Abstract
In the bone marrow of C/EBPε−/− mice, expression of neutrophil secondary and tertiary granule mRNAs is absent for lactoferrin (LF), neutrophil gelatinase (NG), murine cathelinlike protein (MCLP), and the cathelin B9; it is severely reduced for neutrophil collagenase (NC) and neutrophil gelatinase-associated lipocalin (NGAL). In addition, the expression of eosinophil granule genes, major basic protein (MBP), and eosinophil peroxidase (EPX) is absent. These mice express C/EBPα, C/EBPβ, and C/EBPδ in the bone marrow at levels similar to those of their wild-type counterparts, suggesting a lack of functional redundancy among the family in vivo. Stable inducible expression of C/EBPε and C/EBPα in the murine fibroblast cell line NIH 3T3 activated expression of mRNAs for B9, MCLP, NC, and NGAL but not for LF. In transient transfections of C/EBPε and C/EBPα, B9 was strongly induced with weaker induction of the other genes. C/EBPβ and C/EBPδ proteins weakly induced B9 expression, but C/EBPδ induced NC expression more efficiently than the other C/EBPs. The expression of MBP was inefficiently induced by C/EBPε alone and weakly induced with C/EBPε and GATA-1, but the addition of PU.1 resulted in a striking cooperative induction of MBP in NIH 3T3 cells. Mutation of a predicted PU.1 site in the human MBP promoter-luciferase reporter construct abrogated the response to PU.1. Gel-shift analysis demonstrated binding of PU.1 to this site. MBP and EPX mRNAs were absent in a PU.1-null myeloid cell line established from the embryonic liver of PU.1−/− mice. Restitution of PU.1 protein expression restored MBP and EPX protein expression. This study demonstrates that C/EBPε is essential and sufficient for the expression of a particular subset of neutrophil secondary granule genes. Furthermore, it indicates the importance of PU.1 in the cooperative activation of eosinophil granule genes.
Introduction
Transcription factors play a major role in the development of specific lineages in the hematopoietic system.1,2 Mature cells of the myeloid lineage, including peripheral blood monocytes, tissue macrophages, and neutrophilic and eosinophilic granulocytes, derive from a common myeloid precursor. Of the numerous transcription factors involved in myelopoiesis, C/EBPα, C/EBPε, and PU.1 are critical for normal granulocytic differentiation. Targeted inactivation of these genes in mice demonstrates their importance in the development, function, and maturation of neutrophils and eosinophils.2-6
In the 2 murine models for PU.1 deficiency, mice either die at late gestation or survive for approximately 48 hours after birth and succumb to septicemia.5,6 Both models have defects in the myeloid and lymphoid lineages. A lack of mature macrophages, neutrophils, dendritic cells, osteoclasts, B cells, and T cells is observed.5-7 Antibiotic treatment allows mice that survive birth to live up to 2 weeks, during which time some cells with the characteristics of neutrophils develop by day 3.6 These neutrophils appear normal by morphology and express neutrophil markers such as Gr-1 and chloroacetate esterase, but they fail to mature completely as indicated by the lack of secondary granule gene expression.8 In addition, neutrophils from PU.1 knockout mice are functionally impaired with defects in superoxide production, bacterial uptake, and killing.8 Restoration ofPU.1 gene expression in a myeloid cell line derived from the embryonic livers of these mice reinstated expression of the secondary granule genes, and functions of normal terminal neutrophil maturation were acquired.9
Mice lacking the C/EBPα protein displayed abnormalities in liver, adipose tissue, and lungs and died within the first few hours of birth because of impaired glucose metabolism.10,11 Analysis of C/EBPα-deficient mice revealed a loss of mature neutrophils and eosinophils in peripheral blood and in fetal and newborn livers.12 Most white blood cells resembled immature myeloid cells with a block at the myeloblast stage.12 This was associated with a dramatically reduced expression of the granulocyte–colony-stimulating factor (G-CSF) and interleukin-6 (IL-6) receptor mRNAs, whereas mRNA levels of the macrophage-CSF (M-CSF) and granulocyte macrophage-CSF (GM-CSF) receptors were unaffected.12 13
Like C/EBPα-deficient mice, C/EBPε knockout mice displayed defects in granulopoiesis.3 An increase in granulocyte progenitors occurred in the C/EBPε-deficient bone marrow. In addition, the peripheral blood from C/EBPε-deficient mice displayed increased numbers of hyposegmented morphologically atypical neutrophils.3 This indicated a block at a later stage of granulocytic differentiation not observed in the C/EBPα-deficient mice. In contrast to C/EBPα-deficient mice, C/EBPε-deficient mice developed normally and were fertile but eventually died of opportunistic infections likely resulting from neutrophil cell dysfunction.3 Neutrophils from C/EBPε-deficient mice are defective in chemotaxis, disaggregation, receptor up-regulation, superoxide production, and bactericidal activity.3,14 C/EBPε-deficient mice lacked or displayed severely reduced bone marrow expression of mRNAs encoding secondary and tertiary granule proteins. These included lactoferrin (LF), neutrophil collagenase (NC), neutrophil gelatinase (NG), neutrophil gelatinase-associated lipocalin (NGAL), cathelicidin B9/NGP (neutrophil granule protein), and murine cathelinlike peptide (MCLP/CNLP).14,15 C/EBPε-deficient mice also exhibited abnormalities in their eosinophils.3
Phenotypic and functional defects of neutrophils and eosinophils observed in C/EBPε-deficient mice closely paralleled those of neutrophil-specific granule deficiency (SGD), a rare congenital disorder in humans.3,14,15 Determination of the mutational status of the human C/EBPE locus in 2 patients with SGD revealed that it results from an autosomal recessive inheritance of frame-shift mutations in the C/EBPE gene.16-18Because of these numerous deficiencies and functional defects, SGD patients and C/EBPε-deficient mice are immunocompromised and acquire frequent bacterial infections, including infection fromPseudomonas aeruginosa and Staphylococcus aureus.
Together with in vitro studies of gene expression, these murine models aid in the identification of target genes for these transcription factors. A number of myeloid-specific genes contain functional C/EBP- and PU.1 binding sites in their promoters, making them potential targets for C/EBPα or C/EBPε. These include the primary granule proteins neutrophil elastase (NE), proteinase 3, and myeloperoxidase (MPO), the receptors for G-CSF, M-CSF, and GM-CSF, and the secondary granule protein LF.19-24 A number of these genes show reduced or absent expression in the knockout mice.25 26The absence of expression of a particular gene indicates that it may be a target gene directly activated by the transcription factor, but it is necessary to assess whether the gene is directly activated. Alternatively, the block in granulocytic differentiation observed in the knockout mouse models may result in an insufficient number of cells that express the gene in question. On the other hand, ectopic expression of the transcription factor(s) to induce target gene expression may promote differentiation in myeloid cell lines that, in turn, results in the indirect regulation of the “target” gene. In this report, the hypothesis that C/EBPε is essential and sufficient for the expression of neutrophil and eosinophil secondary granule genes was tested. Using a combination of studies on cells from the knockout mice and induction of myeloid-specific gene expression in a nonhematopoietic murine fibroblast cell line, it was demonstrated that C/EBPε is a key transcriptional regulator of neutrophil secondary granule genes. Additionally, C/EBPε cooperates with PU.1 and GATA-1 to activate eosinophil granule gene expression.
Materials and methods
C/EBPε−/− mice, C/EBPβ−/− mice, and PU.1−/− cell lines
Bone marrow cells were derived from wild-type or C/EBPε−/− mice from a 129SV × NIH Black Swiss background at 6 to 8 weeks of age.3 Bone marrow cells were derived from C/EBPβ−/− mice at 6 to 8 weeks of age.4 C/EBPε−/− mice were kindly provided by K. Xanthopolous and J. Lekstrom-Himes (Aurora Biosciences, San Diego, CA; and National Institutes of Health, Bethesda, MD; respectively), and C/EBPβ−/− mice were kindly provided by Shizuo Akira (Department of Biochemistry, Hyogo School of Medicine, Japan). The PU.1−/− myeloid cell line and the PU.1 and M-CSF receptor–transduced derivatives were prepared as described previously.9
Transfections and generation of stable cell lines
NIH 3T3 cells were maintained in Dulbecco modified Eagle medium (DMEM; Gibco/BRL, Gaithersburg, MD) containing 10% adult bovine serum (BS; Gemini BioProducts, Calabasas, CA). For transient transfections, approximately 3 × 105 NIH 3T3 cells were seeded per well of a 12-well plate. For each DNA combination, 3 μg plasmid DNA was mixed with 15 μL GenePORTER reagent (Gene Therapy Systems, San Diego, CA) in 1 mL OptiMEM reduced serum medium (Gibco/BRL) as directed by the manufacturer. One third of the DNA mixture was added per well of a 12-well plate (3 wells per DNA combination). After a 5-hour incubation, 2 mL culture medium was added. For stable transformation, approximately 0.5 × 106 NIH 3T3 cells were plated in a 35-mm well and were transfected with 3 μg plasmid (pMTCB6+, pMT-ε32, or pMT-α) using 10 μL GenePORTER reagent. At 48 hours after transfection, cells were trypsinized and plated into a 100-mm dish in the presence of 700 μg/mL G418 (Gibco/BRL). A polyclonal population of G418-resistant cells was obtained from each transfection. These clones were screened for inducible expression of either C/EBPε32 or C/EBPα by Western blot analysis. Expression vectors were induced by the addition of ZnSO4 (Sigma Chemical, St Louis, MO) to a final concentration of 0.1 mM. Transient transfections in 35-mm plates (6-well dishes) were performed as described for the stable cell lines but were harvested 24 to 48 hours after transfection.
Expression vectors and promoter–reporter gene constructs
The cDNA encoding human C/EBPε30 was subcloned from pcDNAI-C/EBPε30 into pcDNA3 (Invitrogen, Carlsbad, CA).27 The pcDNA3 expression vector containing the human C/EBPε32 isoform was a generous gift from Dr K. Xanthopoulos (Aurora, San Diego, CA). The pMSV-C/EBPα (rat) expression vector was kindly provided by Dr A. Friedman (Johns Hopkins University, Baltimore, MD) and the expression vectors pXM-GATA-128 and pMT2-FOG (friend of gata) (murine)29 were kind gifts from Dr S. Orkin (Harvard University, Boston, MA). The human major basic protein gene(hMBP) luciferase reporter construct containing sequence between positions nucleotide (nt) −117 to +47 of the hMBPP2 promoter region was described previously.30Construction of the zinc-inducible C/EBPε32 vector pMT-ε32 was described previously.31 To construct the zinc-inducible rat C/EBPα expression vector, a 1.1–kilobase pair (kbp) NcoI C/EBPα cDNA fragment was blunt-ended with Klenow and subcloned into EcoRV-cut pMT-CB6+ (kindly provided by F. Rauscher).32 The pCMVSPORT vectors expressing human C/EBPε32, C/EBPε30, and C/EBPα were generated as described previously.33
RNA isolation and analysis
Total RNA was isolated by lysis of cells in TRIzol reagent as described by the manufacturer (Gibco/BRL). For reverse transcription–polymerase chain reaction (RT-PCR) analysis, total RNA was treated with RNase-free DNaseI (Promega, Madison, WI) and synthesized into cDNA with Moloney murine leukemia virus (MMLV) reverse transcriptase in a 50 μL volume as described by the manufacturer (Gibco/BRL). For transiently transfected cells, the entire RNA sample was reverse transcribed; for stably transformed cells, 2 μg RNA was used. PCR was performed with 1 μL cDNA per reaction using HotStar Taq polymerase (Qiagen, Chatsworth, CA). After a 15-minute denaturing step at 94°C, reactions were 94°C for 30 seconds, 55°C, 56°C, or 58°C for 30 seconds, and 72°C for 30 seconds. Cycle numbers for each primer set are described in the figure legends. Products were electrophoresed on 2% agarose gels and blotted for Southern analysis as described previously.17 Primers used for PCR and Southern blot hybridization were described previously15 or are summarized in Table 1. Each primer pair used for RT-PCR was designed to span 1 or more introns; therefore, amplification of contaminating genomic DNA would result in a product much larger than expected for amplification of the cDNA. A region 1 probe (N-terminal transcriptional activation domain, more than 90% similar to murine) of the human C/EBPα gene was used to for Northern hybridization.34
Primers used for RT-PCR or amplification of probes for Northern analysis
| Gene name . | Forward primer . | Reverse primer . | Primer range, nt . | GenBank ID no. . |
|---|---|---|---|---|
| C/EBPβ | agaagacggtggacaagctg | acacgtgtgttgegtagtee | 751-1249 | X62600.1 mRNA |
| C/EBPδ | gagatgcagcagaagctggt | agcttctctcgcagtccagt | 967-1267 | NM_007679.1 mRNA |
| MBP MBP(QRT-PCR) | ggagccagctttgcaaacttg ttccctctacttctggctcttc | gtcaaagcattcagccctcc ctcatccatccatgggcttt | 197-1971 995-1710 | L46768.1 Genomic L46768.1 Genomic |
| EPX | catacatgaaggtggcatcg | tgaaaactccccatttctgc | 1658-2106 | NM_007946.1 Genomic |
| NGAL | gaaaccatggccctgagtgtc | agccacactcaccacccattc | 841-3711 | X81627.1 Genomic |
| B9/NGP | gaagacccctcttccgcctg | ggtatcctctcgactgcaatc | 190-431 | L37297mRNA NW_000359 Genomic |
| NC/MMP8 | cctgaccttcaccgagate | gccattgatatcatcttgagg | 440-773 | Y13342mRNA NW_000350 Genomic |
| MCLP/CNLP | atgcagttccagagggacgtc | gctctggctgaggtacaag | 46-2056 | AF035680Genomic |
| Gene name . | Forward primer . | Reverse primer . | Primer range, nt . | GenBank ID no. . |
|---|---|---|---|---|
| C/EBPβ | agaagacggtggacaagctg | acacgtgtgttgegtagtee | 751-1249 | X62600.1 mRNA |
| C/EBPδ | gagatgcagcagaagctggt | agcttctctcgcagtccagt | 967-1267 | NM_007679.1 mRNA |
| MBP MBP(QRT-PCR) | ggagccagctttgcaaacttg ttccctctacttctggctcttc | gtcaaagcattcagccctcc ctcatccatccatgggcttt | 197-1971 995-1710 | L46768.1 Genomic L46768.1 Genomic |
| EPX | catacatgaaggtggcatcg | tgaaaactccccatttctgc | 1658-2106 | NM_007946.1 Genomic |
| NGAL | gaaaccatggccctgagtgtc | agccacactcaccacccattc | 841-3711 | X81627.1 Genomic |
| B9/NGP | gaagacccctcttccgcctg | ggtatcctctcgactgcaatc | 190-431 | L37297mRNA NW_000359 Genomic |
| NC/MMP8 | cctgaccttcaccgagate | gccattgatatcatcttgagg | 440-773 | Y13342mRNA NW_000350 Genomic |
| MCLP/CNLP | atgcagttccagagggacgtc | gctctggctgaggtacaag | 46-2056 | AF035680Genomic |
Quantitative real-time–PCR (QRT-PCR) for major basic protein (MBP) expression was performed using a second primer set that spanned an intron, as described in Table 1. Triplicate reactions for each cDNA were set up as described in the previous paragraph, and SYBR Green I (Molecular Probes, Eugene, OR) was added at a 1:60 000 dilution. After denaturing the template 15 minutes at 95°C, a 4-step PCR reaction of 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds, and 80°C for 20 seconds was performed. At the last step, measurement of incorporated SYBR Green I was performed at 2°C below the empirically determined melting temperature for the PCR product. This melted all potential nonspecific products while maintaining the specific product and ensuring that nonspecific products were not detected. Gel electrophoresis confirmed that nonspecific amplification was negligible. The balance of the cDNAs was determined by quantifying the relative levels of 18S RNA in the samples. This was performed with a TaqMan probe (FAM-agcaggcgcgcaaattaccc-TAMRA) and amplification using TaqMan Universal PCR Master Mix (PE Biosystems, Foster City, CA). The nucleotide sequence of the PCR primers for 18S was 5′-aaacggctaccacatccaag-3′ (18S-F) and 5′-cctccaatggatcctcgtta-3′ (18S-R). The threshold cycle (Ct) for each was determined, and the expression levels of MBP were normalized to 18S. The fold change (FC) of the expression vector (v)–transfected samples compared with the empty vector (ev)–transfected control was determined by the following equation: FC = 2 − ((CtMBPv − Ct18Sv) − (CtMBPev − Cft18Sev)).
For Northern blot analysis, total RNA was electrophoresed through 1% agarose/formaldehyde gels and was transferred to Hybond N+ membranes (Amersham Life Sciences, Arlington Heights, IL). Probes were synthesized with a Strip-EZ random priming kit (Ambion, Austin, TX) with the incorporation of 5′-[α32P]-dATP (3000 Ci/mmol [111 000 GBq/mmol]; Dupont/NEN, Boston, MA). Blots were hybridized overnight in UltraHYB solution (Ambion) and were washed at a final stringency of 0.1 × SSC (standard saline citrate), 0.1% sodium dodecyl sulfate (SDS) at 68°C for 20 minutes. Blots were exposed to Kodak XO-Mat film with an intensifying screen (Eastman-Kodak, Rochester, NY). The probes used for Northern blot analysis are described in the first paragraph of this section or in Table 1.
Protein isolation and analysis
For the stable cell lines, total cell protein was prepared and Western blot analysis was performed as described previously.17 For transient transfections, total cell protein was prepared from the organic phase of the Trizol lysate as described by the manufacturer (Gibco/BRL). The antiserum against the amino-terminal half of C/EBPε was described previously.35 Commercially available antibodies against C/EBPε (C-22), C/EBPα (14AA), C/EBPβ (C19), and C/EBPδ (M17) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). All antibodies were used at a final concentration of 0.2 μg/mL. The primary antibody was detected with a donkey-antirabbit horseradish peroxidase (HRP) conjugate (Amersham Life Sciences) diluted 1:5000. Antigen–antibody complexes were visualized using the Supersignal chemiluminescence kit (Pierce, Rockford, IL) and exposure to Kodak XO-Mat film.
Electrophoresis mobility shift assays
COS-1 cells were transfected with empty or PU.1 expression vector as described in “Transfections and generation of stable cell lines.” For total cell extract, cells were washed with phosphate-buffered saline (PBS) and were lysed in NP-40 lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% NP-40) containing Complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). Double-stranded oligonucleotides were end labeled with32P-γ adenosine triphosphate (ATP; Dupont/NEN) using T4 polynucleotide kinase (Invitrogen) as described by the manufacturer. Oligonucleotide sequences (sense-strand) were: (1) wild-type consensus PU.1 5′-gggctgcttgaggaagtataagaat-3′; (2) mutant consensus PU.1 5′-gggctgcttgagagagtataagaat-3′; (3) wild-type MBP PU.1 5′-tctccctgggggaagttcctccaaggcc-3′; and (4) mutant MBP PU.1 5′-tctccctgggcaaagtttgtccaaggcc-3′. Core sequences are highlighted as underlined text. Electrophoresis mobility shift assays (EMSAs) were performed as described previously.34
Results
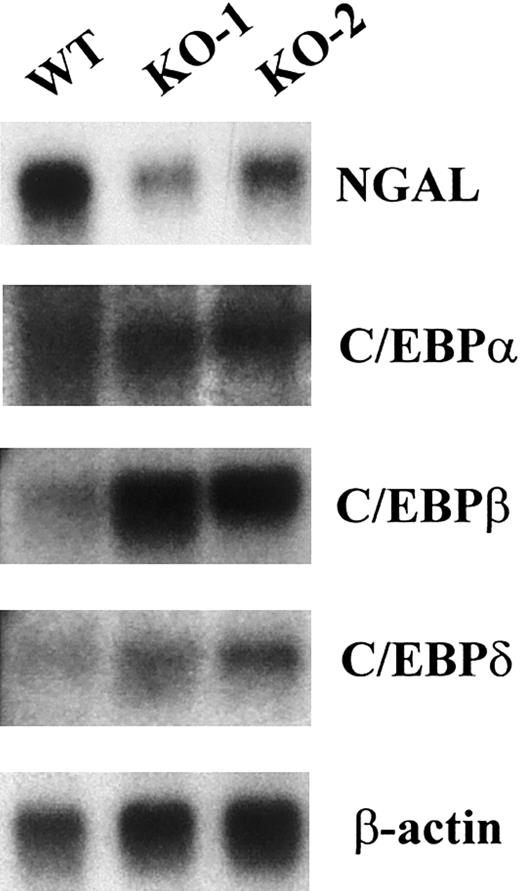
Absence of secondary granule gene mRNA expression in the C/EBPε-deficient mouse occurred in the presence of other C/EBP family members. The expression of secondary granule gene mRNAs in the bone marrow RNA is severely reduced (NC) or absent (LF, NG, MCLP, and B9) in the C/EBPε-deficient mouse.15 In addition, NGAL levels were substantially reduced (Figure 1). Numerous in vitro studies indicate that the different C/EBP family members have the ability to bind the same DNA sequences and to activate transcription. This suggests the possibility of a functional redundancy among family members. To determine whether levels of expression of other members were altered in the bone marrow of C/EBPε-null mice, we performed Northern blot analysis for C/EBPα, C/EBPβ, and C/EBPδ. In each case, these family members were expressed at levels equal to or higher than (for C/EBPβ) those found in the wild-type mouse (Figure1). These results indicate that the presence of other family members is not sufficient to compensate for the loss of C/EBPε. This supports the hypothesis that C/EBPε, and not the other C/EBP family members, is required for the expression of those secondary and tertiary granule genes that are missing in humans and mice lacking functional C/EBPε.
Bone marrow of C/EBPε-deficient mice expresses mRNAs encoding C/EBPα, C/EBPβ, and C/EBPδ but lacks mRNA expression of neutrophil secondary granule proteins.
Northern blot of total RNA (5 μg) from one wild-type (WT) and 2 C/EBPε-null (KO-1 and KO-2) mice. The blot was hybridized sequentially with probes for NGAL, C/EBPα, C/EBPβ, C/EBPδ, and β-actin.
Bone marrow of C/EBPε-deficient mice expresses mRNAs encoding C/EBPα, C/EBPβ, and C/EBPδ but lacks mRNA expression of neutrophil secondary granule proteins.
Northern blot of total RNA (5 μg) from one wild-type (WT) and 2 C/EBPε-null (KO-1 and KO-2) mice. The blot was hybridized sequentially with probes for NGAL, C/EBPα, C/EBPβ, C/EBPδ, and β-actin.
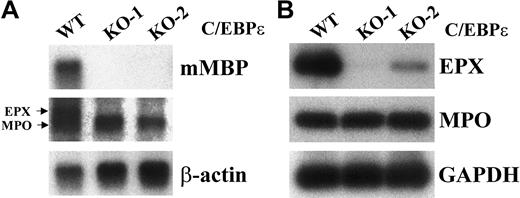
The bone marrow of C/EBPε-deficient mice lacks mRNA expression for eosinophil granule proteins. The severe reduction of eosinophils in the bone marrow and peripheral blood of C/EBPε-deficient mice3 and the lack of eosinophil granule protein expression in SGD patients36 suggested that eosinophil granule gene expression may be impaired in the C/EBPε-deficient mice. Northern blot analysis of total bone marrow RNA from wild-type and C/EBPε-deficient mice revealed an absence of MBP and eosinophil peroxidase (EPX) mRNA expression, but normal expression of myeloperoxidase (MPO) mRNA (Figure 2A). Because the MPO and EPX mRNAs are similar in size and sequence and are detected by the same probe in Northern hybridizations, RT-PCR analysis of the samples was performed with primers specific for either MPO or EPX. As observed by Northern blot analysis, the expression of EPX mRNA was severely reduced or absent in the C/EBPε-deficient mice, whereas MPO expression was not (Figure 2B). The expression of MBP and EPX was not reduced in the bone marrow of C/EBPβ−/− mice (data not shown). These data indicate that C/EBPε is required for eosinophil granule gene expression.
Bone marrow of C/EBPε-deficient mice lacks mRNA expression for the eosinophil granule proteins MBP and EPX.
(A) Northern blot analysis of total RNA (5 μg) from one wild-type (WT) and 2 C/EBPε-null (KO-1 and KO-2) mice. The blot was hybridized sequentially with probes MBP, EPX, and β-actin. (B) RT-PCR analysis of MPO and EPX gene expression in the total bone marrow RNA from one wild-type and 2 C/EBPε-null mice. PCR of cDNAs was performed with primers specific for either MPO or EPX (35 cycles). Products were electrophoresed on a 2% agarose gel, Southern blotted, and hybridized with radiolabeled, gene-specific oligonucleotides (Table 1).
Bone marrow of C/EBPε-deficient mice lacks mRNA expression for the eosinophil granule proteins MBP and EPX.
(A) Northern blot analysis of total RNA (5 μg) from one wild-type (WT) and 2 C/EBPε-null (KO-1 and KO-2) mice. The blot was hybridized sequentially with probes MBP, EPX, and β-actin. (B) RT-PCR analysis of MPO and EPX gene expression in the total bone marrow RNA from one wild-type and 2 C/EBPε-null mice. PCR of cDNAs was performed with primers specific for either MPO or EPX (35 cycles). Products were electrophoresed on a 2% agarose gel, Southern blotted, and hybridized with radiolabeled, gene-specific oligonucleotides (Table 1).
Generation of stable NIH 3T3 cell lines capable of zinc-inducible expression of C/EBPε32 and C/EBPα
The overexpression of C/EBPα or C/EBPε in several leukemia cell lines promotes granulocytic differentiation.31,37This, in turn, leads to induction of a number of myeloid-specific genes that may or may not be direct targets of the transcription factors. In addition, hematopoietic cell lines express transcription factors important in the regulation of myeloid-specific genes, thus making it difficult to discern the contribution of these other transcription factors in cooperative activation. Myeloid-specific gene expression was induced in avian fibroblasts by the coexpression of exogenous NFM (nuclear factor myeloid, also known as avian C/EBPβ) and MYB.38 39 This occurs in the absence of differentiation and eliminates the induction of genes that occurs during this process.
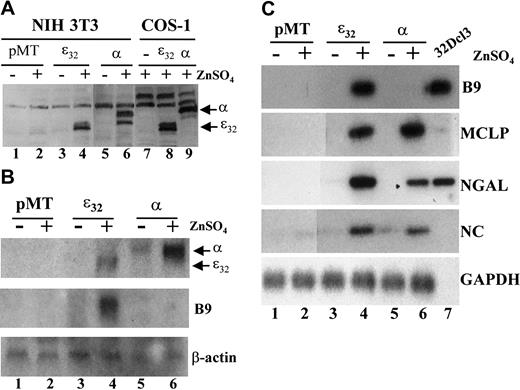
We reasoned that a similar phenomenon would occur in a mammalian fibroblast cell line such as NIH 3T3. To determine whether secondary granule genes are direct targets of C/EBPε or C/EBPα in a mammalian system, we established cells with stable, inducible expression of each transcription factor (Figure 3). Levels of C/EBPε and C/EBPα protein were undetectable in the empty vector (pMT) cells in the presence or absence of zinc induction (Figure 3A, lanes 1-2). In contrast, expression of either the C/EBPε32 or C/EBPα protein was readily detected after 24 hours of zinc treatment in the pMT-ε32 and pMT-α stably transformed cell lines (Figure 3A, lanes 3-6). Northern blot analysis indicated that comparable levels of C/EBPε and C/EBPα mRNA were expressed on incubation with ZnSO4 (Figure 3B). Expression of C/EBPα mRNA levels appeared slightly higher than C/EBPε, with some leaky expression detected in the absence of zinc induction (Figure 3B, lanes 5-6). These results indicate that the stable NIH 3T3 cell lines express comparable levels of C/EBPε or C/EBPα in an inducible fashion.
Induction of neutrophil secondary granule protein mRNAs in stably transformed NIH 3T3 cell lines expressing inducible human C/EBPε32 and rat C/EBPα.
(A) Western blot analysis of NIH 3T3 cell lines stably transformed with either empty (pMT), C/EBPε32, or C/EBPα containing zinc-inducible expression vector. Expression of C/EBPε32and C/EBPα was determined for cells incubated in the absence (−) or presence (+) of 0.1 mM ZnSO4 for 24 hours (lanes 1-6). COS-1 cells transfected with the same expression vectors and incubated with 0.1 mM ZnSO4 for 24 hours were included as negative and positive controls (lanes 7-9). (B) Northern blot analysis of total RNA prepared from the stable NIH 3T3 cell lines incubated in the absence (−) or presence (+) of 0.1 mM ZnSO4 for 48 hours. The blot was hybridized sequentially with probes for C/EBPε and C/EBPα, Β9, and β-actin. (C) Stable NIH 3T3 cell lines were incubated in the absence (−) or presence (+) of 0.1 mM ZnSO4 for 48 hours. Total RNA was harvested and subjected to RT-PCR analysis for the secondary granule genes B9 (33 cycles), MCLP (35 cycles), NGAL (33 cycles), NC (33 cycles), and the control gene GAPDH (25 cycles). Total RNA from undifferentiated 32Dcl3 cells was included as a positive control, but GAPDH expression was not determined (lane 7).
Induction of neutrophil secondary granule protein mRNAs in stably transformed NIH 3T3 cell lines expressing inducible human C/EBPε32 and rat C/EBPα.
(A) Western blot analysis of NIH 3T3 cell lines stably transformed with either empty (pMT), C/EBPε32, or C/EBPα containing zinc-inducible expression vector. Expression of C/EBPε32and C/EBPα was determined for cells incubated in the absence (−) or presence (+) of 0.1 mM ZnSO4 for 24 hours (lanes 1-6). COS-1 cells transfected with the same expression vectors and incubated with 0.1 mM ZnSO4 for 24 hours were included as negative and positive controls (lanes 7-9). (B) Northern blot analysis of total RNA prepared from the stable NIH 3T3 cell lines incubated in the absence (−) or presence (+) of 0.1 mM ZnSO4 for 48 hours. The blot was hybridized sequentially with probes for C/EBPε and C/EBPα, Β9, and β-actin. (C) Stable NIH 3T3 cell lines were incubated in the absence (−) or presence (+) of 0.1 mM ZnSO4 for 48 hours. Total RNA was harvested and subjected to RT-PCR analysis for the secondary granule genes B9 (33 cycles), MCLP (35 cycles), NGAL (33 cycles), NC (33 cycles), and the control gene GAPDH (25 cycles). Total RNA from undifferentiated 32Dcl3 cells was included as a positive control, but GAPDH expression was not determined (lane 7).
Activation of neutrophil secondary granule gene expression in NIH 3T3
To determine the ability of C/EBPε and C/EBPα to activate gene expression of secondary granule genes, the stable cell lines were incubated in the absence or presence of ZnSO4 for 48 hours. Total RNA was subjected to RT-PCR analysis for genes that are deficient in the bone marrow of the C/EBPε-null mice. Both C/EBPε and C/EBPα induced expression of MCLP, NGAL, and NC (Figure 3C). Interestingly, at 30 to 33 cycles, B9 was detected only in the C/EBPε-expressing cell line (Figure 3C); however, at 35 cycles, low levels of B9 were observed in the C/EBPα stable line (data not shown). The induction of B9 by C/EBPε was detected by Northern blot analysis as well (Figure 3B). These results demonstrated that using both RT-PCR and Northern assays, we could detect the expression of neutrophil-specific genes in a nonhematopoietic cell system.
Differential induction of neutrophil secondary granule genes mediated by C/EBP family members
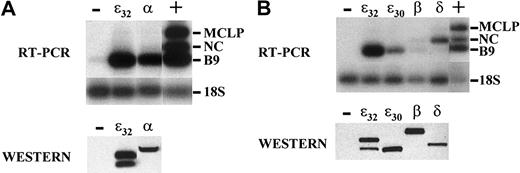
The C/EBP family members are all capable of binding the same DNA site and activating the same promoters in promoter–reporter assays. To compare the ability of the different C/EBP family members to activate secondary granule gene expression in NIH 3T3, cells were transiently transfected with expression vectors either for C/EBPε32, C/EBPε30, C/EBPα, C/EBPβ, or C/EBPδ. RT-PCR analysis for the genes MCLP, NC, or B9 was performed (Figure 4, top panels). Western blot analysis of the proteins extracted from the organic phase of the TRIzol lysate prepared from the transfected cells was performed to demonstrate that the respective proteins were expressed (Figure 4, lower panels). A cDNA prepared from 32Dcl3 cells served as a positive control (+) and a cDNA from empty vector transfected cells was used as a negative control (−).
Differential induction of neutrophil secondary granule genes mediated by C/EBP family members.
(A) NIH 3T3 cells were transiently transfected with 3.0 μg empty expression vector (−), human C/EBPε32, or human C/EBPα. RT-PCR analysis for the genes MCLP, NC, or B9 was performed. A cDNA prepared from 32Dcl3 cells served as a positive control (+). Products were analyzed on 2% agarose gels, Southern blotted, and hybridized with radiolabeled, gene-specific probes (Table 1). The lower panel represents a Western blot of the protein extracted from the organic phase of the TRIzol lysate prepared from the transfected cells. (B) NIH 3T3 cells were transfected with expression vectors for human C/EBPε32, C/EBPε30, C/EBPβ, and C/EBPδ and were analyzed for expression as described above. The lower panel represents a Western blot simultaneously probed for each family member of the protein extracted from the organic phase of the TRIzol lysate prepared from the transfected cells.
Differential induction of neutrophil secondary granule genes mediated by C/EBP family members.
(A) NIH 3T3 cells were transiently transfected with 3.0 μg empty expression vector (−), human C/EBPε32, or human C/EBPα. RT-PCR analysis for the genes MCLP, NC, or B9 was performed. A cDNA prepared from 32Dcl3 cells served as a positive control (+). Products were analyzed on 2% agarose gels, Southern blotted, and hybridized with radiolabeled, gene-specific probes (Table 1). The lower panel represents a Western blot of the protein extracted from the organic phase of the TRIzol lysate prepared from the transfected cells. (B) NIH 3T3 cells were transfected with expression vectors for human C/EBPε32, C/EBPε30, C/EBPβ, and C/EBPδ and were analyzed for expression as described above. The lower panel represents a Western blot simultaneously probed for each family member of the protein extracted from the organic phase of the TRIzol lysate prepared from the transfected cells.
Both C/EBPε and C/EBPα efficiently induced the expression of B9 (Figure 4A) in a dose-response fashion (data not shown) and weakly induced MCLP and NC expression (Figure 4A and data not shown). The expression of MCLP and NC was not as easily detected in the transient transfections as in the stable transformants (Figure 3), probably because of the lower efficiency of transient transfections. The p30 isoform of C/EBPε, which activates reporter gene expression several-fold less efficiently than the p32 isoform,40induced B9 expression less efficiently (Figure 4B). In contrast, C/EBPβ was unable to induce significantly the expression of any of the secondary granule genes (Figure 4B). Interestingly, C/EBPδ was unable to induce either B9 or MCLP, but it induced NC gene expression significantly better than the other C/EBP family members (Figure 4B). All family members induced NGAL expression (data not shown). These results demonstrate differences in the ability of the C/EBP family members to activate particular target genes, suggesting a lack of functional redundancy.
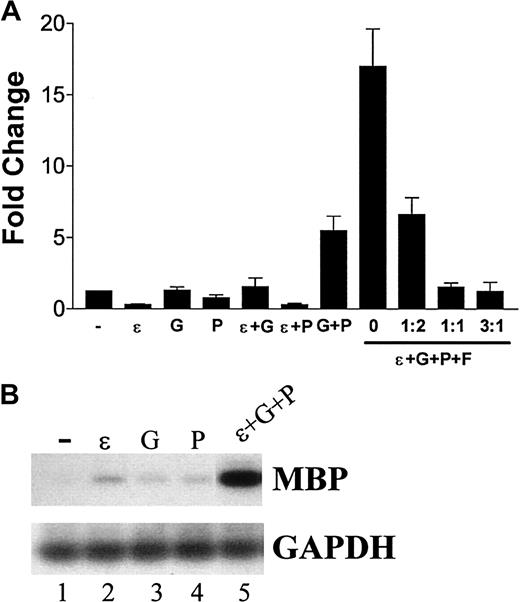
Cooperative transcriptional activation of MBP gene expression in NIH 3T3 cells by C/EBPε32, GATA-1, and PU.1
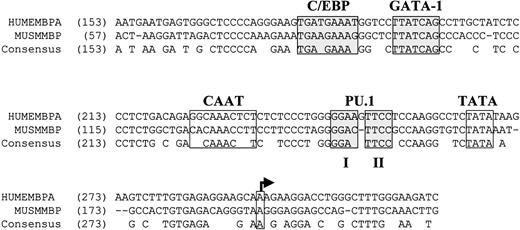
Overexpression of C/EBPε in NIH 3T3 cells alone was unable to induce MBP expression efficiently (Figure5). Previous studies demonstrated that GATA-1 and C/EBPβ synergistically activate the human MBP-P2 promoter.41 Because MBP expression is deficient in the bone marrow cells of the C/EBPε−/− mouse (Figure 2) but not in the C/EBPβ−/− mouse (data not shown), we cotransfected C/EBPε with GATA-1. A slight increase in MBP levels over either C/EBPε or GATA-1 alone was observed (Figure 5A). We hypothesized that an additional factor was required for the induction of high levels of MBP expression. Alignment of the nucleotide sequence of the murine and human MBP promoter regions revealed conservation of the C/EBP and GATA-1 sites (Figure6). In addition, 2 GGAA core sequence motifs (1 and 2) in a purine-rich region of the promoters indicated a potential PU.1 binding site (Figure 6). The PU.1 core element 2 was perfectly conserved between the murine and human promoters, but core element 1 was not. This suggested the possibility that PU.1 may be involved in the regulation of MBP gene expression.
Cooperative transcriptional activation of MBP gene expression in NIH 3T3 cells by C/EBPε32, GATA-1, and PU.1.
(A) NIH 3T3 cells were seeded in 6-well plates at approximately 70% confluence and were transfected with the combinations of expression vectors indicated below the graph. The total amount of expression vector was kept equal in each transfection with the addition of empty vector. The combinations of expression vectors and the ratio of FOG expression vector to GATA-1 expression vector are indicated on the x-axis. At 24 hours after transfection, the cells were lysed in TRIzol reagent, total RNA was isolated and treated with DNaseI, and cDNA was synthesized. Quantitative RT-PCR using SYBR Green was performed in triplicate for each sample. The levels of MBP expression were normalized to 18S. Results are presented as fold changes (± SD) compared with cells transfected with empty vector. (B) Southern blot analysis of the amplification products from single (−, ε, G, and P) or 3 expression vector (ε+G+P)–transfected NIH 3T3 cells. PCR was performed for MBP (33 cycles) and GAPDH (25 cycles). − indicates empty; (ε), C/EBPε; (P), PU.1; (G), GATA-1; (F), FOG.
Cooperative transcriptional activation of MBP gene expression in NIH 3T3 cells by C/EBPε32, GATA-1, and PU.1.
(A) NIH 3T3 cells were seeded in 6-well plates at approximately 70% confluence and were transfected with the combinations of expression vectors indicated below the graph. The total amount of expression vector was kept equal in each transfection with the addition of empty vector. The combinations of expression vectors and the ratio of FOG expression vector to GATA-1 expression vector are indicated on the x-axis. At 24 hours after transfection, the cells were lysed in TRIzol reagent, total RNA was isolated and treated with DNaseI, and cDNA was synthesized. Quantitative RT-PCR using SYBR Green was performed in triplicate for each sample. The levels of MBP expression were normalized to 18S. Results are presented as fold changes (± SD) compared with cells transfected with empty vector. (B) Southern blot analysis of the amplification products from single (−, ε, G, and P) or 3 expression vector (ε+G+P)–transfected NIH 3T3 cells. PCR was performed for MBP (33 cycles) and GAPDH (25 cycles). − indicates empty; (ε), C/EBPε; (P), PU.1; (G), GATA-1; (F), FOG.
Conservation of the C/EBP, GATA-1, and PU.1 binding sites in the human and murine MBP-P2 promoter regions.
Genomic DNA sequences for the human and murine MBP promoter regions (GenBank accession numbers M34462 and L46768, respectively) were aligned using the Align X feature of the Vector NTI 6.0 software program (Informax, Bethesda, MD). Nucleotide numbers are indicated in parentheses. Previously characterized binding sites for C/EBP and GATA-1 are shaded.30 41 The putative TATA box and transcription start site (arrow) are indicated. Conserved core consensus sequences 1 and 2 for PU.1 (GGAA) are shaded. Gaps in the alignment are denoted by −. Conserved nucleotides are indicated by the consensus sequence.
Conservation of the C/EBP, GATA-1, and PU.1 binding sites in the human and murine MBP-P2 promoter regions.
Genomic DNA sequences for the human and murine MBP promoter regions (GenBank accession numbers M34462 and L46768, respectively) were aligned using the Align X feature of the Vector NTI 6.0 software program (Informax, Bethesda, MD). Nucleotide numbers are indicated in parentheses. Previously characterized binding sites for C/EBP and GATA-1 are shaded.30 41 The putative TATA box and transcription start site (arrow) are indicated. Conserved core consensus sequences 1 and 2 for PU.1 (GGAA) are shaded. Gaps in the alignment are denoted by −. Conserved nucleotides are indicated by the consensus sequence.
To determine whether PU.1 cooperates with C/EBPε and GATA-1 in the activation of MBP expression, combinations of the 3 expression vectors were transfected into NIH 3T3 cells and the induction of MBP expression was monitored by RT-PCR. PU.1 alone was unable to induce gene expression efficiently (Figure 5A-B). However, a striking induction was observed when C/EBPε, GATA-1, and PU.1 were cotransfected (Figure 5A-B). FOG, an inhibitor of MBP gene expression,41 repressed this induction in a dose-response fashion (Figure 5A). To determine whether this occurred with the human promoter, a MBP promoter-luciferase construct [pMBP(-117)–LUC]30 was cotransfected with the same combinations of vectors. The induction of luciferase activity reflected the results seen with the induction of the endogenous murine gene (Figure 7). Taken together, these data indicated that the MBP promoter required a combination of C/EBPε, GATA-1, and PU.1 for efficient activation.
Cooperative transcriptional activation of the humanMBP promoter by C/EBPε32, GATA-1, and PU.1.
NIH 3T3 cells were cotransfected with combinations of expression vectors indicated on the x-axis. In addition, a human MBP-P2 promoter-luciferase construct [pMBP(-117)–LUC] was included in each transfection.30 At 24 hours after transfection, lysates were prepared, and luciferase activity (RLU) was measured and normalized to Renilla luciferase to control for transfection efficiency. The graph represents the average (± SD) of 2 experiments performed in triplicate.
Cooperative transcriptional activation of the humanMBP promoter by C/EBPε32, GATA-1, and PU.1.
NIH 3T3 cells were cotransfected with combinations of expression vectors indicated on the x-axis. In addition, a human MBP-P2 promoter-luciferase construct [pMBP(-117)–LUC] was included in each transfection.30 At 24 hours after transfection, lysates were prepared, and luciferase activity (RLU) was measured and normalized to Renilla luciferase to control for transfection efficiency. The graph represents the average (± SD) of 2 experiments performed in triplicate.
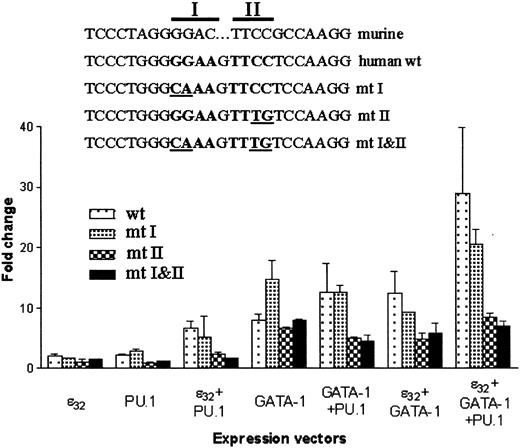
Mutation of the conserved PU.1 site in the human MBP-P2 promoter abolishes cooperative transcriptional activation with PU.1
To determine which core element in the conserved PU.1 site was required for the cooperative activation of the human MBPpromoter, either core 1 (mutant [mt] 1), core 2 (mt 2), or both (mt 1-2) core sequences were mutated in the reporter construct pMBP(-117)–LUC. Wild-type or mutant reporter constructs were cotransfected with expression vectors for C/EBPε, GATA-1, or PU.1 alone or in various combinations. The mt 2 and mt 1-2 reporter constructs lost response to PU.1, but not C/EBPε or GATA-1, compared with the wild-type or mt 1 reporter constructs (Figure8). In addition, the synergistic activation by all 3 factors was abrogated in the mt 2 and mt 1-2 reporter constructs (Figure 8). These results indicate that the highly conserved core 2 sequence is required for activation of theMBP promoter by PU.1. In transfections with expression vectors for C/EBPε or GATA-1, the mt 2 and mt 1-2 reporter constructs activated less efficiently. This may be attributed to the loss of binding by ETS-like factors found in NIH 3T3 cells to the mutated promoters whereas their binding would occur in the wild-type and mt 1 promoter constructs.
Mutation of the conserved PU.1 site in the human MBP-P2 promoter abolishes cooperation with PU.1.
Mutations were introduced to the PU.1 core elements 1 and 2 of the pMBP(−117)–luciferase construct, as indicated in the figure. These were cotransfected with various combinations of the expression vectors for PU.1, GATA-1, and C/EBPε32. At 24 hours after transfection, lysates were prepared and luciferase activity (RLU) was measured and normalized to renilla luciferase to control for transfection efficiency. The graph represents the average (± SD) of 2 experiments performed in triplicate.
Mutation of the conserved PU.1 site in the human MBP-P2 promoter abolishes cooperation with PU.1.
Mutations were introduced to the PU.1 core elements 1 and 2 of the pMBP(−117)–luciferase construct, as indicated in the figure. These were cotransfected with various combinations of the expression vectors for PU.1, GATA-1, and C/EBPε32. At 24 hours after transfection, lysates were prepared and luciferase activity (RLU) was measured and normalized to renilla luciferase to control for transfection efficiency. The graph represents the average (± SD) of 2 experiments performed in triplicate.
The loss of response to PU.1 was predicted to result from the loss of PU.1 binding to the promoter. Electrophoretic mobility shift assay (EMSA) revealed that PU.1 expressed in COS-1 cells was able to bind to its consensus site and that this binding was blocked by the addition of anti-PU.1 antibody (Figure 9, left panel). The binding of PU.1 to the consensus site was competed by the unlabeled wild-type consensus and the wild-type MBP PU.1 site, but not when the PU.1 sites were mutated in either of these oligonucleotides (Figure 9, middle panel). Finally, PU.1 binding to the wild-type MBP PU.1 site was detected in COS-1 extracts expressing PU.1, and this binding was blocked by the addition of anti-PU.1 antibody (Figure 9, right panel). These data demonstrate the binding of PU.1 to the predicted site in the MBP promoter and, together with the reporter construct data, indicate that the loss of promoter activation by PU.1 results from a lack of PU.1 binding to the mutated core 2 site.
PU.1 binds the predicted site in the MBP-P2 promoter.
COS-1 cells were transfected with empty (−) or PU.1 (+) expression vector, and total cell extracts were prepared 24 hours after transfection. Wild-type consensus PU.1 (left and middle panels; 5′-gggctgcttgaggaagtataagaat-3′) or wild-type MBP PU.1 (right panel; 5′-tctccctgggggaagttcctccaaggcc-3′) double-stranded oligonucleotides were end-labeled with [32P]-γ adenosine triphosphate (ATP). Labeled oligonucleotides (0.02 pmol) were incubated with 5 μg total cellular extract in the presence or absence of anti-PU.1 (1 μg) antibody (left and right panels) or unlabeled (0.2 pmol) wild-type or mutant oligonucleotides (consensus, 5′-gggctgcttgagagagtataagaat-3′; MBP PU.1, 5′-tctccctgggcaaagtttgtccaaggcc-3′). Arrows at the right of each panel indicate the positions of the complexes containing PU.1. Wt indicates wild-type; Mt, mutant; CON, consensus PU.1 binding site; MBP, major basic protein promoter PU.1 binding site; Ab, anti-PU.1 antibody (sc-352; Santa Cruz Biotechnology).
PU.1 binds the predicted site in the MBP-P2 promoter.
COS-1 cells were transfected with empty (−) or PU.1 (+) expression vector, and total cell extracts were prepared 24 hours after transfection. Wild-type consensus PU.1 (left and middle panels; 5′-gggctgcttgaggaagtataagaat-3′) or wild-type MBP PU.1 (right panel; 5′-tctccctgggggaagttcctccaaggcc-3′) double-stranded oligonucleotides were end-labeled with [32P]-γ adenosine triphosphate (ATP). Labeled oligonucleotides (0.02 pmol) were incubated with 5 μg total cellular extract in the presence or absence of anti-PU.1 (1 μg) antibody (left and right panels) or unlabeled (0.2 pmol) wild-type or mutant oligonucleotides (consensus, 5′-gggctgcttgagagagtataagaat-3′; MBP PU.1, 5′-tctccctgggcaaagtttgtccaaggcc-3′). Arrows at the right of each panel indicate the positions of the complexes containing PU.1. Wt indicates wild-type; Mt, mutant; CON, consensus PU.1 binding site; MBP, major basic protein promoter PU.1 binding site; Ab, anti-PU.1 antibody (sc-352; Santa Cruz Biotechnology).
Eosinophil granule gene expression is absent in PU.1-deficient cells
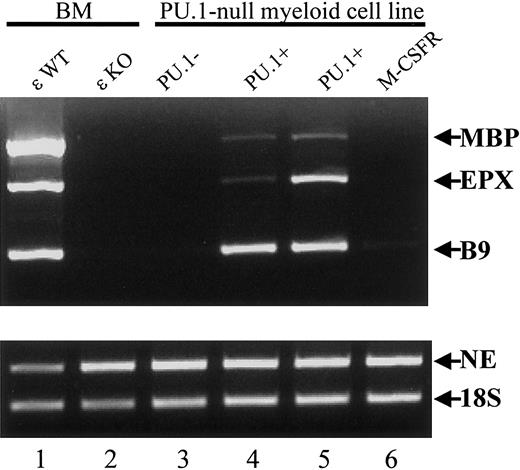
To determine the role of PU.1 in the expression of eosinophil granule genes, levels of MBP and EPX mRNA were determined in a myeloid cell line derived from the embryonic liver cells of PU.1-deficient mice.8,9 Like the bone marrow of C/EBPε-deficient mice, these cells lacked expression of neutrophil secondary granule mRNAs such as LF, NG,8 and B9 (Figure 10, lanes 1-3), but they expressed primary granule mRNAs including MPO and NE8 (Figure 10, lanes 1-3). Expression of the eosinophil granule mRNAs MBP and EPX was deficient in the PU.1-null cell line (Figure 10, lane 3). When PU.1 expression was reinstated by retroviral transduction, neutrophil9 and eosinophil granule gene expression was restored (Figure 10, lanes 4-5). This was not observed on the restoration of M-CSF receptor expression by retroviral transduction (Figure 10, lane 6). These results indicate that both C/EBPε and PU.1 are important for the expression of eosinophil granule proteins in the mouse.
Lack of eosinophil granule gene expression in a PU.1-null myeloid cell line derived from the embryonic liver of the PU.1−/− mouse.
The cDNAs were prepared from total RNA from the bone marrow of wild-type and C/EBP−/− mice, a PU.1-null cell line, 2 PU.1-null cell lines with restored PU.1 expression, and one with restored M-CSFR expression. Primers specific for the primary granule gene NE, the secondary granule gene B9, and the eosinophil granule genes MBP and EPX were used (35 cycles for all). Products were analyzed by electrophoresis through a 2% agarose gel and ethidium bromide staining. The 18S rRNA was amplified (15 cycles) as a control for the balance and integrity of the cDNAs.
Lack of eosinophil granule gene expression in a PU.1-null myeloid cell line derived from the embryonic liver of the PU.1−/− mouse.
The cDNAs were prepared from total RNA from the bone marrow of wild-type and C/EBP−/− mice, a PU.1-null cell line, 2 PU.1-null cell lines with restored PU.1 expression, and one with restored M-CSFR expression. Primers specific for the primary granule gene NE, the secondary granule gene B9, and the eosinophil granule genes MBP and EPX were used (35 cycles for all). Products were analyzed by electrophoresis through a 2% agarose gel and ethidium bromide staining. The 18S rRNA was amplified (15 cycles) as a control for the balance and integrity of the cDNAs.
Discussion
Studies on neutrophil secondary granule gene expression demonstrate an important role for the C/EBP family in granule gene transcriptional regulation. The LF promoter possesses a C/EBP binding site near its transcriptional start site that C/EBP proteins bind to and activate transcription.15,42The overlapping expression patterns of the C/EBP family members during granulopoiesis obscure the roles the different C/EBP family members play in the in vivo regulation of these genes. Neutrophil secondary granule gene expression is severely impaired in C/EBPα−/− and C/EBPε−/−mice,14,15,25 suggesting that they both may regulate expression of these genes. However, because the C/EBPα−/− mice show an earlier block in differentiation than the C/EBPε−/− mice and lack C/EBPε expression,25 we hypothesized that the loss of secondary granule gene expression resulted from a lack of C/EBPε. This hypothesis is supported by the presence of C/EBPα, C/EBPβ, and C/EBPδ at comparable or higher levels in the bone marrow cells of the C/EBPε−/− mice (Figure 1). The presence of these other family members and the lack of secondary granule gene expression indicate that they are unable to compensate functionally for the loss of C/EBPε in the bone marrow cells.
Granulocytes of C/EBPε−/− mice display a block in differentiation at the promyelocyte–myelocyte stage. The loss of secondary granule gene expression may be a consequence of this block. To determine whether endogenous expression of secondary granule genes could be activated in the absence of differentiation and therefore be direct targets of C/EBPs, we tested the ability of exogenous C/EBPε and C/EBPα to induce their expression in NIH 3T3 cells. The aim was to develop a system similar to that of quail fibroblasts in which ectopic expression of NFM, chicken C/EBPβ, and MYB induces the expression of myeloid-specific genes including mim-1 and lysozyme.38,39 Ectopic expression of C/EBP family members in a lymphoblastic B-cell line induced macrophage-specific genes in a lipopolysaccharide (LPS)–inducible fashion,43,44 and C/EBPα induced MPO and LF expression in a pro–B-cell line.45 In addition, ectopic expression of C/EBPε in QT6 cells induced mim-1 gene expression (A.F.G. and H.P.K., unpublished results, 1998). This demonstrates that C/EBP proteins induce potential target genes in heterologous mammalian cell types. NIH 3T3 cells have the capability of differentiating toward adipocytes, myocytes, and neuronal-like cells by the overexpression of transcription factors involved in those processes.46-49 Because of their potential to express genes normally restricted to more specialized cells, NIH 3T3 cells were selected for this study.
Stable and transient expression of the 2 C/EBP family members most critical for granulocytic differentiation, C/EBPα and C/EBPε, induced expression of most of the secondary granule genes tested. These included NC, NGAL, and MCLP (Figure 3). The data in this study are qualitative and do not indicate whether C/EBPε or C/EBPα is a better activator of secondary granule genes. Interestingly, we were unable to induce LF gene expression (data not shown). Forced expression of C/EBPα or C/EBPε in U937 or HL-60 cells induced LF expression.31,37 It is possible in NIH 3T3 cells that the presence or absence of another factor(s) is required for the induction of LF. Indeed, C/EBPα cooperatively activates the LF promoter–reporter construct with Sp1. CCAAT displacement protein (CDP/cut) recognizes a silencer element in the LF promoter, and down-regulation of the protein may be necessary for expression of LF in myeloid cell lines.42,50 With the exception of NGAL, the C/EBPβ protein was unable to activate secondary granule gene expression efficiently, whereas C/EBPδ induced NC and NGAL. Of note, although B9, MCLP, NG, and LF expression are absent in the C/EBPε−/− mice,15 NC and NGAL expression is reduced but not absent (Figure 1).15 This raises the question of the role of C/EBPδ, expressed throughout granulocytic differentiation,51 in regulating NGAL and NC expression. Determination of their levels in the bone marrow of C/EBPδ-deficient mice would address this.
If both C/EBPα and C/EBPε are capable of inducing the same secondary granule genes in the NIH 3T3 cells, why is C/EBPα unable to compensate for the lack of C/EBPε in the knockout mouse? Perhaps additional factors differentially interact with C/EBPs to enhance or repress the activation of target genes. Alternatively, C/EBPα expression may be too low in cells of the late granulocytic lineage in the C/EBPε−/− mouse and, hence, unable to activate significant levels of gene expression. Regarding the first possibility, activation of the MBP promoter by C/EBPε required cooperation with 2 additional transcription factors, GATA-1 and PU.1. Previous studies demonstrated that GATA-1 and C/EBPβ activated the human MBP promoter in a cooperative fashion.30 Abnormalities in the eosinophils of C/EBPβ−/− mice are not reported, and we did not detect aberrant expression of the eosinophil granule genes MBP and EPX in the bone marrow of C/EBPβ−/− mice (data not shown). However, eosinophils are absent in the C/EBPε-deficient mice, and MBP and EPX gene expression is absent (Figure 2).3 The C/EBPε and GATA-1 proteins weakly induced MBP expression, and this prompted a search for an additional factor. Examination of the promoter indicated the presence of a potential PU.1 site (Figure 6). The addition of PU.1 resulted in a dramatic up-regulation of endogenous MBP expression in NIH 3T3 (Figure 5) and activation of a promoter–reporter construct (Figure 7). Mutation of the PU.1 site eliminated this response, demonstrating that PU.1 binding to the promoter is required for activation of the gene (Figures 8 and 9). Most important, the absence of MBP and EPX gene expression was observed in a myeloid cell line derived from the embryonic liver of the PU.1−/−mouse. Restitution of PU.1 in these cells restored the expression of MBP and EPX (Figure 10). Consistent with our results, Du et al52 reported impaired eosinophilopoiesis in the PU.1 knockout mouse. Additionally, a role for PU.1 in regulating expression of the eosinophil-derived neurotoxin (EDN) gene was recently reported.53 The regulation of MBP and EPX by C/EBPε, PU.1, and GATA-1 substantiates the importance of these factors in eosinophil differentiation, function, and gene expression.
Both PU.1 and GATA-1 interact at the protein level and reciprocally inhibit the transcription- and differentiation-inducing functions of each other.54-58 These studies suggest this interaction is important for the decision of stem cells to differentiate down the erythroid versus the myeloid lineage. Our study demonstrates that the consequences of PU.1 and GATA-1, interacting with their adjacent binding sites on a promoter of a target gene, result in the activation rather than the inhibition of gene expression. Similarly, PU.1 and GATA-1 cooperatively stimulated activity of the mast cell–specific intronic enhancer of IL-4.59
In summary, we demonstrated the ability of C/EBP family members to differentially activate granulocytic secondary granule genes in a nonhematopoietic cell line. This suggests that, in addition to C/EBPε, other members such as C/EBPδ may regulate subsets of these genes. Using NIH 3T3 cells removes the complexity of differentiation-related effects on gene expression in much the same way as studies in the avian fibroblast system and provides an additional tool to study mammalian myeloid gene expression. It allows the selective addition of hematopoietic transcription factors and an analysis of their role in regulating the endogenous activation of a particular target gene in the context of its genomic location rather than taking it out of context in a reporter construct. Using this cell line and in vitro studies on the MBP promoter together with verification of expression in a murine knockout model, we demonstrated the importance of C/EBPε, GATA-1, and PU.1 in the expression of eosinophil granule proteins. Similar studies with other granulocyte-specific genes will allow further dissection of the molecular mechanisms involved in their regulation.
We thank Kleanthis Xanthopolous and Julie Lekstrom-Himes for sharing the C/EBPε knockout mice and Shizuo Akira for providing the C/EBPβ knockout mice. We thank Stuart Orkin for generously providing the GATA-1 and FOG expression vectors, Alan Friedman for providing the rat C/EBPα expression vector, and Frank Rauscher for providing the MT-CB6+ zinc-inducible vector.
Prepublished online as Blood First Edition Paper, December 19, 2002; DOI 10.1182/blood-2002-04-1039.
Supported by National Institutes of Health grants CA26038-20 (H.P.K.) and DK54938 (B.E.T.), the Joseph Troy Leukemia Fund, the Horn Foundation, the Lymphoma Research Foundation of America, and the C. and H. Koeffler Fund. A.F.G. is a recipient of a Lymphoma Research Foundation of America Fellowship. H.P.K. holds the Mark Goodson endowed chair for Cancer Research and is a member of the Jonsson Cancer Center.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Adrian F. Gombart, Division of Hematology/Oncology, Cedars-Sinai Medical Center, Davis Bldg 5019, Los Angeles, CA 90048; e-mail: gombarta@csmc.edu.

![Fig. 7. Cooperative transcriptional activation of the humanMBP promoter by C/EBPε32, GATA-1, and PU.1. / NIH 3T3 cells were cotransfected with combinations of expression vectors indicated on the x-axis. In addition, a human MBP-P2 promoter-luciferase construct [pMBP(-117)–LUC] was included in each transfection.30 At 24 hours after transfection, lysates were prepared, and luciferase activity (RLU) was measured and normalized to Renilla luciferase to control for transfection efficiency. The graph represents the average (± SD) of 2 experiments performed in triplicate.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/8/10.1182_blood-2002-04-1039/4/m_h80834157007.jpeg?Expires=1769946811&Signature=ZPHU3fdvEimNqwnTLFeW65DYgfsjvoL0GKsWuLmhgHG2~3ZV0zgc3MmnsZASh1okwYfSWrx0xePWh5zGpNMRluIOprNTYYW39sCRqihGBcsrj~bDyb1Et95576DSslxth19XLYyVjoTbl-Zyqh2PJas6JNx366JBI9jUlrPA1pthuoDCMLvcg-rh2fdcvqV4k~iVsYPvaqeAbOoSEtS~3fMbRPHefBQh~EQBXXjtj6PVhKtY-Ag2uu1Kq1iUPBy0kxPok-rINUD3q8Bmy2u~c8efnfu4nBTx5GdPvj-j6ol5gQ2HBfZewDzrP5WSQigT9T5gyGQLRRtNgzGy3vWP5g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 9. PU.1 binds the predicted site in the MBP-P2 promoter. / COS-1 cells were transfected with empty (−) or PU.1 (+) expression vector, and total cell extracts were prepared 24 hours after transfection. Wild-type consensus PU.1 (left and middle panels; 5′-gggctgcttgaggaagtataagaat-3′) or wild-type MBP PU.1 (right panel; 5′-tctccctgggggaagttcctccaaggcc-3′) double-stranded oligonucleotides were end-labeled with [32P]-γ adenosine triphosphate (ATP). Labeled oligonucleotides (0.02 pmol) were incubated with 5 μg total cellular extract in the presence or absence of anti-PU.1 (1 μg) antibody (left and right panels) or unlabeled (0.2 pmol) wild-type or mutant oligonucleotides (consensus, 5′-gggctgcttgagagagtataagaat-3′; MBP PU.1, 5′-tctccctgggcaaagtttgtccaaggcc-3′). Arrows at the right of each panel indicate the positions of the complexes containing PU.1. Wt indicates wild-type; Mt, mutant; CON, consensus PU.1 binding site; MBP, major basic protein promoter PU.1 binding site; Ab, anti-PU.1 antibody (sc-352; Santa Cruz Biotechnology).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/8/10.1182_blood-2002-04-1039/4/m_h80834157009.jpeg?Expires=1769946811&Signature=vOgHaxRAlu9~5pJwIgi9OBBcy-vRCc57TkV4-rUsvaPMVGAjpIgPTjvftxLZgmUdiuGfepKygI1aJzb9BAIDDk70bsTq73z5GSiJTSg47lFZKR1xN2-ZA2YSlNVzaIj72DBLJ3s3nYd0xZbxXZrO4rUosk6QkpbOyKxnTNj-rCy-TK2jDECwp3rIm0kfUm37IbKUYkif~PtfGD4GCTO1mRn0L-nJiF716qGklho4sL-iT7uS4FOvsmIPZY4xw82ZYdTVto3AVfd1pLMwiUM3~LlbJAd3frRY~v4JPizR-u5zMNg-7WzEkACtDqllSR-wEXDwFhqjD9ZmAx7zzj3zPA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal