Abstract
Chronic lymphocytic leukemia (CLL) comprises 2 subsets, distinguished by expression of unmutated or mutated VHgenes, with the former having a worse prognosis. Biased usage of the V1-69 gene is found in unmutated cases and is combined with selected D gene segments and JH6. It is controversial whether this is a CLL-associated feature or mirrors the normal B-cell pattern. Since CLL is a disease of the elderly, where changes in the B-cell repertoire may occur, we have analyzedV1-69 usage in the elderly (older than 75 years) population. Using monoclonal antibody (MoAb) G6, specific for 51p1-related V1-69 alleles, we found no increased expression with age. In 51p1-encoded immunoglobulin M (IgM), complementarity-determining region 3 (CDR3) length and frequency of D and JH genes were similar to those in the healthy young and distinct from those in CLL. These findings support the concept that CLL arises from B cells driven by antigen/superantigen and is not a stochastic event in the elderly B-cell population.
Introduction
Analysis of morphology, immunophenotype, and gene expression of cases of chronic lymphocytic leukemia (CLL) has indicated considerable disease homogeneity.1,2 However, investigation of the clonal immunoglobulin V gene sequences has revealed 2 subsets, derived from B cells either before or after somatic mutation.3-6 Since there is a powerful prognostic difference between the 2 subsets, with the unmutated being worse,5-7 insight into the pathogenesis of each may be relevant for understanding malignant behavior.
CLL was known to show biased usage of the V1-69 gene segment, ranging from 10% to 21%,3-11 and this lies within the unmutated subset.3-5 In contrast, bias to other VH genes, including V4-34, lies in the mutated subset.3-5 Since bias may reflect response to a superantigen,12 these differences point to distinct influences on pathogenesis of the subsets.
The 51p1-related subset of the V1-69 locus is frequently expressed in CLL5,7,11 and, in that setting, is commonly associated with D3-3 and D3-10 genes,JH6, and a long complementarity-determining region 3 (CDR3).5,8 There is controversy as to whether these features are disease-associated8,13 or simply reflect the pattern of usage in normal B cells.9Comparisons have generally been made with the healthy adult repertoire derived from relatively young individuals.9,13 However, CLL presents at a mean age of approximately 60 years, and the B-cell repertoire may change with age.14,15 Asymmetries in VH gene usage could contribute a stochastic element to the pattern in CLL. To resolve this question, we have investigated expression of the 51p1 gene in a healthy elderly population (older than 75 years) using monoclonal antibody G6 (MoAb G6).16 We have also analyzed51p1-derived sequences from the immunoglobulin M (IgM) of B cells of 4 elderly individuals.
Materials and methods
Immunophenotypic analysis of healthy elderly and young adult blood lymphocytes
Blood was obtained from 35 young (younger than 35 years; 17 male, 18 female) and 42 elderly individuals (older than 75 years; 14 male, 28 female). Subjects were excluded if they had a history of lymphoproliferative disease, acute infection, or autoimmune disease or if they were taking immunosuppressive agents. The MoAb G6 was used to determine the frequency of 51p1-related alleles.16 G6 reacts with an idiotypic determinant derived from CDR2 in the five 51p1-like alleles, but not in the 8 hv1263-like variants at theV1-69 locus.11,17 18 Peripheral blood mononuclear cells (PBMCs) were stained with G6 followed by sheep antimouse–fluorescein isothiocyanate (FITC) and subsequently with CD5-phycoerythrin (PE)/CD19–R-phycoerythrin (RPE)–cyanin 5(Cy5) (Dako, Glostrup, Denmark). Red cells were lysed (FACSLyse; BD, Cowley, Oxford, United Kingdom), and an initial 900 000 cells were acquired in a BD Facscalibur flow cytometer.
Analysis of 51p1 gene sequences
VH51p1-encoded genes were amplified by touchdown polymerase chain reaction (PCR) (QIAGEN, Crawley, United Kingdom) of cDNA from PBMCs from 4 elderly individuals by means of a 51p1-variant CDR2-specific primer13 and a Cμ primer.19Gel-purified PCR products were cloned in pGEM T-vector (Promega, Southampton, United Kingdom). The 26 sequences determined in this study can be found in GenBank under accession numbers AY179836-AY179861. Twelve sequences from 5 healthy elderly subjects all 65 years or older14 were added to those in this study. Included were 44 sequences from CLL patients4,5,8 and 44 from healthy young adults9 13 expressing unmutated51p1-related IgM.
Results and discussion
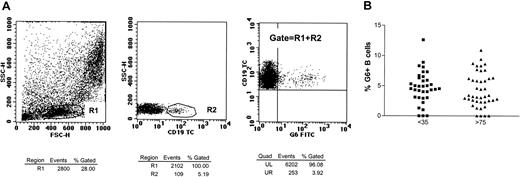
The 35 young and 42 elderly individuals had similar B-cell numbers, and percentages of CD5+ B cells (20.5% and 16.7%, respectively). G6 expression correlates with the germ-line copy number of 51p1-related genes and is absent in approximately 11% of healthy individuals (Sasso et al20). A wide range of G6+ B cells was found in both groups (Figure 1), with 1% to 12.6% (mean, 4.7% ± 0.45%) in the young and 1% to 10% (mean, 4.3% ± 0.44%) in the elderly, and approximately 10% of both cohorts being G6−. Mean levels are higher than a previous report (1.7%), probably owing to low numbers (7 individuals) studied (Shokri et al21). Heterogeneity in expression ofV1-69 in healthy people was also found by means of IgM cDNA libraries and single-cell analysis.14 That study pointed to a possible increased expression in the elderly (older than 65 years) (6%) as compared with the young (0.8%). However, libraries from only 5 elderly individuals were assessed, again a small sample. Clearly, although there is a reported dysregulation of B-cell responses in older populations of both humans15 and mice,22probably owing to failing T-cell help, this does not affect the number of IgM+51p1-expressing B cells.
Percentages of G6+ B cells in the peripheral blood.
(A) Peripheral blood was stained with mouse MoAb G6 followed by sheep antimouse-FITC and subsequently with CD5-PE/CD19-RPE-Cy5. On an initial 10 000-cell acquisition, the B cells were defined as shown by the gates R1 (lymphocytes) and R2 (CD19+ lymphocytes). Approximately 900 000 total events per tube were acquired in an R1-plus-R2 live gate to give percentages for G6 reactivity. (B) Comparison of the percentage of G6+ lymphocytes in 35 young (▪) and 42 (▴) elderly individuals.
Percentages of G6+ B cells in the peripheral blood.
(A) Peripheral blood was stained with mouse MoAb G6 followed by sheep antimouse-FITC and subsequently with CD5-PE/CD19-RPE-Cy5. On an initial 10 000-cell acquisition, the B cells were defined as shown by the gates R1 (lymphocytes) and R2 (CD19+ lymphocytes). Approximately 900 000 total events per tube were acquired in an R1-plus-R2 live gate to give percentages for G6 reactivity. (B) Comparison of the percentage of G6+ lymphocytes in 35 young (▪) and 42 (▴) elderly individuals.
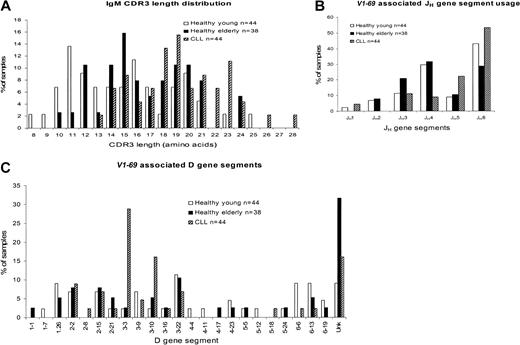
Sequence analysis of the available CDR2–framework region 4 (FR4) regions of the 51p1-related genes from the G6+ elderly group revealed all to be unmutated. A previous study of one individual from a cohort of healthy laboratory volunteers and blood donors, assumed to be younger than 60 years, had found heterogeneity of CDR3 length (mean, 14.6 ± 4.3 codons); use of a range of D genes; and frequent use of JH4 andJH6.13 These, compiled with a further 6 sequences from “healthy young”9 individuals (Figure 2A) are clearly different from cases of CLL that have a longer CDR3 (mean, 19.3 ± 3.5 codons) and common involvement of JH6.4,5 8Superimposition of the sequence data from the elderly cohort indicates a length distribution (mean, 16.6 ± 3.5 codons) similar to that of the young group (mean, 15.1 ± 4.1 codons; P = .074), and distinct from CLL sequences (mean, 19.3 ± 3.5 codons;P = .0009) (Figure 2A). In fact, both healthy groups appear heterogeneous in length, with a possibility of 2 subsets of long and short sequence. Although numbers are small, the subsets with longer CDR3 appear to overlap the major fraction of CLL sequences.
Comparison of V1-69 (51p1) CDR3 length distribution and associated JH and D gene segments in healthy young people, healthy elderly people, and cases of CLL.
(A) CDR3 length distribution. (B) JH gene segment usage. (C) D gene segment usage. Unk indicates unknown.
Comparison of V1-69 (51p1) CDR3 length distribution and associated JH and D gene segments in healthy young people, healthy elderly people, and cases of CLL.
(A) CDR3 length distribution. (B) JH gene segment usage. (C) D gene segment usage. Unk indicates unknown.
The CDR3 sequence includes JH-derived amino acids, and involvement of JH6 often leads to a longer CDR3.13 Analysis of JH genes combined with the51p1 gene in the elderly shows JH4 to be most common (Figure 2B), further distinguishing normal B cells from CLL, where JH6 is preferred.4,5,8The young cohort is again similar to the elderly, apart from a higher use of JH6 in the former. In the 22 sequences that have a CDR3 of 17 or fewer amino acids, only 4 (18.2%) useJH6. In contrast, in the 16 sequences in which the CDR3 had 18 or more amino acids, 7 (43.8%) useJH6. Therefore, there appears to be an association, although not complete, between JH6usage and longer CDR3 sequences, as indicated previously.13
The D gene segment was identified in the elderly cohort for 26 of 38 sequences. Figure 2C indicates that the D3 (8 of 38) and D2 (8 of 38) families were the most frequently used. The young frequently used the D6 and D3 families, with both frequently using D3-22. In contrast, CLL sequences have a strikingly common use ofD3-3, followed by D3-10 (Figure2C).4,5,8 In cases of CLL involving D3-3, 8 of 13 tend to have CDR3 derived from the second reading frame with the motif (Y)DFWSGY(Y)(P).13 Since selection of D3-3 is not a feature of normal B cells in either young or elderly repertoires, this may be a disease-associated feature. However, other sequence motifs evident in51p1-encoded CLL may not be disease-associated. Thus, a motif encoded by the third (hydrophobic) reading frame of theD2-2 gene (VVPAA), found frequently in CLL, was also found in 2 of the D2-2–derived sequences in the elderly group. Interestingly, this was rare in the young 51p1repertoire.13
Bias to 51p1 together with selected CDR3 sequences points to a role for antigen/superantigen in driving the cell of origin of this subset of unmutated CLL. We found no evidence for an expanded pool of B cells accumulating with age that would act as precursors for transformation. Although dysregulation of immunity occurs in the elderly, with emergence of endogenous viruses23 and increasing autoreactivity,15 perturbation apparently does not occur in 51p1-encoded immunoglobulin. This supports the concept that development of CLL is not a simple stochastic event, but derives from cells driven by specific antigens, some of which are recognized by 51p1-encoded IgM. If antigen is persistent, as for certain pathogens, or autoantigens, it could contribute to tumor growth via surface IgM (sIgM)–mediated signals, known to be transmitted in the unmutated subset.2 24
We acknowledge Gavin Babbage for DNA sequencing. We are grateful to Professor Roy Jefferis (University of Birmingham Medical School, Birmingham, United Kingdom) for the kind gift of monoclonal antibody G6.
Prepublished online as Blood First Edition Paper, December 12, 2002; DOI 10.1182/blood-2002-08-2432.
Supported by Tenovus, United Kingdom.
K.N.P. and J.O. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kathleen N. Potter, Tenovus Laboratory, Tremona Rd, Southampton University Hospitals Trust, Southampton, SO16 6YD, United Kingdom; e-mail: kp1@soton.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal