Abstract
β2-Microglobulin (β2m) is a chaperone of major histocompatibility complex (MHC) class I (–like) molecules that play a central role in antigen presentation, immunoglobulin transport, and iron metabolism. It is therefore of importance that β2m is adequately expressed in cells that perform these functions, such as hematopoietic cells. In this study, we investigated the transcriptional regulation of β2m in lymphoid and myeloid cell lines through a promoter containing a putative E box, Ets/interferon-stimulated response element (ISRE), and κB site. Here we show that upstream stimulatory factor 1 (USF1) and USF2 bind to the E box and regulate β2m transactivation. The nuclear factor κB (NF-κB) subunits p50 and p65 bind to the κB box and p65 transactivates β2m. Interferon regulatory factor 1 (IRF1), IRF2, IRF4, and IRF8, but not PU.1, bind to the Ets/ISRE, and IRF1 and IRF3 are strong transactivators of β2m. Together, all 3 boxes are important for the constitutive and cytokine-induced levels of β2m expression in lymphoid and myeloid cell types. As such, β2m transactivation is under the control of important transcriptional pathways, which are activated during injury, infection, and inflammation.
Introduction
β2-Microglobulin (β2m) is a ubiquitously expressed 12-kDa glycoprotein that associates with major histocompatibility complex (MHC) class I (–like) molecules that are of great importance in antigen presentation, IgG transport, and iron metabolism.1 As such, β2m is linked to a variety of human diseases because of its association with immunologically and hematologically relevant molecules. β2m is best known for its association with the MHC class I heavy chain, which is essential for the stable expression of these antigen-presenting molecules. Classical MHC class I molecules (HLA-A, -B, and -C) are ubiquitously expressed in most somatic cells. They are essential in the immune response because they present antigen-derived peptides to cytotoxic T lymphocytes and are important in protection against natural killer (NK) cell–mediated cytotoxicity.2β2m also associates with MHC class Ib or class I–like molecules, such as HLA-E, -F, -G, and CD1, which have a more restricted tissue distribution and have more specialized functions in antigen presentation.3-5 β2m is also a partner of HFE (formerly called HLA-H), an MHC class I–like molecule that is important for transferrin-mediated iron uptake.6-9Patients suffering from hereditary hemochromatosis have been found to bear a mutation in the HFE gene that specifically disrupts its association with β2m, resulting in a strongly compromised function.6,7 This is characterized by iron accumulation in parenchymal cells in various organs but a paucity of iron in Kupffer cells and macrophages.6-9 Clinical consequences include liver cirrhosis, diabetes, arthritis, and heart failure. Furthermore, β2m is also able to form a dimer with the neonatal Fc receptor, which is important for fetomaternal transport of IgG.10,11 Finally, a role for β2m has also been suggested in amyloidosis. Patients undergoing long-term dialysis often develop amyloidosis, which in turn affects bone cell metabolism by inducing bone mineral dissolving and enhancing osteoblast proliferation.12,13 This is thought to be related to the binding of β2m with α2-macroglobulin and heparin sulfate.14 15
Because β2m is essential for the functioning of molecules central in antigen presentation, IgG transport, and iron metabolism, a tight control of β2m expression is essential to secure the expression in a variety of cell types, such as hematopoietic, parenchymal, and syncytiotrophoblast cells. The basal level of β2m expression can be enhanced by cytokines to meet local requirements for an adequate immune response and possibly also in fulfilling any of its other functions. The transcriptional regulation of β2m is thought to be similar to that of MHC class I genes. Both β2m and MHC class I genes possess the SXY module, a set of regulatory elements shared with MHC class II genes, and are regulated through an MHC-specific enhanceosome.16,17 This multiprotein complex, containing RFX, CREB/ATF, and NF-Y transcription factors, is the basis for transactivation driven by the class II transactivator (CIITA).17 Other potential regulatory elements that could mediate the cytokine-induced transactivation have been identified further upstream in the human β2m promoter, but these elements have never been fully characterized. Therefore, we investigated the transcriptional regulation through these upstream promoter elements that could control the β2m expression in lymphoid and myeloid cell lines, central in immunologic and hematologic functions.
Materials and methods
Cell culture
The cell lines used in this study were the acute T-cell leukemia Jurkat, the Burkitt lymphoma B-cell line Raji, the acute lymphoblastic leukemia B-cell line SB, the monocytic cell line THP-1, the cervical carcinoma cell line HeLa, the teratocarcinoma cell line Tera-2 (all from American Type Culture Collection, Manassas, VA), and Epstein-Barr virus (EBV)–transformed B cells MSH. These cell lines were grown in Iscove modified Dulbecco medium (IMDM; Life Technologies, Paisley, Scotland) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum (Life Technologies), penicillin (100 IU/mL), and streptomycin (100 μg/mL). Where indicated, cells were treated with tumor necrosis factor α (TNF-α; 500 U/mL; Bender Medsystem, Vienna, Austria), interferon α2c (IFN-α2c; 500 U/mL; Bender Medsystem), IFN-β1a (500 U/mL; Avonex/Biogen, Cambridge, MA), or IFN-γ (500 U/mL; Boehringer Ingelheim, Alkmaar, the Netherlands).
Preparation of nuclear extracts and electrophoretic mobility shift assay
Nuclear extracts were prepared as previously described.16 Nuclear extracts (about 5 μg protein) were incubated in DNA/protein-binding buffer (20 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.9, 50 mM KCl, 10% vol/vol glycerol, 0.5 mM dithiothreitol [DTT], 0.1 mM EDTA [ethylenediaminetetraacetic acid]), with 200 ng poly(dI·dC), 200 ng sonicated single-stranded herring sperm DNA, and 1 ng [32P]-radiolabeled probe for 30 minutes at 4°C. The samples were run on a 6% nondenaturing polyacrylamide gel in 0.25 × TBE (Tris-borate EDTA) buffer at 200 V for 2 hours. The gels were fixed with a 10% methanol and 10% acetic acid solution, dried onto Whatmann 3M paper and exposed to an x-ray film.
The following ds-oligonucleotides were used as probes for the putative E box, Ets/interferon-stimulated response element (ISRE), and κB site of β2m: E: 5′-AAACATCACGAGACTCT-3′; Ets/ISRE: 5′-TAAGAAAAGGAAACTGAAAACG-3′; κB: 5′-ACGGGAAAGTCCCTC-3′. The following probe was used as control Ets/ISRE site: 5′-CAGTCCACAGTAGGAAGTGAAATTA-3′.
For supershift assays, 1 μg of each antibody (Ab) specifically directed against the different transcription factors was added 20 minutes after the nuclear extract had been incubated with the probe and this mixture was incubated for an extra 30 minutes at 4°C. The antibodies used were directed against upstream stimulatory factor 1 (USF1; sc-229), USF2 (sc-861), E47 (sc-763), interferon regulatory factor 1 (IRF1; sc-497), IRF2 (sc-498), IRF3 (sc-9082), IRF4 (sc-6059), IRF7 (sc-9083), IRF8 (sc-6058), PU.1 (sc-352), Ets1/2 (sc-112), Spi-B (sc-5944), signal transducer and activator of transcription 1 (STAT1; sc-345), p50 (sc-114), p65 (sc-109), c-Rel (sc-71), RelB (sc-226); all were from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmids
Luciferase reporter plasmids used were generated by cloning genomic promoter fragments into pGL3-Basic (Promega, Madison, WI). These constructs contain a polymerase chain reaction (PCR)–generated promoter fragment of β2m of, respectively, 302 bp (pGL3-β2m), 193 bp (pGL3-β2m193), 180 bp β2m (pGL3-β2m180), 157 bp β2m PCR (pGL3-β2m157), 145 bp (pGL3-β2m145), and a 269-bp AspI-AhaII HLA-B7 promoter fragment (pGL3-HLA-B). The mutant promoter constructs of β2m containing a mutation either in the E box, ISRE, or κB site were generated by overlap extension PCR.16 These mutant promoter constructs are identical to the wild-type constructs (pGL3-β2m) except for a 2- to 3-bp mutation in the core sequence of the individual boxes (Figure 1). The Renillaluciferase constructs pRL-actin was used as internal control for transfection efficiency. pRL-actin was generated by cloning a PCR-generated 1-kb human β-actin promoter fragment into pRL-null (Promega).17
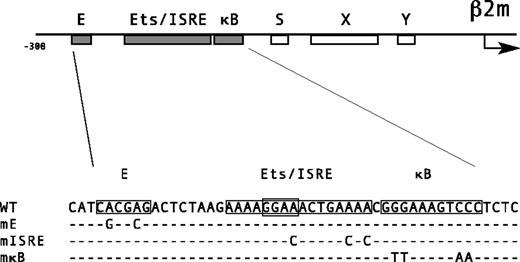
Promoter structure of human β2m.
Schematic representation of the promoter of β2m, depicting the position and order of the E box, ISRE, and κB site. Underneath, the nucleotide sequence of the upstream region of β2m containing the 3 boxes is shown, indicating the mutations in the putative E box, Ets/ISRE, and κB site that are introduced for the mutant promoter constructs. Human β2m promoter region accession numberAF092744.
Promoter structure of human β2m.
Schematic representation of the promoter of β2m, depicting the position and order of the E box, ISRE, and κB site. Underneath, the nucleotide sequence of the upstream region of β2m containing the 3 boxes is shown, indicating the mutations in the putative E box, Ets/ISRE, and κB site that are introduced for the mutant promoter constructs. Human β2m promoter region accession numberAF092744.
The expression vectors pSG5-USF1 and pSG5-USF2 were a kind gift of Dr M. Sawadogo.18 The pRc/RSV expression vectors of IRF1, IRF2, IRF4, and IRF8 were generated by cloning the PCR-amplified cDNAs of IRF1, IRF2, IRF4, and IRF8 into pRc/RSV. Also PU.1 was cloned into pRc/RSV (kindly provided as pCDNA3-PU.1 by Dr M. Fenton). The expression vectors pCMVBL/IRF3, pCMVBL/IRF3ΔN, pCMVBL/IRF3(5D), pCMVBL/IRF7 and pCMVBL/IRF7(2D) were a kind gift of Dr J. Hiscott,19 20 and these inserts were also cloned into the expression vector pRc/RSV.
Transient transfection
Adherent cells were transfected by the calcium phosphate coprecipitation method as described previously.16 In each of 4 wells of a 6-well plate, 0.2 × 106 cells were transfected with a DNA mix containing 1 μg firefly luciferase pGL3 reporter plasmid and 0.2 μg Renilla luciferase pRL-SV40 control plasmid (Tera-2). For cotransfection 1 μg pRc/RSV expression vector was used. For cytokine induction experiments, cells were treated with 500 U/mL TNF-α, IFN-α, IFN-β, or IFN-γ for 48 hours after transfection. Nonadherent cells (Jurkat, Raji, THP-1) were transfected by electroporation,17 with 10 μg firefly luciferase pGL3 reporter plasmid and 1 μg Renilla luciferase pRL-actin control plasmid. To measure promoter activity, cells were harvested 3 days after calcium phosphate transfection or 2 days after electroporation. Luciferase activity was determined using the dual-luciferase reporter assay system (Promega) and a luminometer (Tropix, Bedford, MA).
Results
Promoter structure of the human β2m gene
The proximal promoter region of β2m contains a set of regulatory elements that form the SXY regulatory module, a module that is shared with MHC class I and class II genes.16 The SXY module is the basis for an MHC-specific enhanceosome that is important in the CIITA route of transactivation.17
Computer-aided inspection of the human β2m promoter region upstream of the SXY module revealed the presence of a putative E box, ISRE, and κB site (Figure 1). This is similar to the promoter structure of the β2m promoter in the mouse except that the E box, located upstream of the ISRE, is not found in the mouse β2m promoter.21 22 The ISRE and κB boxes are also found in MHC class I promoters, but the order of these boxes is reversed, that is, the ISRE in the β2m promoter is positioned 5′ of the κB site. Furthermore, unlike MHC class I promoters, the ISRE region of β2m consists of 2 overlapping ISREs including a putative Ets-binding site (GGAA), which therefore classifies as a potential combined Ets/IRF-response element (Figure 1).
The E box, ISRE, and κB site are important for the constitutive and cytokine-induced β2m promoter activity
To test for the importance of the potential E box, ISRE, and κB site for a constitutive level of promoter activity, we generated promoter constructs, which were mutated in each of the 3 regulatory sites (Figure 1). Transient transfection of these β2m promoter constructs in Raji B cells, Jurkat T cells, and THP-1 monocytes revealed that mutation of the E box, ISRE, or κB site significantly reduced the basal promoter activity (Figure2A). Mutation of the ISRE resulted in the most dramatic reduction in basal promoter activity (reduced to about 10%-15%), whereas mutation of the E box or κB site resulted in reductions to 20% to 50% of wild-type. Therefore, all 3 potential regulatory sites play a role in the basal promoter activity in lymphoid and monocytic cells.
The importance of the E box, ISRE, and κB site in the constitutive and cytokine-induced β2m promoter activity.
(A) Transient transfection of wild-type β2m and E box-, ISRE-, or κB-mutated reporter constructs in Jurkat, Raji, and THP-1 cells revealing the importance of each regulatory site to the constitutive promoter activity in lymphoid and monocytic cells. (B) Transient transfection of the β2m reporter construct in Tera-2 cells induced with TNF-α, IFN-α, IFN-β, or IFN-γ (each 500 U/mL) for 48 hours. β2m is induced by all cytokines of which IFN-γ is the most potent. The induction ratios are indicated above the histogram. (C) Transient transfection of wild-type β2m and E box-, ISRE-, or κB-mutated reporter constructs in Tera-2 cells induced with TNF-α or IFN-γ (each 500 U/mL) for 48 hours. All boxes are important in the TNF-α– and IFN-γ–induced β2m promoter activity. The induction ratios are indicated above the histogram. The luciferase activity values were normalized with the Renilla luciferase activity values and are expressed as mean ± SD of 4 experiments. RLU indicates relative light units.
The importance of the E box, ISRE, and κB site in the constitutive and cytokine-induced β2m promoter activity.
(A) Transient transfection of wild-type β2m and E box-, ISRE-, or κB-mutated reporter constructs in Jurkat, Raji, and THP-1 cells revealing the importance of each regulatory site to the constitutive promoter activity in lymphoid and monocytic cells. (B) Transient transfection of the β2m reporter construct in Tera-2 cells induced with TNF-α, IFN-α, IFN-β, or IFN-γ (each 500 U/mL) for 48 hours. β2m is induced by all cytokines of which IFN-γ is the most potent. The induction ratios are indicated above the histogram. (C) Transient transfection of wild-type β2m and E box-, ISRE-, or κB-mutated reporter constructs in Tera-2 cells induced with TNF-α or IFN-γ (each 500 U/mL) for 48 hours. All boxes are important in the TNF-α– and IFN-γ–induced β2m promoter activity. The induction ratios are indicated above the histogram. The luciferase activity values were normalized with the Renilla luciferase activity values and are expressed as mean ± SD of 4 experiments. RLU indicates relative light units.
Because the ISRE and κB site are mediators of cytokine-induced transactivation, we tested whether β2m promoter activity could be up-regulated by IFN and TNF-α. In transient transfection experiments in the cytokine-responsive teratocarcinoma cell line Tera-2, promoter activity of β2m was induced in response to TNF-α and to IFN-α, IFN-β, and IFN-γ (Figure 2B). Of these cytokines IFN-γ was the most potent inducer of β2m transactivation. Using the mutant constructs, we demonstrated that mutation not only of the ISRE or κB site, but also of the E box, in general compromised the IFN-γ and TNF-α induction of β2m promoter activity (Figure 2C). Together, these results strongly suggest an important regulatory role for the E box, ISRE, and κB site in constitutive and cytokine-induced β2m transactivation.
Transactivation of β2m is controlled by upstream stimulatory factors binding to the E box
The binding capacity of the putative regulatory sites was investigated by electrophoretic mobility shift assay (EMSA) analysis. Transcription factor binding to putative E box was tested using nuclear extracts of the B-cell line Raji, T-cell line Jurkat, and monocytic cell line THP-1. The probe encompassing the E box bound a complex, which supershifted with Abs directed against USF1 and USF2. We also tested for the presence of the lymphoid/myeloid-specific factor E47, but did not detect E47 with the Ab used (Figure3A). Furthermore, using the Ab for either USF1 or USF2, the complex was almost completely supershifted, which implies that the complex consists almost entirely of the USF1/USF2 heterodimer. It should also be noted that in the 3 cell lines the intensity of the bands representing the complex was similar, indicative of a comparable protein content in the different nuclear extracts. Similar results were found using nuclear extract of nonlymphoid cells such as HeLa or Tera-2 cells, underscoring the ubiquitous expression and binding of these USF transcription factors to the E box.
Transcription factor binding and transactivation capacity of the E box of β2m.
(A) EMSA showing binding of complexes to the E box of β2m. Using specific Abs, the complex binding to the E box was shown to contain USF1 and USF2 in Jurkat, Raji, and THP-1 cells. The presence of E47 was not detected. Arrowheads indicate the USF1/USF2 complex; *, supershifted complex(es). (B) Transient transfection of wild-type β2m- and E box–mutated reporter constructs with USF1 and USF2 expression vectors (1 μg) in Tera-2 cells. The luciferase activity values were normalized with the Renillaluciferase activity values and are expressed as mean ± SD of 4 samples. The induction ratios are indicated above the histogram. RLU indicates relative light units.
Transcription factor binding and transactivation capacity of the E box of β2m.
(A) EMSA showing binding of complexes to the E box of β2m. Using specific Abs, the complex binding to the E box was shown to contain USF1 and USF2 in Jurkat, Raji, and THP-1 cells. The presence of E47 was not detected. Arrowheads indicate the USF1/USF2 complex; *, supershifted complex(es). (B) Transient transfection of wild-type β2m- and E box–mutated reporter constructs with USF1 and USF2 expression vectors (1 μg) in Tera-2 cells. The luciferase activity values were normalized with the Renillaluciferase activity values and are expressed as mean ± SD of 4 samples. The induction ratios are indicated above the histogram. RLU indicates relative light units.
Because the E box was shown to be a binding site for USF1 and USF2, we tested whether β2m promoter activity was controlled by USFs with transient transfection experiments in Tera-2 cells. Cotransfection of the β2m promoter construct with exogenous USF1 or USF2 enhanced promoter activity of β2m (Figure 3B), demonstrating that USF1 and USF are positive regulators of β2m promoter activity. Mutation of the E box reduced the elevated promoter activity of β2m, revealing that the E box contributes to the USF-mediated transactivation of β2m (Figure 3B).
Transactivation of β2m is regulated by NF-κB through its κB site
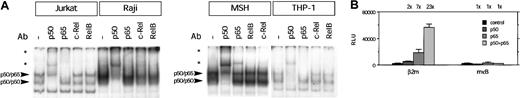
Next, the binding capacity of the κB site was investigated. The κB site was shown to bind protein complexes containing p50 and p65 using nuclear extracts of Jurkat and THP-1 cells. This was similar to nonlymphoid cells, where p50 and p65 were supershifted in EMSA using nuclear extract from HeLa cells induced with TNF-α (data not shown). However, supershift analysis using Raji nuclear extracts revealed that the Ab against p50 gave a strong supershifted complex, whereas the supershift with the p65 Ab was weak. Using Abs against c-Rel and RelB resulted in a slight reduction of the protein/DNA complex, which could suggest that c-Rel and RelB are also present in the complex (Figure4A). Similarly, in other B-cell lines a predominance of p50 binding to the κB site was found combined with a weak supershift for p65, c-Rel, and RelB (Figure 4A; data not shown).
Transcription factor binding and transactivation capacity of the κB site of β2m.
(A) EMSA showing binding of complexes to the κB site of β2m. Using specific Abs, the complex binding to the κB was shown to contain p50 and p65. The presence of c-Rel and RelB was weakly detectable in Raji and MSH B cells. Note the difference in the quantity of complex formation with an equal loading as in Figure3. Arrowheads indicate NF-κB complexes binding the κB site; *, supershifted complex(es). (B) Transient transfection of wild-type β2m- and κB site–mutated reporter constructs with p50 and p65 expression vectors (1 μg) in Tera-2 cells. The luciferase activity values were normalized with the Renilla luciferase activity values and are expressed as mean ± SD of 4 samples. The induction ratios are indicated above the histogram. RLU indicates relative light units.
Transcription factor binding and transactivation capacity of the κB site of β2m.
(A) EMSA showing binding of complexes to the κB site of β2m. Using specific Abs, the complex binding to the κB was shown to contain p50 and p65. The presence of c-Rel and RelB was weakly detectable in Raji and MSH B cells. Note the difference in the quantity of complex formation with an equal loading as in Figure3. Arrowheads indicate NF-κB complexes binding the κB site; *, supershifted complex(es). (B) Transient transfection of wild-type β2m- and κB site–mutated reporter constructs with p50 and p65 expression vectors (1 μg) in Tera-2 cells. The luciferase activity values were normalized with the Renilla luciferase activity values and are expressed as mean ± SD of 4 samples. The induction ratios are indicated above the histogram. RLU indicates relative light units.
The transactivation of β2m by nuclear factor κB (NF-κB) was tested in transient transfection experiment in Tera-2 cells. Cotransfection of the β2m promoter construct with p50 and p65 resulted in enhanced promoter activity (Figure 4B). Mutation of the κB site strongly impaired the induced promoter activity by NF-κB (Figure 4B). Together this demonstrates that this site is a functional κB site.
IRFs regulate β2m transactivation through the ISRE
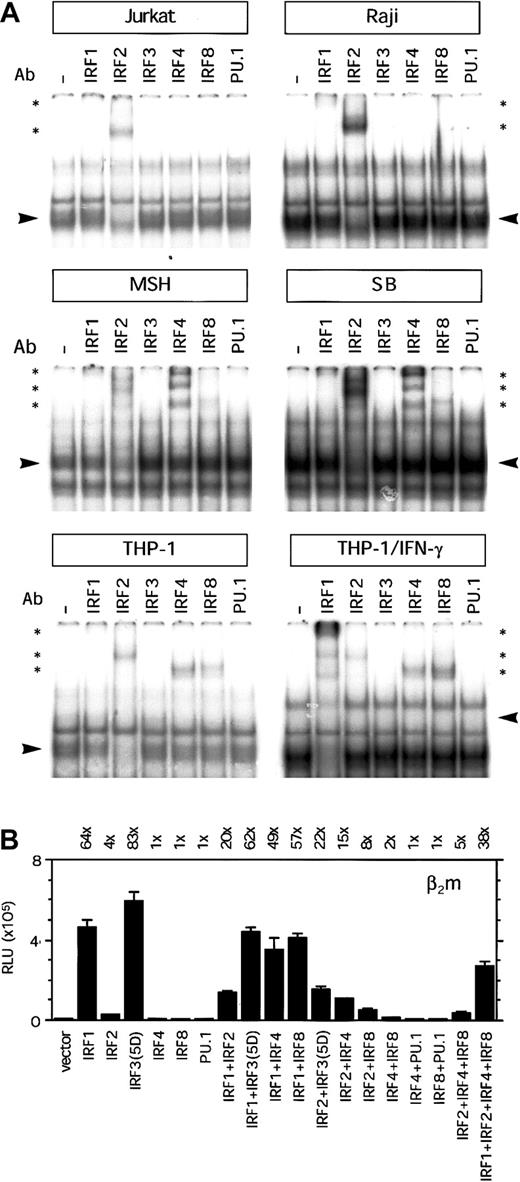
Binding activity to the ISRE region was tested in EMSAs, using nuclear extract of Raji, Jurkat, and THP-1 cells. Several protein/DNA complexes were observed in EMSAs with nuclear extracts from Jurkat, Raji, and THP-1 cells. The supershift with the Ab directed against IRF2 was strongest (Figure 5A), indicating that this was the predominant factor in the complex. Only a weak supershift was observed with the Ab directed against IRF1, indicating that the presence was less abundant in this transcription factor complex binding the ISRE region (Figure 5A). In Jurkat and Raji cells we could not detect binding of lymphoid/myeloid-specific factors IRF4 and IRF8. In contrast to Raji B cells, in EMSAs with nuclear extract from MSH and SB we observed a strong supershift with the IRF4-specific Ab and a weak supershift with IRF8-specific Ab (Figure 5A). In addition, IRF4 and IRF8 were also present in the complex binding to the ISRE region in THP-1 cells. Moreover, induction of THP-1 cells with IFN-γ resulted in a strong increase in IRF1 binding and a mild increase in IRF8 and reduction of IRF2 binding (Figure 5A). In these experiments, we could not detect the binding of IRF3 or IRF7 in any of the cell lines (Figure 5A and data not shown). It is not clear whether this is due to the absence of these factors or caused by a poor quality of these Abs. Although the ISRE contains a potential Ets-binding site (GGAA), we could not detect any binding of PU.1, Ets1/2, or Spi-B in THP-1 nuclear extracts (Figure 5A and data not shown). This was an unexpected finding because IRF4 and IRF8 are considered to form a complex with PU.1/Ets factors for optimal binding. The presence of PU.1 in the nuclear extracts and the quality of the PU.1 Ab were verified with another ISRE/Ets probe that served as control (data not shown). Because the supershift with the IRF Abs reduced only marginally the protein/DNA complex, it is possible that other transcription factors are also present in the complex. It is of interest to note that there was more complex formation with nuclear extracts from Raji cells and IFN-γ–induced THP-1 cells than Jurkat and noninduced THP-1 cells, which is not due to loading difference (compare binding to the E probe in Figure 3A). In nonlymphoid cells, the binding of IRFs to the ISRE was more restricted. Using nuclear extract from HeLa cells, IRF2 and little IRF1 were present in the complex binding to the ISRE, whereas upon IFN-γ treatment we observed a strong increase of IRF1 in the complex (data not shown). No STAT1 binding to the ISRE of β2m was detected in any of the cell lines (data not shown). Together, this indicates that IRF2 and in certain B cells also IRF4 and IRF8 are important factors binding to the ISRE in lymphoid cells. Furthermore, IRF2, IRF4, and IRF8 are important factors binding to the ISRE in nonactivated monocytic cells, whereas IRF1 becomes important after IFN-γ induction of monocytic cells.
Transcription factor binding and transactivation capacity of the ISRE of β2m.
(A) EMSA showing binding of complexes to the ISRE of β2m. Using specific Abs, the complex binding to the ISRE was shown to contain in IRF2, and little IRF1, in Jurkat T and Raji B cells. In the B cells MSH and SB there was, in addition, a strong presence of IRF4 and a weak presence of IRF8. IRF4 and IRF8 were also found in THP-1 cells, whereas in THP-1 cells induced with IFN-γ, there was an additional strong presence of IRF1. Arrowheads indicate the complex containing IRF factors; *, supershifted complex(es). (B) Transient transfection of the β2m reporter construct with IRF1, IRF2, IRF3(5D), IRF4, IRF8, and PU.1 expression vectors (1 μg) in Tera-2 cells, as indicated. IRF1 or IRF3(5D) could barely transactivate the ISRE-mutated reporter construct in Tera-2 cells (3-fold by IRF1 and 2-fold by IRF3(5D); data not shown). The luciferase activity values were normalized with the Renilla luciferase activity values and are expressed as mean ± SD of 4 samples. The induction ratios are indicated above the histogram. RLU indicates relative light units.
Transcription factor binding and transactivation capacity of the ISRE of β2m.
(A) EMSA showing binding of complexes to the ISRE of β2m. Using specific Abs, the complex binding to the ISRE was shown to contain in IRF2, and little IRF1, in Jurkat T and Raji B cells. In the B cells MSH and SB there was, in addition, a strong presence of IRF4 and a weak presence of IRF8. IRF4 and IRF8 were also found in THP-1 cells, whereas in THP-1 cells induced with IFN-γ, there was an additional strong presence of IRF1. Arrowheads indicate the complex containing IRF factors; *, supershifted complex(es). (B) Transient transfection of the β2m reporter construct with IRF1, IRF2, IRF3(5D), IRF4, IRF8, and PU.1 expression vectors (1 μg) in Tera-2 cells, as indicated. IRF1 or IRF3(5D) could barely transactivate the ISRE-mutated reporter construct in Tera-2 cells (3-fold by IRF1 and 2-fold by IRF3(5D); data not shown). The luciferase activity values were normalized with the Renilla luciferase activity values and are expressed as mean ± SD of 4 samples. The induction ratios are indicated above the histogram. RLU indicates relative light units.
Next, we investigated the ability of IRF factors to transactivate β2m in Tera-2 cells, because this cell line does not express IRFs constitutively. IRF1 was shown to be an important factor binding to the ISRE in IFN-γ–induced THP-1 cells. In transient transfection assays, IRF1 strongly enhanced β2m promoter activity (64-fold induction; Figure 5B). In addition, IRF2, which constitutively binds to the ISRE in monocytic cells, moderately enhanced β2m promoter activity (4-fold induction; Figure5B). Cotransfection of IRF1 with IRF2 did not further enhance β2m promoter activity but rather tempered the IRF1 induction. Although in EMSA we could not detect any binding activity of IRF3 or IRF7, we also tested their capacity to transactivate β2m. These IRFs posses an autoinhibitory domain and are present in a nonactive form. Therefore, we used the constitutively active forms IRF3(5D) and IRF7(2D), which bear a mutation in their autoinhibitory domain.19,20 Transient transfection assays with these mutant constructs showed that IRF3(5D), but not IRF7(2D), was a potent inducer of β2m promoter activity (83-fold induction by IRF3(5D) versus 1-fold induction by IRF7(2D); Figure 5B and data not shown). Cotransfection of IRF3(5D) with IRF7(2D) did not change the strength of β2m transactivation by IRF3(5D) (data not shown). The wild-type forms of IRF3 and IRF7 did not transactivate β2m and served as controls (data not shown). Furthermore, transactivation of ISRE-mutated β2m promoter with IRF1 or IRF3(5D) was negligible (3-fold and 2-fold, respectively; data not shown). Next, we tested the lymphoid and myeloid-specific factors IRF4 and IRF8 because in some B and monocytic cell lines the protein complex binding to the ISRE also contained IRF4 and IRF8. Neither IRF4 nor IRF8 alone was able to enhance β2m transactivation (Figure 5B). Interestingly, the combination of IRF2 with IRF4 or IRF8 enhanced the weak induction β2m by IRF2 alone (Figure 5B). Because these IRFs often require PU.1 or other Ets factors as binding partner for their activity, we tested their transactivation potential in combination with PU.1. However, in EMSA we had not detected PU.1 binding to the ISRE. In line with this finding, IRF4 or IRF 8 was not able to transactivate β2m in combination with PU.1 (Figure 5B). For genes such as ISG15, it has been described that IRF4 and IRF8 can also bind in a complex with IRF1 and IRF2, which positively regulates gene activation.23 To test this possibility we determined the joint transactivation capacity of IRF1, IRF2, IRF4, and IRF8. However, IRF2, IRF4, and IRF8 tempered the IRF1-induced β2m transactivation, indicating that there was no obvious additive or synergistic effect of the joint expression and binding of these IRFs (Figure 5B).
Discussion
As chaperone of MHC class I (-like) molecules, β2m is essential for the functioning of molecules central in antigen presentation, IgG transport, and iron metabolism. This necessitates a tight control of β2m transactivation to secure an adequate expression in a variety of tissues and cell types. In particular, hematopoietic cells fulfill an important role in these functions.2,5,9 The clinical consequences of an aberrant β2m expression are exemplified in studies with β2m knock-out mice that have a greatly compromised immune response due to the lack of antigen presentation and have a disturbed iron metabolism similar to patients with hereditary hemochromatosis.6-8
In the present study, we investigated the regulatory elements upstream of the SXY regulatory module that could provide for the constitutive and cytokine-induced transactivation of β2m. In the human β2m promoter, this region includes a (putative) E box, an Ets/ISRE, and a κB box. It is noteworthy that the mouse β2m promoter differs from the human promoter in that it lacks the E box and the Ets site within the ISRE.21,22 24
All 3 sites were important for the constitutive levels of β2m transactivation because mutation of either regulatory site strongly reduced the basal level of β2m promoter activity in lymphoid and monocytic cell lines. Based on the level of reduction, the ISRE is the strongest contributor to the basal promoter activity of β2m. In addition, β2m transactivation is up-regulated by several cytokines, such as TNF-α, IFN-α, IFN-β, and IFN-γ. This is of biologic importance for the coordinate cytokine-regulated expression of all genes involved in the MHC class I antigen presentation pathway, including β2m and MHC class I molecules, during inflammation or infection.25-28 In this context, it would be of interest to investigate whether the expression of other molecules that β2m associates with, such as HFE and the neonatal Fc receptor, are also regulated by cytokines. As predicted, mutation of the ISRE almost entirely abolished the transactivation induced by IFN-γ, as did the mutation of the κB site for the transactivation induced by TNF-α. Interestingly, mutations of the ISRE compromised the TNF-α–induced promoter activity and, conversely, mutation of the κB site compromised that by IFN-γ in absolute numbers. This can be explained by the induction of NF-κB by IFN-γ and the induction of IRF1 by TNF-α.29-32 In addition, mutation of the E box compromised β2m transactivation by both IFN-γ and TNF-α. This suggests cooperation between the different boxes, which could be brought about by interactions between transcription factors of the different families or through joint recruitment of a general coactivator.
More detailed analysis revealed that the E box, positioned upstream of the ISRE in the β2m promoter region, is almost exclusively bound by USF1 and USF2. These 2 USFs, which most likely form a heterodimer, are transactivators of β2m and mutation of the E box diminished the USF-enhanced level of β2m transactivation. The fact that USF1 and USF2 could still moderately enhance β2m promoter activity when the E box was mutated may be due to an additional E box positioned in the X box in the SXY module. This second E box was able to bind USF1 and USF2 (S.J.P.G. et al, unpublished results, January 2002) and may also contribute to the β2m promoter activity controlled by USFs. Because USF1 and USF2 are ubiquitously expressed factors, they fulfill a more general rather than a lymphoid/myeloid-specific role in the regulation of β2m transcription.
The κB site, flanking the ISRE at the 3′ site, is the binding site for both p50 and p65 in T cells and monocytic cells. These NF-κB subunits most likely form a heterodimer,33 and together strongly induced β2m transactivation. B cells displayed a slightly different binding profile in that they bind predominantly p50 and less p65. In addition, the binding of RelB and c-Rel was also detected but was very weak. Nevertheless, this opens the possibility that in B cells different heterodimers are formed and bind the κB site of β2m, although it remains to be determined whether also other dimers are important for the transactivation of β2m in this cell type. NF-κB is a crucial transcriptional regulator of genes, which products are essential in the immune functions fulfilled by lymphoid and myeloid cells.33 In this context it is of relevance that also β2m is under the control of NF-κB.
Perhaps the most important regulatory site of the upstream region of the β2m promoter is the ISRE because mutation of this site dramatically reduced the basal and cytokine-induced promoter activity in lymphoid and monocytic cells. The ISRE of β2m is in fact built up of 2 overlapping ISREs and contains a putative Ets-binding site. The ISRE is bound by several general and lymphoid/myeloid-specific IRFs. Based on supershift analysis we observed a difference in presence and redundancy of IRFs in the complex binding the ISRE. In B and monocytic cell lines, IRF2, IRF 4, and IRF8 were detected in the complex binding to the ISRE. However, in Jurkat T and Raji B cells we could not detect the binding of IRF4 or IRF8. In the case of Jurkat cells, this is most likely due to the fact that expression of these lymphoid/myeloid-specific IRF factors requires an activation step.34,35 Furthermore, little IRF1 was present in the ISRE-binding complex using nuclear extracts of the different cell lines. However, IFN-γ induction of THP-1 cells greatly increased the presence of IRF1 in the complex. The binding of IRF3 and IRF7 was not detected, although we could not exclude the possibility that this was due to the quality of the Abs. The binding of several IRF factors allows the formation of different protein complexes. In transient transfection assays, IRF1 and IRF3 were found to be strong transactivators of β2m. Combinations of IRF1 or IRF3 with other IRFs tempered their transactivation potential. IRF2 alone weakly induced β2m promoter activity, but in combination with IRF1 it compromised the activation strength of IRF1. IRF1 is a general transactivator of genes bearing a conventional ISRE.36,37These include MHC class I genes, which are also regulated by IRF1.26,38 Similar to the promoters of RANTES and ISG56, β2m possesses an extended ISRE with several GAAA tandem-repeats and is also regulated by IRF3.39,40 In contrast, IRF7 did not induce β2m promoter activity, which may be due to the fact that the 3 GAAA repeats in the ISRE (GAAAA, GAAAC, and GAAAA,respectively) are preferentially or specifically bound by IRF3 and not IRF7.41 The MHC class I gene HLA-B, which does not posses an extended ISRE, is only marginally activated by IRF3 (5-fold) and not by IRF7 (S.J.P.G. et al, unpublished results, July 2002). Genes that have a combined Ets/ISRE are cooperatively bound and regulated by the lymphoid/myeloid-specific factors IRF4, IRF8, PU.1, and Spi-B.35,42-44 Because the ISRE of β2m also contained a potential Ets site (GGAA), we investigated binding of PU.1 and other Ets factors to this ISRE.43,45 Despite a perfect Ets core sequence, we could not detect the binding of PU.1, Ets1/2, or Spi-B. In the case of PU.1, this was not related to the quality of the Ab, because the Ab could supershift PU.1 bound to a control Ets/ISRE probe. It is possible that the lack of binding of PU.1 to this Ets site is related to the flanking sequences.46 This excludes PU.1 as binding partner for IRF4 or IRF8, but it is possible that other Ets factors could bind the Ets site and form a complex with the IRF factors. Despite the binding of IRF4 and IRF8 to the ISRE of β2m, they did not activate the β2m promoter, although the combinations of IRF2 with IRF4 or IRF8 weakly induced β2m promoter activity. However, this was tested in a nonlymphoid/myeloid cell line and it is therefore possible that IRF4 and IRF8 require lymphoid/myeloid-specific partners to be able to control β2m transactivation. Alternatively, it is also possible that the observed transactivation by IRF4 and IRF8 is mediated by IRF2, which is expressed in all lymphoid/myeloid cells types. It is therefore of importance to determine the exact role of IRF4 and IRF8 in the transactivaiton of β2m in lymphoid and myeloid cells. Interestingly, functional interactions of IRF4 and IRF8 with E47 are reported for the Ig κ 3′ enhancer, which contains a flanking ISRE and E box.47 48 It is tempting to speculate that IRF4 and IRF8 could interact with E box-binding proteins, although in our experiments and with the Ab used we could not detect E47 binding.
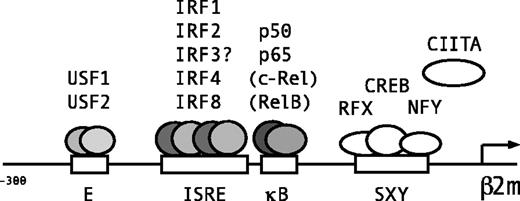
In conclusion, 3 juxtaposed regulatory sites, the E box, ISRE, and κB site in the upstream promoter region of β2m mediate the constitutive and cytokine-induced regulation of β2m transactivation (Figure 6). The E box is a binding site for the ubiquitous factors USF1 and USF2 (Figure 6). The κB box is bound by the NF-κB subunits p50 and p65, and specifically in B cells weakly bound by c-Rel and RelB (Figure 6). The ISRE was bound by the ubiquitous factors IRF1 and IRF2. The binding of IRF3 was not detected but both IRF1 and IRF3 are strong transactivators of β2m (Figure 6). The lymphoid/myeloid-specific factors, IRF4 and IRF8 (but not PU.1) bound to the ISRE, but their role in β2m transactivation is not clear. Thus, all 3 boxes are important for the constitutive and cytokine-induced levels of β2m expression in lymphoid and myeloid cell types. Thus, similar to MHC class I molecules, β2m transactivation is under the control of important transcriptional pathways that are activated during injury, infection, and inflammation.25,33,34,49 50 This is of general physiologic importance for adequate antigen presentation, IgG transport, and iron metabolism during these circumstances.
Lymphoid/myeloid-specific binding of transcription factors to the E box, ISRE, and κB site of β2m.
Schematic representation of the β2m promoter and transcription factors binding to the 3 adjacent regulatory sites in the promoter region upstream of the SXY regulatory module. The E box is bound by ubiquitous factors USF1 and USF2. The ISRE is a binding site for IRF1 and IRF2 and the lymphoid/myeloid factors IRF4 and IRF8. Because IRF3 is also a potent transactivator of β2m, also IRF3 is likely to bind the ISRE. The κB site is bound by the NF-κB subunits p50 and p65, and in B cells also marginally by c-Rel and RelB.
Lymphoid/myeloid-specific binding of transcription factors to the E box, ISRE, and κB site of β2m.
Schematic representation of the β2m promoter and transcription factors binding to the 3 adjacent regulatory sites in the promoter region upstream of the SXY regulatory module. The E box is bound by ubiquitous factors USF1 and USF2. The ISRE is a binding site for IRF1 and IRF2 and the lymphoid/myeloid factors IRF4 and IRF8. Because IRF3 is also a potent transactivator of β2m, also IRF3 is likely to bind the ISRE. The κB site is bound by the NF-κB subunits p50 and p65, and in B cells also marginally by c-Rel and RelB.
The authors thank Dr M. Fenton, Dr J. Hiscott, and Dr M. Sawadogo for providing expression plasmids.
Prepublished online as Blood First Edition Paper, December 12, 2002; DOI 10.1182/blood-2002-09-2924.
Supported by the Netherlands Foundation for the Support of Multiple Sclerosis Research no. 96-248 MS and the Dr Gisela Thier Foundation. S.J.P.G. is a research fellow of the Royal Netherlands Academy of Arts and Sciences.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Peter J. Van den Elsen, Department of Immunohematology and Blood Transfusion, Leiden University Medical Center, Albinusdreef 2, 2333 ZA Leiden, the Netherlands; e-mail:pjvdelsen@lumc.nl.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal