Circulating CD34+ cells are used in reparative medicine as a stem cell source, but they contain cells already committed to different lineages. Many think that B-cell progenitors (BCPs) are confined to bone marrow (BM) niches until they differentiate into B cells and that they do not circulate in blood. The prevailing convention is that BCP transit a CD34+CD19−10+early-B→CD34+CD19+CD10+B-cell progenitor (pro-B)→CD34−CD19+CD10+ B-cell precursor (pre-B) differentiation pathway within BM. However, populations of CD34+CD10+ and CD34+CD19+ cells circulate in adult peripheral blood and neonatal umbilical cord blood (CB) that are operationally taken as BCPs on the basis of their phenotypes, although they have not been submitted to a systematic characterization of their gene expression profiles. Here, conventional CD34+CD19+CD10+ and novel CD34+CD19+CD10− BCP populations are characterized in CB by single-cell sorting and multiplex analyses of gene expression patterns. Circulating BCP are Pax-5+cells that span the early-B, pro-B, and pre-B developmental stages, defined by the profiles of rearranged V-D-JH, CD79, VpreB, recombination activating gene (RAG), and terminal deoxynucleotidyl transferase (TdT) expression. Contrary to the expectation, circulating CD34+CD19−CD10+ cells are essentially devoid of Pax-5+ BCP. Interestingly, the novel CD34+CD19+CD10− BCP appears to be the normal counterpart of circulating preleukemic BCPs that undergo chromosomal translocations in utero months or years before their promotion into infant acute lymphoblastic B-cell leukemia after secondary postnatal mutations. The results underscore the power of single-cell analyses to characterize the gene expression profiles in a minor population of rare cells, which has broad implications in biomedicine.
Introduction
Circulating blood CD34+ cells are proposed as a totipotent stem cell source in organ and gene regenerative medicine.1-5 CD34+ cells are a heterogeneous population, however, because they also contain early progenitors already committed to distinct lineages.5-9Early in life, a major subset of bone marrow (BM) CD34+cells are B-cell progenitors (BCPs).8,10 In contrast, CD34+ BCPs were undetectable in fetal and term umbilical cord blood (CB) in many independent phenotypic studies.6-8The latter reports support the accepted idea that BCPs are retained in the BM and fetal liver until they acquire the surface immunoglobulin (sIg), IgM/CD79 antigen receptor complex, and that B-lineage emigrants to peripheral blood are solely immature sIgM+ B cells selected for tolerance to self-antigens.11-13 The paradigm that BCPs do not emigrate to the periphery has been challenged by authors who report that rare populations of CD34+ cells that express either CD10 or CD19 develop in blood.7,14Although gene expression profiles of the CD34+CD10+ and CD34+CD19+ blood cells have not been characterized, as was done for BM and in vitro–differentiated BCPs,8-10,15,16 they are often operationally defined as circulating BCPs on the basis of their surface phenotypes.2,17 (The prevailing convention is that B-lineage–committed cells pass through a CD34+CD19−CD10+early-B→CD34+CD19+CD10+pro-B→CD34−CD19+CD10+pre–B-cell development pathway; reviewed in LeBien9.) The use of surface phenotypes to unambiguously define the lineage affiliation and differentiation stage of early CD34+progenitors may, however, be an oversimplification.9 The CD34+CD19−CD10+ phenotype is not B-lineage specific,9 and blood CD34+CD19+ cells markedly outnumber the CD34+CD10+ subset,14 suggesting that yet uncharacterized CD34+CD19+CD10− BCP occurs. Single-cell analyses of the expression pattern of genes and proteins implicated in the developmental specification of the B-lineage,8-10,15-17 conducted by fluorescence activated cell sorting (FACS) and multiplex reverse transcription–polymerase chain reaction (RT-PCR) study of the gene expression profiles in the individually separated cells, has allowed us to unambiguously characterize BM CD34+CD19+CD10+ cells as pro-B and pre-BI cells.10 Here, we address whether CB CD34+ cells contain BCP and define their phenotype and differentiation stage by single-cell molecular analyses.
B-cell differentiation is characterized by a commitment event and a series of specification steps.18-20 The commitment event requires expression of the Pax5 gene. Experiments usingPax5−/− mouse cells have shown that considerable progress down the B-cell specification pathway is possible in the absence of commitment.18 CD19 expression depends strictly on Pax-5,20 making its surface expression a natural reporter for the transcription of a gene essential in B-lineage commitment.19 We are not aware of single-cell studies on CD34+CD19− human early–B-cell candidates. The expression of surface CD10 and sterile transcripts of the IgH locus or CD79, terminal deoxynucleotidyl transferase (TdT), or VpreB mRNA have been used to define early-B cells in bulk CD34+CD19− BM populations.9,21-23In Pax-5− cells, however, these are considered priming events in the specification of hemopoietic progenitors because they do not impose a B-cell fate, a role reserved for the Pax-5 gene product.18,19 The BM pro-B cell is the earliest BCP stage defined by RT-PCR in single cells.10CD34+CD10+CD19+ pro-B cells do not bear rearranged IgH or L gene products, but they express enzymes belonging to the immunoglobulin gene rearrangement and diversification machinery (RAG-1, RAG-2, TdT) and the surrogate light chain subunits (VpreB and λ5, ψL).8-10,15,16 Progression to the pre–B-cell stage is marked by the synthesis of a rearranged IgH product. V-D-JH rearrangement is an imprecise process, and the IgH products are nonfunctional in most pre–B-cell clones and are retained within the cytoplasm (cμH+). However, pre-BI cells that bear in-frame productive V-D-JH transport the μH chains to the membrane associated with ψL and CD79, which signals several rounds of division.15,16 The large cycling pre-BI cells that bear functional μH down-regulate CD34, TdT, RAG-1, and RAG-2, and maintain expression of ψL, CD10, and CD19 genes.10,15,16 To allow for Ig κ or λ L gene recombination, RAG-1 and RAG-2 expression are reinduced in small resting CD34−CD19+CD10+ pre-BII cells.10 Functional IgL recombination leads to the surface expression of Ig(H+L)2/CD79 complexes that mark the pre-BII to newly formed B-cell transition.8-10
Here, single-cell multiplex RT-PCR analyses show that the CD34+ population circulating in CB includes sizable Pax-5+ subsets with early-B, pro-B, and pre-B cell genotypes. Surface expression of CD19 and CD10 is markedly asynchronous in CB CD34+ cells. Notably, the circulating CD34+CD19+sIgH− cell subset, independently of its CD10 expression status (CD34+CD19+CD10+sIgH−or CD34+CD19+CD10−sIgH−), includes most circulating CD34+ BCPs, whereas the CD34+CD10+CD19−sIgH−cells are essentially devoid of Pax-5+ BCP.
Materials and methods
Mononuclear cell purification and CD34+ progenitor cell enrichment
Neonatal umbilical CB, obtained after informed consent from 49 donors at 38- to 41-weeks' gestation, was placed in sterile heparin-containing tubes (10 U/mL) and processed within 24 hours. Mononuclear cells were isolated by Ficoll-Hypaque gradient (density, 1.077 g/mL) (Pharmacia, Uppsala, Sweden). CD34+ progenitors from mononuclear cells were enriched using the immunomagnetic CD34 Progenitor Cell Selection System (Dynal, Oslo, Norway), according to the manufacturer's instructions. Cells detached from the paramagnetic beads were typically 85% to 95% CD34+ by flow cytometry. Circulating CD34− T-cell precursors were purified as reported.24
Flow cytometry and single-cell sorting
Surface staining10,25 of CB mononuclear cells and CD34+-enriched cells was performed using a panel of monoclonal antibodies including fluorescein isothiocyanate (FITC)–labeled anti-CD19, cychrome–anti-CD34 (both from Pharmingen, San Diego, CA), phycoerythrin (PE)–anti-CD10 (Caltag, San Francisco, CA), and biotin–anti-human μH (SA-DA4; Southern Biotech, Birmingham, AL) combined with streptavidin–allophycocyanin (APC) (Becton Dickinson, San Jose, CA). FITC-labeled anti-CD3, -CD14, -CD16, and -CD56 and PE–anti-CD4 antibodies were all from Becton Dickinson, and CD79a was from DAKO (Glostrup, Denmark). Three- and 4-color immunofluorescence and flow cytometry analyses were performed on an EPICS-XL (Coulter-Beckman, Hialeah, FL) and a FACSCalibur flow cytometer (Becton Dickinson), respectively. Four-color immunofluorescence and single-cell sorting was performed on a dual-laser FACStar Plus cell sorter (Becton Dickinson), using its automated cell deposition unit (ACDU). Only cells exhibiting low forward-angle and low right-angle scatter properties (lymphoid gate) were analyzed and sorted when needed. Routinely, 2 × 105events within the lymphoid gate were acquired for flow cytometry analyses. Isotype-matched antibody controls (Pharmingen) were used to set backgrounds. Data from all flow cytometers were displayed and analyzed using FlowJo-3 software (San Carlos, CA). There is a major difference between the single-cell analyses performed here in CB and in our previous study in BM.10 Here the expression profiles of 4 CD34+sIg− CB subsets were studied (ie, CD19+CD10+, CD19+CD10−, CD19−CD10+, or CD19−CD10−), whereas only the CD10+ subpopulation was sorted in the BM.10That experimental design difference reveals the novel CD34+CD19+CD10− BCP population and the distinct gene expression profile of CD34+CD19+CD10+ and CD34+CD19−CD10+ cells.
Multiplex RT-PCR on single sorted cells
A total of 216 individual cells were analyzed in each population, obtained from 8 different blood samples (range, 18-36 cells per donor and population). Single cells from the selected populations were placed by the ACDU into 0.2 mL PCR tubes containing 2.5 μL phosphate-buffered saline solution (PBS) and immediately frozen on dry ice. All reactions were performed on a GeneAmp PCR System 9700 (PE Applied Biosystems, Branchburg, NJ). A 2-step strategy was used to detect mRNA from a single cell.10 This involves an initial one-step multiplex RT-PCR, followed by a second PCR in individual aliquots of the initial PCR reaction that use the panel of primers specific for each gene separately. Samples were heated (2 minutes, 65°C), then chilled on ice before the addition of the multiplex RT-PCR reaction mix. RT-PCR reactions were performed using SuperScript One-Step RT-PCR System (Life Technologies, Paisley, United Kingdom). Each RT-PCR reaction consisted of one cycle of reverse transcription (50°C, 30 minutes) and a denaturation step (94°C, 2 minutes) linked to 30 cycles of PCR amplification, each at 94°C for 20 seconds, 60°C for 30 seconds, 72°C for 30 seconds, and a final extension cycle at 72°C for 7 minutes. The multiplex RT-PCR reaction mix included up to 8 primer pairs, specific for each gene amplified: GAPDH, Pax-5, VpreB, TdT, RAG-1, mb-1, VH, and preTα. Oligonucleotides used for the multiplex RT-PCR were GAPDH sense, 5′-GAAGGTGAAGGTCGGAGTC-3′, GAPDH antisense, 5′-GAAGATGGTGATGGGATTTC-3′; Pax5 primers, designed by Ryan et al,22 VpreB sense, 5′-TTTGTCTACTGCACAGGTTGTGG-3′, VpreB antisense, 5′-TGCAGTGGGTTCCATTTCTTCC-3′; RAG-1 sense, 5′-CCAAATTGCAGACATCTCAAC-3′, RAG-1 antisense, 5′-CAACATCTGCCTTCACATCGATCC-3′; TdT sense, 5′-GCCGTCAGTGTGCTGGTTAAAGAGG-3′, TdT antisense, 5′-TCTGCTTTGAGGAATATCCTCTTGG-3′; mb-1 sense, 5′-TCCAAGCTCTGCCTGCCACCAT-3′, mb-1 antisense, 5′-GACTGCTGGTATGACTCGTTGC-3′; VH sense (VHframework III consensus), 5′-GACACGGCCGTGTATTACTG-3′, VHantisense (CHμ), 5′-GGAATTCTCACAGGAGACGAG-3′; preTα sense, 5′-GGCACACCCTTTCCTTCTCTG-3′, preTα antisense, 5′-GCAGGTCCTGGCTGTAGAAGC-3′. A second PCR amplification was performed with 1 μL first RT-PCR reaction, using the primer pair specific for each gene in individual reactions. The second PCR consisted of 30 cycles at 94°C for 20 seconds, 60°C for 30 seconds, 72°C for 30 seconds, and a final extension cycle at 72°C for 7 minutes. Nested amplifications were used for VpreB, RAG-1, TdT and mb-1, using the following internal oligonucleotides: VpreB antisense, 5′-GTAATACATAGCCTCGTCCTCAGG-3′; RAG-1 antisense, 5′-ACCATCCACAGGACCATGGACTGG-3′; TdT antisense, 5′-AGAATCATCTTCCGCTCATGTGTGG-3′; mb-1 antisense, 5′-AGAACTCAGGGGGCCACGTGTA-3′. PCR primers were designed to allow for discrimination between cDNA and contaminating DNA amplification. The method for the detection of specific DNA or RNA in single cells is an established one.10 The RT-PCR end point of mRNA detection was checked by limiting-dilution analyses of riboprobes (cRNA) for 8 different genes, and we found that single RNA copies are indeed detected after optimization of the primers, RNA reverse transcription, and DNA polymerase amplification conditions in preliminary experiments. RNA from riboprobes and cell lines were next mixed to find that the sensitivity was maintained in the presence of whole-cell lysates. Finally, the primer pairs were incorporated one by one into the reaction, and we checked that sensitivity was preserved in the multiplex RT-PCR. Here, the GAPDH housekeeping gene was the control for cell-sorting yield rather than the B-cell–specific geneCD79b, used for that purpose in BM.10 When single cells were manually deposed into tubes, under visual control all tubes gave a positive GAPDH reaction. We programmed the FACS to depose the single droplets calculated to contain the desired cells into the reaction tubes under conditions optimized for purity and “sort-abort” when neighbor droplets contained cells. Rare, occasional tubes can, however, receive cell-free droplets, leading to a negative reaction for GAPDH and all other genes. All PCR products were sequenced to confirm the specificity of the gene amplifications using dye terminator technology and an automated DNA sequencer as indicated.10
Results
Cord blood CD34+ progenitor subsets express CD19, CD10, and VpreB markers
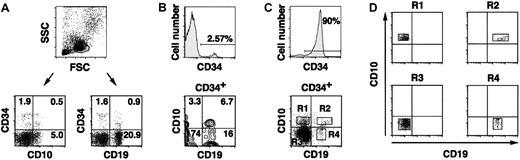
The CD34+ population of human CB cells was analyzed using antibodies against 2 surface antigens classically associated with the CD34+ B-lineage progenitors, CD19 and CD10. After isolation of CB mononuclear cells by Ficoll-Hypaque gradient, 3-color staining, and gating for lymphoid cells, analyses of the CD34+ cells (range, 0.7%-3.5%; median, 2.58%) show that minor subpopulations bear CD10 or CD19 molecules (Figure1A). We also examined VpreBgene transcription in CB cell subpopulations, purified by FACS using lineage-specific surface markers and submitted to a sensitive RT-PCR able to detect mRNA and single-copy cDNA from single cells.10 VpreB mRNA was readily detectable in CD34+ cells (approximately 2 × 10−2 cells) and was rare in the CD19+ B-cell lineage pool (5 × 10−3 to 2 × 10−4 cells). It was undetectable, however, in replicates of 104 T (CD3+) cells, natural killer (CD16+ or CD56+) cells, or monocytes (CD14+). VpreB was originally considered a BCP-specific gene, but a population of mature B cells (ie, sμH/κ or λ positive) was recently shown to coexpress VpreB and other genes previously used to characterize BCP.9,26 Therefore, 4-color staining with CD34, CD19, CD10, and IgM-specific antibodies was performed to determine the coexpression pattern of CD10 and CD19 molecules on the surfaces of CD34+ cells and to exclude the latter sIg+ B cells that are editing their immunoglobulin genes26 from our BCP analyses. The contour-plot analyses of CD19 versus CD10 expression (Figure 1B) were obtained after gating of the lymphoid cells, selection of the surface μH-negative cell region that excluded the sμHdull or bright populations present in CB, and selection of the CD34+ cells.
B-cell progenitors circulate in cord blood: flow cytometry identification and purification of candidate populations.
(A) Three-color surface immunofluorescence studies of lymphoid cells (forward scatter × side scatter gate) show minor CB subsets that coexpress CD34 and either CD10 (0.5%) or CD19 (0.9%), above the background indicated by the quadrant bars, set using isotype-matched antibodies (less than 0.01%; see “Materials and methods”). (B) After gating on the IgM− CD34+ cells (2.57%, top panel), the contour plot correlated distribution of CD19 versus CD10 is shown (bottom panel). Four hemopoietic progenitor populations (CD34+) are found in CB: CD34+CD19−CD10+ (3.3%), CD34+CD19+CD10+ (6.7%), CD34+CD19−CD10− (74%), and CD34+CD19+CD10− (16%). Please note that Coulter flow cytometers display contour plots in a different format than FACS analyzers. The data are not overcompensated. (C) Single cells in the 4 populations are purified using the R1-R4 sorting regions indicated in the bottom panel. CD34+ cells are enriched before the sorting step (90%; top panel). (D) On reanalyses to define the sort purity, most cells (more than 99.5%) in every population fall within the predefined sort regions.
B-cell progenitors circulate in cord blood: flow cytometry identification and purification of candidate populations.
(A) Three-color surface immunofluorescence studies of lymphoid cells (forward scatter × side scatter gate) show minor CB subsets that coexpress CD34 and either CD10 (0.5%) or CD19 (0.9%), above the background indicated by the quadrant bars, set using isotype-matched antibodies (less than 0.01%; see “Materials and methods”). (B) After gating on the IgM− CD34+ cells (2.57%, top panel), the contour plot correlated distribution of CD19 versus CD10 is shown (bottom panel). Four hemopoietic progenitor populations (CD34+) are found in CB: CD34+CD19−CD10+ (3.3%), CD34+CD19+CD10+ (6.7%), CD34+CD19−CD10− (74%), and CD34+CD19+CD10− (16%). Please note that Coulter flow cytometers display contour plots in a different format than FACS analyzers. The data are not overcompensated. (C) Single cells in the 4 populations are purified using the R1-R4 sorting regions indicated in the bottom panel. CD34+ cells are enriched before the sorting step (90%; top panel). (D) On reanalyses to define the sort purity, most cells (more than 99.5%) in every population fall within the predefined sort regions.
The results demonstrate that a CD34+CD19+CD10+“triple-positive” population circulates in CB (range, 0.3%-18% of the CD34+ cells; median, 12%). Two populations of cells that bear CD34 but express either CD10 or CD19 in a mutually exclusive manner (Figure 1B) are also evident in CB (CD34+CD19+CD10−; range, 7.7%-21%; median, 15% of CD34+ cells; and CD34+CD19−CD10+; range, 1.2%-16%; median, 3.4% of CD34+ cells).
CD79, VpreB, RAG-1, and TdT mRNA expression patterns in CD34+CD19+CD10+/−μH−cells allow for the characterization of circulating early-B– and pro-B–cell subsets
Here we addressed the correlated study of recombined V-D-JH, VpreB, TdT, RAG-1, CD79a, and preTα mRNA expression in individual cells belonging to 4 CD34+SμH− populations that were defined by their CD19 and CD10 expression patterns (Figure2; Table1). In these experiments, CB mononuclear cells were enriched in CD34+ cells before sorting (Figure 1C) with the use of magnetic beads coated with CD34 antibodies. The sort regions, boxes labeled R1 to R4 in the dot-plot graphs of CD19 versus CD10 expression (Figure 1C), were drawn after gating on the lymphoid cells, selection of the surface μH− cell region, and selection of the CD34+ cell region, similar to the analyses shown in Figure1B. The 4 sorted subsets, CD34+CD19−CD10+ (R1), CD34+CD19+CD10+ (R2), CD34+CD19−CD10−(R3), and CD34+CD19+CD10− (R4) cells, were 99.5% to 99.9% pure upon reanalysis (Figure 1D). No surface μH+, surface κL+, or surface λL+ cells were present in the 4 CD34+subpopulations thus analyzed (data not shown).
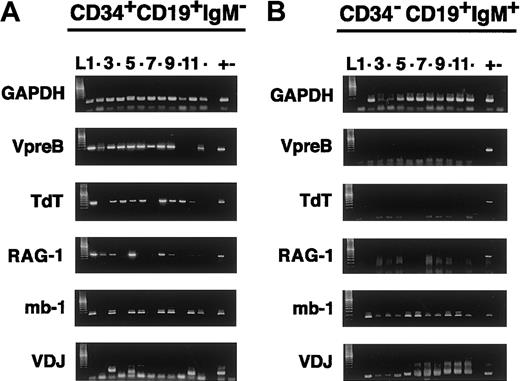
Gene expression profiles in single cells from distinct B-lineage stages.
Surface CD34+CD19+IgM− (A) and CD34−CD19+IgM+ (B) sorted cells are analyzed for mRNA expression of the indicated genes after multiplex RT-PCR. In the left panel, the bands in each of the twelve tracks (numbers 1-12), aligned in the 6 electrophoresis gels, show correlated amplification of gene products from individual CD34+CD19+IgM− cells (ie, cell 1 is VpreB+, TdT+, RAG-1+, mb-1/CD79+, recombined V-D-J). The right panel shows electrophoresis of the gene products amplified from tubes containing titrated numbers of CD34−CD19+IgM+ B cells: tracks 1 to 6, one cell; tracks 7 to 9, 10 cells; tracks 10 to 12, 100 cells. Similarly processed positive controls (+) correspond to individual Nalm-6 pre-B leukemia single cells, whereas no cells are added in the negative controls (−). The identity of the amplified gene products is ascertained by direct sequencing of the cDNA in the excised bands and by molecular weight estimation (ladders). Note that the lower bands in the V-D-J gel in panel A correspond to primer artifacts and do not contain V-D-JH products, as determined by sequencing. L indicates ladder.
Gene expression profiles in single cells from distinct B-lineage stages.
Surface CD34+CD19+IgM− (A) and CD34−CD19+IgM+ (B) sorted cells are analyzed for mRNA expression of the indicated genes after multiplex RT-PCR. In the left panel, the bands in each of the twelve tracks (numbers 1-12), aligned in the 6 electrophoresis gels, show correlated amplification of gene products from individual CD34+CD19+IgM− cells (ie, cell 1 is VpreB+, TdT+, RAG-1+, mb-1/CD79+, recombined V-D-J). The right panel shows electrophoresis of the gene products amplified from tubes containing titrated numbers of CD34−CD19+IgM+ B cells: tracks 1 to 6, one cell; tracks 7 to 9, 10 cells; tracks 10 to 12, 100 cells. Similarly processed positive controls (+) correspond to individual Nalm-6 pre-B leukemia single cells, whereas no cells are added in the negative controls (−). The identity of the amplified gene products is ascertained by direct sequencing of the cDNA in the excised bands and by molecular weight estimation (ladders). Note that the lower bands in the V-D-J gel in panel A correspond to primer artifacts and do not contain V-D-JH products, as determined by sequencing. L indicates ladder.
Gene expression profiles in individual cells from defined CD34+ subpopulations circulating in neonatal cord blood
| Gene . | Peripheral blood CD34+ cell subset . | |||
|---|---|---|---|---|
| CD34+CD19−CD10−% . | CD34+CD19−CD10+% . | CD34+CD19+CD10−% . | CD34+CD19+CD10+ % . | |
| GAPDH | 97.2 (210 of 216) | 98.6 (213 of 216) | 100 (216 of 216) | 98.6 (213 of 216) |
| VpreB | 0 (0 of 210) | 0 (0 of 213) | 58.3 (126 of 216) | 54.9 (117 of 213) |
| TdT | 0 (0 of 210) | 0 (0 of 213) | 91.6 (198 of 216) | 54.9 (117 of 213) |
| RAG-1 | 0 (0 of 210) | 0 (0 of 213) | 37.5 (81 of 216) | 38 (81 of 213) |
| mb-1 | 3.3 (7 of 210) | 19.2 (41 of 213) | 51.3 (111 of 216) | 53.5 (114 of 213) |
| preTα | 1.4 (3 of 210) | 4.2 (9 of 213) | 0 (0 of 216) | 0 (0 of 213) |
| Gene . | Peripheral blood CD34+ cell subset . | |||
|---|---|---|---|---|
| CD34+CD19−CD10−% . | CD34+CD19−CD10+% . | CD34+CD19+CD10−% . | CD34+CD19+CD10+ % . | |
| GAPDH | 97.2 (210 of 216) | 98.6 (213 of 216) | 100 (216 of 216) | 98.6 (213 of 216) |
| VpreB | 0 (0 of 210) | 0 (0 of 213) | 58.3 (126 of 216) | 54.9 (117 of 213) |
| TdT | 0 (0 of 210) | 0 (0 of 213) | 91.6 (198 of 216) | 54.9 (117 of 213) |
| RAG-1 | 0 (0 of 210) | 0 (0 of 213) | 37.5 (81 of 216) | 38 (81 of 213) |
| mb-1 | 3.3 (7 of 210) | 19.2 (41 of 213) | 51.3 (111 of 216) | 53.5 (114 of 213) |
| preTα | 1.4 (3 of 210) | 4.2 (9 of 213) | 0 (0 of 216) | 0 (0 of 213) |
Indicated IgM−CD34+ hemopoietic progenitor populations were purified from neonatal cord blood from 8 donors, as shown in Figure 1. The correlated expression of the gene panel, measured at the single-cell level, was performed as in Figure 2 (18-36 cells/donor). Results indicate the percentage of individual cells showing amplification of the indicated genes among the total number of single cells analyzed in each subpopulation. Only tubes showing GAPDH amplification (97.2%-100%) were considered in this study. Numbers in parentheses indicate the number of cells showing specific amplification divided by the number of GAPDH+ cells analyzed in each subset.
The results indicate that VpreB is expressed in a large percentage of CD34+CD19+ circulating hemopoietic progenitors, whether they are CD10+ or CD10−. Although there is a clear trend toward coexpression of VpreB, RAG-1, TdT, and mb-1 in these populations, their mRNA expression is not synchronous in every single CD34+CD19+ cell. It is noteworthy that one third of CD34+CD19+CD10−cells express TdT but lack VpreB, RAG-1, and mb-1 (Table 1), which is consistent with their phenotype as early-B cells.9 The TdT+VpreB−RAG-1−mb-1−early-B–cell subset is, however, rare and is not readily evident among the CD34+CD19+CD10+ cells. Despite the heterogeneity, the largest portion of circulating CD34+CD19+ cells, both in the CD10+and the CD10− subset, are BCPs that coexpress the recombination and diversification machinery enzymes (RAG-1+and TdT+), VpreB, and the immunoglobulin receptor transduction subunit CD79a (mb-1), although most (82%) of them do not yet express mRNA for recombined V-D-J μH genes; all are features of pro-B cells.9,10,15 16 In control experiments, functional μH mRNA of different lengths is readily amplified in 100% of mature IgM+ B cells (Figure 2B), which are homogeneously CD79a+ but typically do not express VpreB, RAG-1 or TdT. Intracellular staining of the CD34+CD19+CD10+/− cells revealed that 47% to 53% of the cells in the 2 populations bound a CD79a antibody but that none (less than 0.5%) revealed an isotype-matched CD3 antibody (not shown). Altogether, the results indicate that subsets of circulating CD34+CD19+CD10+/−cells that do not express μH mRNA have features of early-B and pro-B cells.
VH-D-JH μH mRNA analyses in blood CD34+CD19+CD10+/−μH−cells show a circulating pre–B-cell subpopulation
We also characterized the minor subset of circulating CD34+CD19+IgM− cells that express recombined VH-D-JH μH mRNA, but not surface IgM H or L chains (Figure 2A; range, 15%-28%; median, 18% CD34+CD19+IgM− cells from individual CB samples). The result of the μH sequence analyses in individual CD34+CD19+IgM− cells shows that two thirds of VH-D-JH μH rearrangements are out-of-frame, rendering the μH product nonfunctional because of stop codons. This is a feature of nonselected pre-B cells.9,16 Five percent to 10% of CD34+CD19+IgM− cells express cytoplasmic μH protein, but none bear Ig L protein (κ−and λ−) in immunofluorescence analyses of permeabilized cells. These results are consistent with the translation of μH in the 5% to 10% of CD34+CD19+IgM−cells containing VH-D-JH mRNA from in-frame rearrangements, and they define this subset as early cμH+CD34+ pre-BI cells.9,10 The analyses of D and JH usage indicates that the cells belong to independent clones, and the D-JH and VH-D coding joint N diversity shows TdT contribution to IgH hypervariable region 3 diversity.27 There is preferential JH4 usage (data not shown), as found after birth, but not the JH or D usage bias found in B cells with edited receptors26 or in fetal B-cell development.28 We conclude that the CB CD34+CD19+CD10+/− cells bearing V-D-JH transcripts represent pre-B cells and that they include a cμH+ cell subset as well as a subpopulation carrying out-of-frame rearrangements that do not bear cμH protein.
CD34+CD19−CD10+μH−circulating cells show a distinct gene expression profile: most cells in this population do not bear features typical of early-B–committed progenitors
One fifth of the CD34+CD19−CD10+ cells express themb-1 gene (CD79a), but, in contrast to the CD34+CD19+CD10+/− subsets, they do not express VpreB, RAG-1, or TdT mRNA (Table 1). CD34+CD19−CD10− cells do not express VpreB, RAG-1, or TdT mRNA either, and only 3.3% are CD79a+ (Table 1). To address whether the CD34+CD19−CD10+ cells that express CD79a are committed to the B lineage, we further incorporated the study of Pax-5 mRNA expression to the multiplex RT-PCR analyses of the circulating CD34+sIg− cells. Interestingly, most circulating CD34+CD19−CD10+ cells do not express Pax-5 (Figure 3; 95% or more CD34+CD19−CD10+ CB single cells are Pax-5−). In addition, unlike the BCP subsets, they are largely VpreB−, RAG-1−, and TdT−, as shown in Table 1. The expression of Pax-5 is also rare in single CD34+CD19−CD10− cells. The latter results are in stark contrast to the expression of Pax-5 in most cells from either the CD34+CD19+CD10+ or the CD34+CD19+CD10− BCP populations (Figure 3). Our results show that CD34+CD19−CD10+ cells show a distinct gene expression profile, regarding the expression of B-cell specification and commitment genes, from CD34+CD19+CD10+/−Pax-5+ BCP subsets. Only a minor subset (less than 5%) appears to be committed to the B lineage, as ascertained byPax5 gene expression.
Pax5 expression in circulating CD34+sIg−subsets correlates with CD19 but not with CD10 surface expression.
Surface IgM−CD34+CD19−CD10−, IgM−CD34+CD19−CD10+, IgM−CD34+CD19+CD10−, and IgM−CD34+CD19+ CD10+sorted single cells were analyzed for mRNA expression ofPax5 after multiplex RT-PCR. For simplicity, the results for GAPDH, VpreB, TdT, RAG-1, and CD79a genes, covered in Figure 2 and Table 1, are not included. In each of the 4 IgM−CD34+ subsets, 19 of 20 (95%) tracks showed a neat GAPDH+ amplification. RT-PCR tubes receiving one FACS droplet were labeled numbers 1 to 20. Similarly processed positive controls (+) corresponded to individual Nalm-6 pre-B leukemia single cells, whereas all reagents without cells were added in the negative controls (−). Results are representative of 5 similar experiments that analyzed the 4 CB populations from independent donors. Approximately 80% of the cells showed neat Pax5 mRNA amplifications in the IgM−CD34+CD19+CD10+/−subset, but less than 5% (less than 1 of 19) cells in the CD34+CD19−CD10+/− subsets bore Pax-5 mRNA. RT-PCR fidelity was ascertained by sequencing of the cDNA in the excised bands and by molecular weight estimation. L indicates ladder.
Pax5 expression in circulating CD34+sIg−subsets correlates with CD19 but not with CD10 surface expression.
Surface IgM−CD34+CD19−CD10−, IgM−CD34+CD19−CD10+, IgM−CD34+CD19+CD10−, and IgM−CD34+CD19+ CD10+sorted single cells were analyzed for mRNA expression ofPax5 after multiplex RT-PCR. For simplicity, the results for GAPDH, VpreB, TdT, RAG-1, and CD79a genes, covered in Figure 2 and Table 1, are not included. In each of the 4 IgM−CD34+ subsets, 19 of 20 (95%) tracks showed a neat GAPDH+ amplification. RT-PCR tubes receiving one FACS droplet were labeled numbers 1 to 20. Similarly processed positive controls (+) corresponded to individual Nalm-6 pre-B leukemia single cells, whereas all reagents without cells were added in the negative controls (−). Results are representative of 5 similar experiments that analyzed the 4 CB populations from independent donors. Approximately 80% of the cells showed neat Pax5 mRNA amplifications in the IgM−CD34+CD19+CD10+/−subset, but less than 5% (less than 1 of 19) cells in the CD34+CD19−CD10+/− subsets bore Pax-5 mRNA. RT-PCR fidelity was ascertained by sequencing of the cDNA in the excised bands and by molecular weight estimation. L indicates ladder.
Our multiplex RT-PCR studies of CD34+ hemopoietic cells included the study of surrogate TCRα chain (preTα, Table 1) and VpreB mRNA expression to monitor early-T versus early-B lymphoid lineage specification, and they rule out illegitimate gene transcription. PreTα expression occurs in CD10+ early T/DC multilineage progenitors devoid of B-lineage potential.24,29,30 Notably, our single-cell analyses show that the frequency of preTα+ cells is low in the CD34+CD19−CD10+ cell subset (4.2% of individual cells), very low (1.4%) in the CD34+CD19−CD10−cells, and undetectable in the CD34+CD19+CD10+/− cells. Although this study is intended to characterize CD34+ BCP, the results are consistent with those of previous studies in bulk circulating preTα+ “pre-T cells,” which found scarce or no expression of preTα mRNA in circulating CB and adult CD34+ cells, respectively.24,29 Notably, the major circulating T-cell precursor population, surface CD34−TCRβ− CB cells that uniformly co-express preTα, TdT, RAG-1, and CD3 transduction molecules,24 do not bear VpreB mRNA (E.S., A. de la H., unpublished results, June 2000). Hence, VpreB and preTα expression occur in separate cell subsets in both CD34+ and CD34− cells. The 19% CD79a+ and 4% pre-Tα+CD34+CD19−CD10+ cells (Table 1) are also in different subsets, because CD79a and preTα expression occur in separate individual cells. We conclude that CD34+CD19−CD10+/− cells are a heterogeneous population and that most cells (more than 80%-95%) are not B-lineage–committed progenitors by available gene expression criteria.9
Discussion
Here we identify CD34+ BCPs circulating in CB and characterize their gene expression profiles with unprecedented resolution using a combination of FACS and single-cell multiplex RT-PCR. These BCPs are CD34+CD19+CD10+/− cells that span early-B, pro-B, and pre-B differentiation stages. B-lineage commitment in the CB CD34+ populations is indicated by Pax5gene expression, which correlates with CD19 but not CD10 surface expression. The specification of B-cell development stages was ascertained by the expression patterns of the rearranged V-D-JH, CD79a, VpreB, RAG-1, and TdT gene products, according to widely accepted conventions.8-20
Our results show that CD34+ BCPs from BM and CB differ in the distribution of CD19 and CD10. In BM, approximately 95% of CD34+CD10+ cells are pro-B/pre-B cells that coexpress CD19. A minor CD34+CD19−CD10+ subset has been characterized as early-B cells, but the CD34+CD19+CD10− subset is not evident in BM.9 In a phenotypic comparison of healthy BM and circulating CD34+ subpopulations, Bender et al14 already noted that the proportion of CD34+CD19+ cells tripled that of CD34+CD10+ cells in adult peripheral blood (ie, 10% vs 3% of CD34+ cells), whereas the CD34+CD10+ cells outnumbered the CD34+CD19+ cells in BM (17% vs 14% of CD34+ cells). In our analyses, more than 95% of CB cells in the novel CD34+CD19+CD10−population are Pax-5+ early-B, pro-B, or pre-B cells. Notably, early-B cells represent approximately 30% to 40% of CD34+CD19+CD10− cells but are not evident in the CD34+CD19+CD10+population. In BM, CD10 decreases as early-B cells progress down the B-cell development pathway,9 an additional indication that CD10 surface levels are not a good surface marker for early BCP in CB. In BM, CD34+CD19−CD10+ cells express Pax-5, CD79, RAG-1, and TdT,9,21,22 but in CB they are heterogeneous, lack Pax-5, RAG-1, and TdT, and express CD79a and preTα in only one fifth and one twentieth of the cells, respectively. The latter genetic profile does not appear to be a consequence of the separation of circulating CD34+ cells from BM stroma. In cultures of CD34+ cells with BM stroma,15 more than 95% of the single CD19−CD10+ CB cells express neither Pax-5 nor VpreB, TdT, CD79, RAG-1, or preTα (E.S., A. de la H., unpublished results, January 2001). Early-B cells are proposed to be the progeny of CD34+ multilymphoid progenitors, which express CD10 in BM.9,31 Galy et al31 described a CD34+Lin−CD38+CD10+progenitor population in BM that had T/B/NK/DC potential but was severely depleted of myelo-erythroid potential. However, Hao et al32 recently reported that CD10 expression alone does not discriminate between progenitors with lymphoid and myeloid potential in CD34+CD38−/lowLin−CD10+cells from CB. It is noteworthy that the populations used in the latter report differ not only in the tissue origin (CB vs BM) but in the selection for primitive, CD38−, CD34+progenitors (cells are Lin−, CD19− in both reports).31,32 Interestingly enough, Hao et al showed that 60% to 70% of CD34+CD38−/lowLin−CD10+ cells, which lack CD7, Pax-5, and TdT, contain progenitors that retain both myelo-erythroid and lymphoid potential in clonal analyses of single cells. Herein we study CD34+ CB cells that are more than 95% CD38+. Our preliminary analyses of the distribution of CD38 show that it is heterogeneous in distinct CD34+ CB subsets. Whereas approximately 95% of CD34+CD19+CD10+/− cells in our samples were CD38+, two thirds of the CD34+CD19−CD10+ cells were CD38−/low and one third were CD38+ (not shown). Further studies are needed to more extensively readdress the gene expression profiles and multipotency of CB CD34+CD19−CD10+ cells in single-cell assays. The CD34+Lin−CD10+ phenotype has been operationally used before to identify BCPs, such as circulating and BM-mobilized BCPs.2 It appears now safe to consider that CD34 and CD10 alone do not discriminate among multilineage, multilymphoid, or B-lineage progenitors in cord blood and that CD10 will have to be combined with several other surface markers to define the putative hemopoietic stage and fate.32
The finding of circulating early-B, pro-B, and pre-B cells appears striking because it is in stark contrast to 3 concepts that have dominated the thinking about B-cell development.11-13 One postulates that BCPs are retained in the BM until they acquire surface immunoglobulin and that the emigrants to peripheral blood are immature IgM+ B cells. The second idea is that BCPs may gradually die if they are deprived of the “protective” signals delivered by microenvironment niches such as BM. The third notion is that such protective BM niches play an essential role during IgM receptor editing and selection for tolerance to self in pre-B/immature B cells. Recent experimental evidence partly challenges the paradigm, however, because of the following. First, mouse BM CD19+Vpre-B+Pro-B/pre-BI cells placed in single-cell cultures to isolate them from the BM environment proliferate spontaneously and differentiate efficiently into sIg+ immature B cells.33Second, when BCPs are mobilized into blood in CXCR4−/− KO mice, they survive and continue to generate B cells.11Similarly, analyses of sorted CD34+ CB cells cocultured with BM stroma15 show that early-B CD19+CD10− cells differentiate into CD19+CD10+ pro-B/pre-B cells that generate B cells (E.S., A. de la H., unpublished data, July 2001). Third, the expression of VpreB, RAG, and TdT has been reported in circulating B cells, which undergo secondary V(D)J rearrangements to edit their sIg/CD79 antigen receptor specificity.26 The latter mature B cells are different from the CB BCPs in that they lack CD34 and CD10 and that they homogeneously express productive μH mRNA message and surface and cytoplasmic H and L.26 34 Taken together, available evidence indicates that circulating BCPs may be biologically relevant. The presence of BCPs in CB poses the question on the origin and sIg repertoire selection mechanisms for circulating RAG+ B-lineage cells.
The mosaic expression of genes (variegation) reported here in CD34+ early-B and pro-B cells are consistent with current stochastic/selective and hybrid selective/instructive models of lymphoid development.19,35,36 As individual cells progress down the differentiation pathway, their B-lineage specification pattern shows a progressive fit with the genotype proposed after bulk population analyses of BCP stages,9,15-19 as best exemplified by mature B cells that show a homogeneous gene expression profile consistent with the prevalent deterministic patterns. Notably, the CD34+CD19+CD10+ CB population reported here resembles the population referred to as pro-B/pre-BI cells after single-cell analyses in BM.10 We characterize a novel CD34+CD19+CD10− BCP not considered in normal B-cell development schemes8-10,15,21-23 but long ago implicated in BCP cancer.37-40 Acute lymphoblastic leukemias (ALLs) are classified in subtypes considered to indicate the stage of development at which tumor transformation occurs.9,38 Notably, the CD10− progenitor B-ALL subtype occurs in infants and typically bears the CD34+CD19+CD10− genotype/phenotype discovered here in CB.39,40 We show that healthy BCP counterparts for the common CD10+CD19+ pro-B, pre-B, and B-cell ALL9,25,40 also circulate in CB. Interestingly, we find that CD34+CD19−CD10+ early-B cells are rare or do not circulate in CB, and a clonogenic leukemic counterpart for healthy CD34+CD19−CD10+early-B cells characterized in fetal liver and BM9,41 was not found among malignant cells from B-ALL patients either.41 In B-ALL, BCPs experience an initiating translocation event in utero, circulate in newborn blood as preleukemic cells, and can remain in the circulation several years before a second mutation event promotes the conversion of the fetal preleukemic clone into a leukemia.37,38 The observation that healthy BCPs circulate in the CB of healthy persons may provide a physiologic mechanism for the blood-borne dissemination of fetal preleukemic cells.42 B-ALL represents 25% of all childhood cancers, and chromosomal translocations are implicated in the ALL pathogeny.37-40 Here we show that a significant fraction of healthy CB CD34+ cells express RAG. RAG can mediate translocation by transpositional mechanisms,41 which raises concern regarding the potential risk for chromosomal translocation36-38,43 in reparative medicine protocols that involve gene manipulation in CD34+cells.4 Studies are under way to experimentally address this possibility.
We thank M. A. R. Marcos and M.-L. Gaspar for critical review of the manuscript, J. Monserrat for superb sortings, V. Parrillas and the nursing staff at the San Francisco de Ası́s Hospital for assistance with cytoplasmic staining and for obtaining cord blood samples, and C. Mark for editorial assistance.
The Department of Immunology and Oncology was founded and is supported by the CSIC and by Pharmacia Corporation.
Prepublished online as Blood First Edition Paper, November 21, 2002; DOI 10.1182/blood- 2002-07-2244.
Supported by the Comisión Interministerial de Ciencia y Tecnologı́a grants 2FD97-2226, SAF-2001-2453 and GEN2001-4856-C13. E.S. is supported by the Ministry of Science and Technology as a Ramón y Cajal Research Scientist.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Antonio de la Hera, Laboratory of Immunology and Oncology, CSIC Associated Unit, Facultad de Medicina, Universidad de Alcalá, Alcalá de Henares, E-28871 Madrid, Spain; e-mail: adelahera@cib.csic.es.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal