquitin mRNA levels, with an associated increase in proteasome 26S transcripts (Figure 1-2; Table 1). These oligonucleotide microarray results were validated by Northern blot analysis of MM.1S, MM.1R, and 2ME2-treated MM.1R cells. As shown in Figure 6D (upper panel), MM.1R cells demonstrated significantly higher ubiquitin mRNA levels than MM.1S cells, and treatment with 2ME2 reduced ubiquitin mRNA without any changes in GAPDH levels (Figure 6D, lower panels). Our prior demonstration that 2ME2 induces apoptosis in Dex-resistant MM cells,4 coupled with the present finding that 2ME2 decreases ubiquitin, suggests that 2ME2 may overcome Dex resistance in MM cells via modulation of ubiquitins.
Effect of 2ME2 on genes encoding Hsps and ubiquitin in Dex-sensitive (MM.1S) and Dex-resistant (MM.1R) MM cells.
MM.1R cells were treated with 3 μM 2ME2 for the indicated times. (A-C) Total cell lysates were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti-Hsp70, anti-Hsp90, or anti-Hsp27 Abs (upper panels) or anti-SHP2 (lower panels) Abs. (D) Total cellular RNA from MM.1S, MM.1R, and 2ME2-treated MM.1R cells was hybridized to 32P-labeled ubiquitin (UBC) and GAPDH cDNA probes.
Effect of 2ME2 on genes encoding Hsps and ubiquitin in Dex-sensitive (MM.1S) and Dex-resistant (MM.1R) MM cells.
MM.1R cells were treated with 3 μM 2ME2 for the indicated times. (A-C) Total cell lysates were separated by 12.5% SDS-PAGE and analyzed by immunoblotting (IB) with anti-Hsp70, anti-Hsp90, or anti-Hsp27 Abs (upper panels) or anti-SHP2 (lower panels) Abs. (D) Total cellular RNA from MM.1S, MM.1R, and 2ME2-treated MM.1R cells was hybridized to 32P-labeled ubiquitin (UBC) and GAPDH cDNA probes.
Collectively, we have in this study identified 2ME2-responsive genes in MM cells using oligonucleotide microarray analysis. Our results demonstrate a differential expression pattern of genes in 2ME2-treated cells, which may both identify molecular targets of sensitivity and resistance and allow for improved therapeutic use of 2ME2. Targeting molecules regulating the UPR pathway, for example, using antisense oligonucleotide to XBP-1 may block the survival signaling and thereby sensitize cells to drug-induced cytotoxicity. Moreover, drugs that target Hsps or genes involved in ubiquitin-proteasome pathway may be combined with 2ME2 to treat Dex-resistant MM cells. A comparative analysis of drug-specific gene profile with patient-derived gene profiles may allow for more effective and individualized design of treatment regimens. Ongoing comparison of the signature gene profiles in MM cell lines and MM patient cells will predict patient responses to 2ME2 therapy and provide a rationale for targeted 2ME2 combination treatment approaches in MM.
Prepublished online as Blood First Edition Paper, December 12, 2002; DOI 10.1182/ blood-2002-10-3146.
Supported by the National Institutes of Health grants 50947 and CA 78373, a Multiple Myeloma Research Foundation grant (D.C., T.H., N.M., C.M.), VA Merit review and Leukemia and Lymphoma Society Scholar in translational research award, a Doris Duke Distinguished Clinical Research Scientist Award (K.C.A.), The Myeloma Research Fund, and The Cure Myeloma Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kenneth C. Anderson, Kraft Family Professor of Medicine, Dana Farber Cancer Institute, 44 Binney St, Boston, MA 02215; e-mail: kenneth_anderson@dfci.harvard.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal