Abstract
SFT, a stimulator of iron (Fe) transport, has been described as a transmembrane protein that facilitates the uptake of ferrous and ferric iron in mammalian cells. This study was initiated to investigate the 5′ regulatory region of SFT and its role in the etiology of hereditary hemochromatosis. Sequence analyses of the putative 5′ regulatory region revealed that the SFT cDNA sequence corresponds to intron 6/exon 7 of UbcH5A, a member of E2 ubiquitin-conjugating enzymes, which is involved in the iron-dependent ubiquitination of the hypoxia-inducible factor (HIF) by the von Hippel-Lindau tumor suppressor (pVHL) E3 ligase complex. Further mRNA expression studies using a sequence-specific reverse transcriptase–polymerase chain reaction (RT-PCR) assay showed that UbcH5A is significantly up-regulated in the liver of iron-overloaded patients with hereditary hemochromatosis, as previously published for SFT. However, in vitro studies on HepG2 cells failed to demonstrate any significant UbcH5A regulation in response to iron loading or iron chelation. In conclusion, in vivo mRNA expression data previously obtained for SFT might be attributed to UbcH5A. The role of UbcH5A and the ubiquitination pathway in the etiology of hereditary hemochromatosis remains to be elucidated further.
Introduction
In mammalian cells, the major route of iron uptake is the receptor-mediated endocytosis of diferric transferrin (Tf) bound to the transferrin receptor (TfR). Following internalization and acidification of the endosome, iron is released from Tf and transported across the endosomal membrane. This step, as well as transport of iron across the intestinal epithelial layer, requires a Tf-independent pathway.1-4 Recently, several putative transporters involved in the Tf-independent uptake of iron have been identified. DMT1 (formerly called Nramp2, DCT1) is a transmembrane divalent cation transporter with a high affinity for ferrous Fe(II) iron.5Whereas DMT1 is thought to mediate the rapid uptake of dietary iron from the intestinal lumen,5-8 IREG1 (also known as ferroportin, MTP1) is localized to the basolateral membrane of the enterocyte and functions as an Fe(II) exporter.9-11Another iron transport protein, SFT (stimulator of Fe transport), has been found to stimulate transport of both ferrous Fe(II) and ferric Fe(III) iron and shows high specifity for iron in Tf-independent uptake.12-14 Accordingly, Northern analysis indicated that SFT expression is up-regulated in iron-deficient HeLa cells exposed to the iron chelator deferoxamine and down-regulated in iron-supplemented HeLa and HepG2 cells.15
Remarkably, hepatic SFT expression was found to be significantly increased in patients with hereditary hemochromatosis (HH) despite tissue iron overload.15 In most cases, this frequent autosomal recessive disorder is associated with a missense mutation (Cys282Tyr) in the hemochromatosis geneHFE, which codes for a protein that is homologous to class I major histocompatibility complex (MHC) molecules and requires β2-microglobulin (light chain) for surface presentation.16-18 However, there is still some evidence that other genetic factors influence expression of the disease.19,20 The observation that SFT expression is down-regulated in iron supplemented HeLa and HepG2 cells but up-regulated in the liver of hemochromatosis patients15may lead to the speculation that a paradoxical regulation of SFT plays a central role in the etiology of HH. Therefore, the aim of this study was to obtain further 5′ sequence information on SFT in order to identify regulatory elements involved in the regulation of SFT and its paradoxical up-regulation in hemochromatosis patients. Surprisingly, during this study the authors retracted21 some of their original results on the nucleotide sequence of SFT.12 Due to the correction of sequencing errors, SFT lost its open reading frame.21Consequently, the question arises whether putative SFT represents an important protein involved in iron metabolism or a cloning artifact.
The present work sheds some light on the rather controversial SFT protein. Herewith, we demonstrate that the previously published nucleotide sequence of SFT indeed corresponds to a genomic fragment of the ubiquitin-conjugating enzyme UbcH5A. In addition, we evaluate whether previously published data on SFT regulation in response to iron loading or iron chelation might be attributed to UbcH5A mRNA expression.
Materials and methods
Cells
HepG2 cells were obtained from DSMZ (Braunschweig, Germany) and grown in RPMI 1640 medium containing 10% fetal calf serum, penicillin (100 U/mL), and streptomycin (100 μg/mL). Incubation with 65 μM Fe-NTA (1:1 ratio) was performed for 72 hours and with 50 μM deferoxamine (DFO) for 18 hours.
Liver biopsies
To evaluate the influence of iron overload on gene expression, liver biopsy samples were obtained from 7 iron-overloaded HH patients homozygous for the C282Y mutation in HFE prior to phlebotomy. All patients had serum ferritin levels above 2000 μg/L. Control samples derived from 4 patients with isolated γ-glutamyl transferase elevation and normal liver histology and from 3 patients with chronic hepatitis C and mild histological activity. All controls had serum ferritin levels below 300 μg/L, showed no significant fibrosis, were negative for stainable iron in liver biopsy, and did not show any abnormalities in serological iron parameters. To evaluate the influence of inflammation on gene expression, 13 iron stain–negative liver biopsy samples from patients with chronic hepatitis C were obtained. Histological grading of activity according to the METAVIR score22 was A1 in 7 patients and A2 in 6 patients. METAVIR fibrosis scores were F1 to F4. Control samples were from 9 patients without chronic hepatitis (γ-glutamyl transferase elevation). Control liver biopsies revealed no significant histological signs of inflammation or fibrosis. All samples were stored at −20°C in RNAlater solution (Ambion, Austin, TX) prior to RNA isolation. The study was approved by the local ethics committee. Informed consent was obtained from all patients.
Chemicals
RPMI 1640 medium, penicillin, and streptomycin were from Life Technologies (Paisley, United Kingdom). Ferric nitrate nonahydrate and nitrilotriacetic acid disodium salt (NTA) were obtained from Sigma-Aldrich (Steinheim, Germany). Deferoxamine was from Novartis (Nürnberg, Germany).
Identification of SFT/UbcH5A genomic DNA sequence
In order to identify 5′ sequence information of SFT, the Human Genome Walker Kit (Clontech, Palo Alto, CA) was used as described in the manufacturer′s manual. The oligonucleotide sequences for the gene-specific primer GSP 1 (5′-CAA TTA CTG GGT CAA TGT GAC TTA GGT TC) and the nested GSP 2 (5′-GCC AAT CCT GAA GGA GCA TAA TCT AC) were derived from SFT cDNA (GenBank accession no. AF020761.1). The polymerase chain reaction (PCR) product generated from theEcoRV library (approximately 1 kb) was reamplified using adaptor primer 2 (provided in the kit) and nested GSP 3 (5′-CCT ACA GTA CAA GCA AGA TGA AGC AGC), subcloned in pUC18 (Sure Clone Ligation Kit; Amersham Pharmacia Biotech, Freiburg, Germany), and sequenced (ABI 310 Genetic Analyzer; Applied Biosystems, Weiterstadt, Germany). From the derived sequence information, a primer pair (SFTPro501 5′-GCA AAA GAG TGT ATG AGC ACA GTA TTC, SFTPro301 5′-CTG GTA CTA AGG GGT CAT CTG G) was chosen in order to obtain a P1 genomic clone by PCR-based screening (GenomeSystems, St Louis, MO) followed by sequencing analysis. The complete exon-intron structure of SFT/UbcH5A was identified by sequence alignment (ClustAl) with the cDNA sequence of an EST clone (IMAGE:1939023; GenomeSystems) containing full-length UbcH5A cDNA.
RNA expression analysis
A multiple-tissue Northern blot was obtained from Clontech. Probes specific for the UbcH5A coding sequence were generated by PCR using the sense primer 5′-ATG GGG CCT CCT GAT AGC, the antisense primer 5-′CTG GTA CTA AGG GGT CAT CTG G, and IMAGE clone 1939023 (UbcH5A cDNA) as a template. The probes were 32P-labeled by random priming; hybridization was performed at 68°C, according to the manufacturer's recommendations.
Quantitative RT-PCR
Total RNA was isolated from liver biopsies and from cell culture using the RNAeasy Mini Kit (Qiagen, Hilden, Germany) including DNAse digestion according to the manufacturer's instructions. Real-time quantification of mRNA transcripts was performed with a 2-step reverse transcriptase (RT) PCR using the LightCycler system and Relative Quantification Software Version 1.0 (Roche Molecular Biochemicals, Mannheim, Germany). In a first step, cDNA synthesis was performed with the First Strand cDNA Synthesis Kit for RT-PCR (Roche Molecular Biochemicals) according to the manufacturer's instructions. In a second step, UbcH5A transcripts were amplified in duplicates with sense primer UbcH5A-501 (5′-GTG ATC CTA ATC CAG ATG ACC C) and antisense primer UbcH5A-301 (5′-TCA ATG CAT ACA AAA TGA TAA ATG C) and detected by sequence-specific hybridization probes UbcH5A-LCA (5′-CAT TGC ATA TTT CTG AGT CCA TTC TCT TGC-fluorescein) and UbcH5A-LCM (5′-LightCycler-Red640-TGT CTG TTG TAT TTT TCT TTG TCT GAT TTA TAG-p). This approach prevented detection of genomic DNA as well as mRNAs homologous to UbcH5A including “SFT mRNA.” To provide a positive control in all experiments, transferrin receptor (TfR) expression was determined quantitatively, as follows. TfR transcripts were amplified in duplicate with sense primer TFR1-502 (5′-TAT AGA AGG TTT GGG GGC TGT G) and antisense primer TFR1-302 (5′-GAG ACC CTA TGA ACT TTT CCC TAG) and detected using SYBR Green I (Roche Molecular Biochemicals) according to the manufacturer's instructions. Both UbcH5A and TfR transcripts were normalized to actin (β-actin) as an internal control. Actin transcripts were amplified in duplicate with sense primer ACTB-502 (5′-AGG ATG CAG AAG GAG ATC ACT G) and antisense primer ACTB-302 (5′-GGG TGT AAC GCA ACT AAG TCA TAG) and detected using SYBR Green I. UbcH5A/actin and TfR/actin ratios were calculated using LightCycler Relative Quantification Software Version 1.0 (Roche Molecular Biochemicals), which provides a fully automated, efficiency-corrected, relative quantification normalized to calibrators. According to the manufacturer's instructions, calibrators for UbcH5A, TfR, and actin were generated from EST clones (IMAGE:1939023, GenomeSystems, for amplification of UbcH5A; IMAGE:841703 from RZPD, Berlin, Germany, for amplification of TfR; IMAGE:2518720, RZPD, for amplification of actin). In addition, standard curves were prepared based on accurately determined dilutions of the plasmids containing cDNA of UbcH5A (IMAGE:1939023), TfR (IMAGE:841703), and actin (IMAGE:2518720) as templates. Plasmid dilutions covered a dynamic range of 5 logarithmic orders. Statistical analysis of quantitative variables was performed by means of the nonparametric Mann-Whitney test. P < .05 was considered significant.
Results
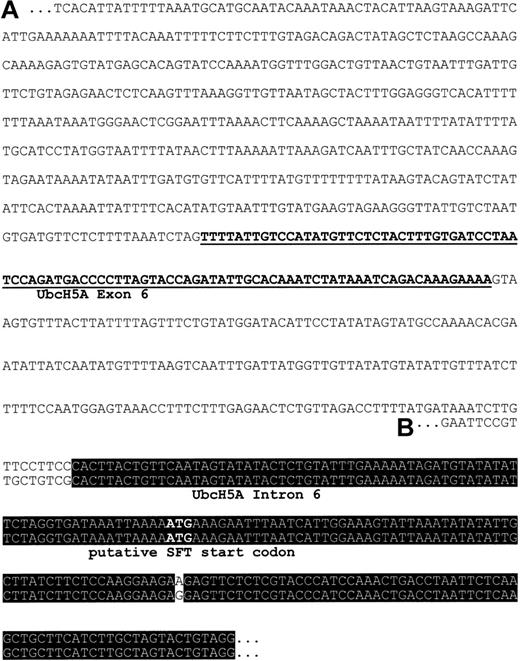
SFT mRNA shows high sequence homology with intron 6 and exon 7 of UbcH5A
Using oligonucleotide primers specific for SFT, a 1-kb PCR product was generated from the EcoRV library of the Human Genome Walker Kit (Clontech). Sequence analysis of the subcloned PCR product was expected to reveal further 5′ sequence information of SFT, including the promotor and regulatory elements. In contrast, we found a nucleotide sequence that had no homology to a previously published short fragment of the SFT 5′ regulatory region.23Therefore, a genomic clone was isolated by PCR-based screening with primers specific for our 1-kb PCR product. Sequence analysis of the genomic clone confirmed the nucleotide sequence of the 1-kb PCR product and demonstrated the reliability of our results. A BlastN2 search of our putative 5′ regulatory region of SFT revealed a 94-nucleotide fragment showing 100% identity with nucleotides 305 through 398 of UbcH5A (ubiquitin-conjugating enzyme E2) mRNA coding sequence (GenBank accession no. NM_003338.1; Figure 1). This observation strongly suggests that the fragment represents an exon of the UbcH5A gene. To prove this hypothesis, full-length mRNA sequence of UbcH5A was obtained by sequencing an EST clone with high homology to the published UbcH5A coding sequence (GenBank accession no. NM_003338.1). Sequence analysis revealed full-length UbcH5A mRNA including the 5′ and 3′ untranslated regions (submitted to GenBank, accession no. AJ272367). The exon/intron organization of theUbcH5A gene was then determined with the information being submitted to GenBank (chromosome 10 genomic clone RP11-373P23; GenBank accession no. AC023170). According to this, the UbcH5A gene is located on chromosome 10, spans 35 kb, and contains 7 exons (submitted to GenBank, accession no. AJ293565). In addition, sequence alignment demonstrates 99% identity between intron 6/exon 7 ofUbcH5A and the published SFT cDNA (GenBank accession no.AF020761, now designated as unknown RNA sequence).
Putative 5′ regulatory region of SFT.
(A) Sequence represents the 1-kb PCR product generated from theEcoRV library of the Human Genome Walker Kit (Clontech) with primers specific for SFT. Underlined nucleotides share 100% identity with exon 6 of the UbcH5A gene, a member of E2 ubiquitin-conjugating enzymes. (B) Sequence corresponds to the 5′ region of SFT RNA (GenBank accession no. AF020761, now designated as unknown RNA sequence). Nucleotides within the black box are identical in both sequences.
Putative 5′ regulatory region of SFT.
(A) Sequence represents the 1-kb PCR product generated from theEcoRV library of the Human Genome Walker Kit (Clontech) with primers specific for SFT. Underlined nucleotides share 100% identity with exon 6 of the UbcH5A gene, a member of E2 ubiquitin-conjugating enzymes. (B) Sequence corresponds to the 5′ region of SFT RNA (GenBank accession no. AF020761, now designated as unknown RNA sequence). Nucleotides within the black box are identical in both sequences.
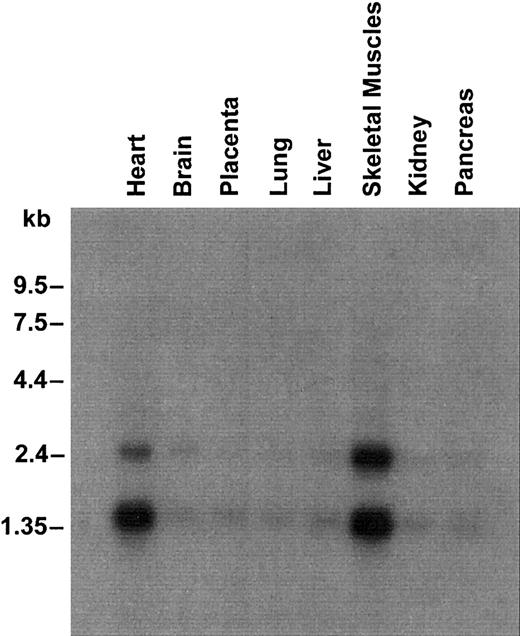
UbcH5A mRNA tissue expression
UbcH5A was ubiquitously expressed with a high level of expression in heart and skeletal muscles (Figure 2). As demonstrated for SFT,12 Northern analysis revealed 2 transcripts of approximately 1.5 kb and approximately 2.4 kb. Both transcripts were observed, although the probes used were specific for UbcH5A mRNA coding sequence and did not show any homology to the published SFT nucleotide sequence.
Northern blot containing 2.5 μg poly(A) RNA from several human tissues.
Hybridization was performed with a 32P-labeled probe specific for UbcH5A coding sequence. Molecular sizes are shown on the left.
Northern blot containing 2.5 μg poly(A) RNA from several human tissues.
Hybridization was performed with a 32P-labeled probe specific for UbcH5A coding sequence. Molecular sizes are shown on the left.
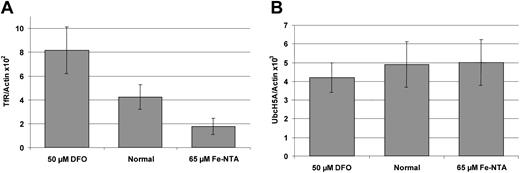
UbcH5A transcript levels are not regulated in response to cellular iron content in HepG2 cells
In all previous studies on SFT mRNA expression, probes matching the 3′ untranslated region (UTR) were used.12,15 24 Since the UbcH5A 3′ UTR (GenBank accession no. AJ272367) is almost identical to the previously published SFT 3′ UTR (GenBank accession no.AF020761.1), those studies might have detected UbcH5A transcripts instead of SFT transcripts. Therefore, we analyzed UbcH5A mRNA expression in response to cellular iron content, using a highly specific assay. UbcH5A transcript levels were determined quantitatively with sequence-specific hybridization probes on the LightCycler PCR system (real-time RT-PCR), and were normalized to actin transcript levels (UbcH5A/actin ratio). To provide a positive control, transferrin receptor (TfR) mRNA expression in response to cellular iron content was measured and normalized to actin (TfR/actin ratio). As demonstrated in Figure 3, HepG2 cells treated with 50 μM deferoxamine (DFO) showed an increased TfR/actin ratio and those treated with 65 μM Fe-NTA a decreased TfR/actin ratio (P < .01). In contrast, neither iron chelation (50 μM DFO) nor iron loading (65 μM Fe-NTA) induced significant changes in the UbcH5A/actin ratio in HepG2 cells.
Iron-dependent expression of transferrin receptor (TfR) and UbcH5A mRNA normalized to actin mRNA levels in HepG2 cells.
Panel A shows expression of transferrin receptor (TfR); panel B shows expression of UbcH5A mRNA. Incubation was performed with 50 μM deferoxamine (DFO) for 18 hours and with 65 μM Fe-NTA for 72 hours. For expression analyses, a 2-step, real-time RT-PCR on the LightCycler system was used. TfR/actin and UbcH5A/actin ratios were calculated using the LightCycler Relative Quantification Software Version 1.0 (Roche Molecular Biochemicals), which provides a fully automated efficiency-corrected relative quantification. The mean values from 8 independent experiments are shown. The error bars represent 95% confidence intervals. The differences between TfR/actin ratios are statistically significant (P < .01). UbcH5A/actin ratios do not differ significantly.
Iron-dependent expression of transferrin receptor (TfR) and UbcH5A mRNA normalized to actin mRNA levels in HepG2 cells.
Panel A shows expression of transferrin receptor (TfR); panel B shows expression of UbcH5A mRNA. Incubation was performed with 50 μM deferoxamine (DFO) for 18 hours and with 65 μM Fe-NTA for 72 hours. For expression analyses, a 2-step, real-time RT-PCR on the LightCycler system was used. TfR/actin and UbcH5A/actin ratios were calculated using the LightCycler Relative Quantification Software Version 1.0 (Roche Molecular Biochemicals), which provides a fully automated efficiency-corrected relative quantification. The mean values from 8 independent experiments are shown. The error bars represent 95% confidence intervals. The differences between TfR/actin ratios are statistically significant (P < .01). UbcH5A/actin ratios do not differ significantly.
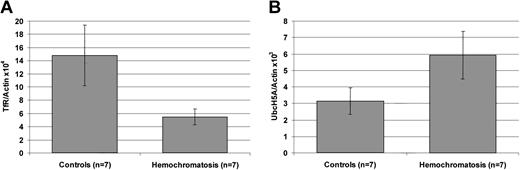
UbcH5A expression is enhanced in the liver of patients with hereditary hemochromatosis
To investigate the role of UbcH5A in hereditary hemochromatosis, levels of UbcH5A transcripts normalized to actin (UbcH5A/actin ratio) were measured in liver biopsies from iron-overloaded patients homozygous for the Cys282Tyr mutation in HFE. In the liver of these patients, the UbcH5A/actin ratio was approximately 2-fold higher compared with liver biopsies from controls with negative iron staining and serum ferritin levels within normal limits (P < .05; Figure 4). In contrast, the TfR/actin ratio used as a positive control was significantly decreased (P < .01) in patients with untreated hemochromatosis. To exclude a significant impact of inflammation on these observations, levels of UbcH5A transcripts normalized to actin (UbcH5A/actin ratio) were measured in liver biopsies from patients with inflammation due to chronic HCV infection and were compared with those in liver biopsies from patients without intrahepatic inflammation. The mean UbcH5A/actin ×103 ratios (± SD) did not differ significantly between the 2 groups (2.60 ± 0.64 vs 2.83 ± 0.89;P = .9468).
Transferrin receptor (TfR) and UbcH5A mRNA expression normalized to actin mRNA levels in liver biopsy samples from 7 control subjects and 7 patients with untreated
HFE-associated hemochromatosis. Panel A shows expression of transferrin receptor (TfR); panel B shows expression of UbcH5A mRNA. For expression analyses, a 2-step, real-time RT-PCR on the LightCycler system was used. TfR/actin and UbcH5A/actin ratios were calculated using LightCycler Relative Quantification Software Version 1.0 (Roche Molecular Biochemicals), which provides a fully automated, efficiency-corrected, relative quantification. Mean values are shown. The error bars represent 95% confidence intervals. The differences between TfR/actin and UbcH5A/actin ratios are statistically significant (P < .01 for TfR/actin ratio; P < .05 for UbcH5A/actin ratio).
Transferrin receptor (TfR) and UbcH5A mRNA expression normalized to actin mRNA levels in liver biopsy samples from 7 control subjects and 7 patients with untreated
HFE-associated hemochromatosis. Panel A shows expression of transferrin receptor (TfR); panel B shows expression of UbcH5A mRNA. For expression analyses, a 2-step, real-time RT-PCR on the LightCycler system was used. TfR/actin and UbcH5A/actin ratios were calculated using LightCycler Relative Quantification Software Version 1.0 (Roche Molecular Biochemicals), which provides a fully automated, efficiency-corrected, relative quantification. Mean values are shown. The error bars represent 95% confidence intervals. The differences between TfR/actin and UbcH5A/actin ratios are statistically significant (P < .01 for TfR/actin ratio; P < .05 for UbcH5A/actin ratio).
Discussion
Recently, several genes involved in iron metabolism, including the hemochromatosis gene HFE, have been isolated.5,9-12,16,25-28 From these, the apical iron transporter DMT1 and the basolateral iron exporter IREG1 mediate iron absorption in the duodenal enterocyte.2,3 Interestingly, both iron transport molecules are up-regulated in the duodenum during iron deficiency as they are in the duodenum of HH patients.29 This paradoxical up-regulation of DMT1 and IREG1 might explain progressive iron accumulation in HH. However, the exact mechanism by which defective HFE leads to these phenomena is not yet clear. Most authors favor the hypothesis that the absence of functional HFE results in an erroneous sensing of body iron stores in immature duodenal crypt cells.1-4 In addition, signaling molecules such as the putative iron store regulator hepcidin,28 or proteins such as SFT, a stimulator of Fe transport,12-15 may contribute to the etiology of HH.
SFT was first described in 1997 as a transmembrane protein stimulating Tf-independent and Tf-dependent iron uptake.12 Although functional data suggest that SFT stimulates the uptake of of ferrous Fe(II) and ferric Fe(III) iron in mammalian cells,13Northern analyses indicate an up-regulation of SFT in response to iron loading and a down-regulation in response to iron chelation.15 In contrast, SFT expression studies in iron-overloaded HH patients demonstrated an up-regulation in liver by Northern analysis15 and in the duodenum by immunohistochemistry.30 This paradoxical up-regulation, despite iron overload conditions, strongly suggests an important role of SFT in the etiology of HH, although these data are not reproducible in the mouse model of HH.31 32
In order to analyze transcriptional regulation of SFT, we began to investigate 5′ sequence information of SFT. In contrast to the previously published short segment of the putative SFT promotor,23 our 5′ sequence data revealed a 94-nucleotide fragment sharing 100% identity with UbcH5A, a member of the E2 ubiquitin-conjugating enzymes. After obtaining an UbcH5A genomic clone and UbcH5A full-length cDNA, sequence alignment demonstrated that the SFT cDNA published in 1997 is almost identical with intron 6 and exon 7 of the UbcH5A gene. Subsequently, the authors retracted some of their original results12 due to sequencing errors and noted that their sequence matches a part of a ubiquitin-conjugating enzyme.21 As a consequence, the original SFT cDNA GenBank entry was deleted and replaced by a sequence now designated as “unknown RNA involved in iron metabolism” (GenBank accession no.AF020761). Identity between this corrected sequence and intron 6/exon 7 of the UbcH5A gene is higher than 99%. Therefore, our data strongly suggest that the putative SFT nucleotide sequence corresponds to a fragment of UbcH5A genomic DNA. Such a hypothesis is also supported by the fact that the putative SFT lost its previously reported open reading frame due to the correction of sequencing errors.21
It is unclear how the previously published data on SFT, particularly the protein and functional data,12-14,30 should be interpreted. Regarding mRNA expression analyses, all previous studies used hybridization probes matching the putative SFT 3′ untranslated region (UTR).12,15,24 As the UbcH5A 3′ UTR is almost identical to the previously published SFT 3′ UTR, those studies most likely detected UbcH5A instead of SFT transcripts. This hypothesis is supported by our finding that Northern analysis for UbcH5A reveals 2 transcripts of approximately 1.5 kb and approximately 2.4 kb. Interestingly, 2 transcripts of the same length have been described for SFT.12 In our study, both transcripts were observed, although the probes used were specific for UbcH5A mRNA coding sequence and did not show any homology to the published SFT nucleotide sequence.
Based on this observation, it might be conceivable that UbcH5A mRNA expression is iron-regulated as previously demonstrated for SFT.15 However, we were not able to demonstrate significant changes in UbcH5A mRNA expression in response to iron loading or chelation in HepG2 cells. On the other hand, UbcH5A transcripts are significantly up-regulated in the liver of iron-overloaded HH patients. This up-regulation of UbcH5A in HH patients is statistically significant but is only 2-fold. Therefore, the question arises whether UbcH5A plays an important role in iron metabolism or hemochromatosis as suggested for SFT.12-14Classical iron-related genes such as DMT1 or IREG1 are also up-regulated only by a factor of 2 in the duodenum of HH patients.29,33 In addition, we observed a less than 3-fold down-regulation of TfR in the liver of HH patients. These data demonstrate that the level of UbcH5A up-regulation in HH patients is in good agreement with other iron-related genes. On the other hand, it might be conceivable that the up-regulation of UbcH5A transcripts in the liver of HH patients is unspecific. Fully expressed HH is associated with intrahepatic inflammation and fibrosis.1,19 20 As these factors might lead to an increased expression of UbcH5A, we evaluated the influence of intrahepatic inflammation or fibrosis on the expression of UbcH5A using liver biopsies from patients with chronic HCV infection and patients showing regular liver histology. These data do not suggest an up-regulation of UbcH5A in response to an inflammatory reaction or to fibrosis.
Nevertheless, it is unclear whether UbcH5A plays a significant role in iron metabolism or hemochromatosis. UbcH5A is a member of the ubiquitin-conjugating enzymes (E2) involved in ubiquitination of specific proteins. In general, ubiquitination requires sequential actions of activating (E1), conjugating (E2), and ligase (E3) enzymes. First, ubiquitin is activated to form a thiol ester intermediate with the E1. Activated ubiquitin is then transferred to an E2 from E1. In the presence of an E3, the E2, such as UbcH5A, transfers ubiquitin to the specific substrate.34 UbcH5A itself is known to be involved in the E6-AP-dependent degradation of p5335 and the ubiquitination of the α subunit of the hypoxia-inducible factor (HIF-α) by the von Hippel-Lindau tumor suppressor (pVHL) E3 ligase complex.36 The ubiquitination of HIF by pVHL plays a central role in the cellular response to changes in oxygen availability, as HIF is a master regulator of oxygen homeostasis that controls angiogenesis, erythropoiesis, and glycolysis via transcriptional activation of target genes.36,37Interestingly, Jaakkola et al38 demonstrated that targeting of HIF to the pVHL ubiquitination complex is regulated through hydroxylation of a proline residue in HIF by the enzyme HIF-prolyl-hydroxylase (HIF-PH). HIF-PH function requires dioxygen as a cosubstrate and iron as a cofactor and, therefore, suggests HIF-PH as a cellular oxygen sensor.39 Experimental data also revealed that iron loading results in increased targeting of HIF to the pVHL ubiquitination complex.38 The fact that UbcH5A functions as an E2 in this process may explain its up-regulation in the liver of iron-overloaded hemochromatosis patients. As recently demonstrated for Ubc9, overexpression of an E2 can lead to increased ubiquitination of its specific target proteins by the pVHL complex.40Therefore, increased expression of UbcH5A might effect iron metabolism by ubiquitin-dependent degradation of specific targets, including HIF. Identification of additional targets would be helpful to assess the exact role of UbcH5A. The observation that a central regulator of iron metabolism, iron regulatory protein 2 (IRP2), is also subject to ubiquitin-dependent degradation41 emphasizes the relevance of this pathway in the iron machinery.
Prepublished online as Blood First Edition Paper, December 12, 2002; DOI 10.1182/blood-2002-07-2192.
Supported by grants from the Deutsche Forschungsgemeinschaft (STR 216/10-1) and the Dietmar Hopp Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Wolfgang Stremmel, University Hospital Heidelberg, Department of Internal Medicine IV, Bergheimer Strasse 58-69115 Heidelberg, Germany; e-mail:stremmel@medizin-online.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal