Type I interferons (IFNs), pleiotropic cytokines with antiviral, antiproliferative, apoptotic, and immunoregulatory functions, are efficacious in the treatment of malignancies, viral infections, and autoimmune diseases. Binding of these cytokines to their cognate receptor leads to activation of the Jak-signal transducers and activators of transcription (STAT) signaling pathway and altered gene expression. This signal pathway has been intensely studied using human IFN-α2 and IFN-β. However, there are over 14 human IFN-α subtypes and over 10 murine IFN-α subtypes, with a single IFN-β subtype in both species. J2E cells are immortalized at the proerythroblast stage of development and produce a rapid and fatal erythroleukemia in vivo. These cells retain the ability to respond to erythropoietin in vitro by proliferating, differentiating, and remaining viable in the absence of serum. Here, we show that J2E cells are also functionally regulated differentially by IFN subtype treatment in vitro. A novel finding was the selective activation of STAT and mitogen-activated protein kinase (MAPK) molecules by different subtypes binding the IFN receptor. These findings indicate distinct effects for individual type I IFN subtypes, which are able to differentially activate members of the STAT and MAPK family. Finally, we investigated the efficacy of IFN naked DNA therapy in treating J2E-induced erythroleukemia in athymic nude mice. IFN subtypes differentially regulated the onset of erythroleukemia with delayed onset and increased survival, possibly via a reduction in cell viability, and enhanced antiproliferative and apoptotic effects observed for IFNA6 and IFNA9treatment, respectively. Moreover, these data highlight the necessity to choose the best IFN subtype in disease treatment.
Introduction
Erythroleukemias represent the highly malignant M6 subtype of acute myeloid leukemia.1 The J2E cell line, immortalized at the proerythroblast/basophilic erythroblast stage of erythroid development,2 induces a rapid and fatal erythroleukemia in mice.3 However, while these cells are immortalized, they are still capable of normal signaling in response to erythropoietin (Epo) stimulation. They maintain all biologic responses to Epo by differentiating biochemically with hemoglobin production and undergoing cellular alterations with a percentage of cells enucleating to form mature reticulocytes.2,4-7 J2E cells also display increased proliferation and enhanced viability in the absence of serum as a result of Epo stimulation.2,4 5
Type I interferon (IFN) subtypes are clinically effective in the treatment of disease conditions such as hepatitis, hairy cell leukemia, condyloma acuminatum, multiple sclerosis, and Kaposi sarcoma.8-11 These IFNs have also proven effective in the treatment of both myelogenous and metastatic tumors,12,13and IFN-α has the ability to normalize hemoglobin levels in anemic patients.14 Subtype diversity for treatment is limited however to human IFN-α2 (huIFN-α2)15 and huIFN-β (IFN-β-1b; IFN-β-1a).
The type I IFNs have been attributed to multiple and diverse functions in the immune response. This multigene family has over 14 IFN-α subtypes in humans and 10 IFN-α subtypes in mice, with a single IFN-β subtype for each. The murine and human IFN gene families are highly analogous.16 Induced early in the immune response, IFNs stimulate antiviral,17,18antiproliferative,19 and apoptotic activity,20 as well as have numerous immunomodulatory effects. Type I IFNs regulate dendritic cell major histocompatibility complex (MHC) expression and maturation,21-24activate natural killer (NK) cells,25 induce bystander T-cell proliferation,26 selectively induce clonal CD8+ T-cell expansion,26 and skew the immune response to a Th1-like response.27 28
At the cellular level, type I IFN binds the IFN-α receptor 2 (IFNAR2) subunit29 instigating its association with the IFNAR1 subunit.30 The preassociated Jak kinases, Jak1 and Tyk2, are subsequently cross-phosphorylated.31,32Phosphorylation of signal transducers and activators of transcription 2 (STAT2) occurs, which forms a heterodimer with STAT1 and phosphorylation of the latter. Phosphorylation of other STATs in response to IFN-α include STAT3,33 STAT4,34STAT5, and STAT6.35,36 Phosphorylation of STAT4 however is not generally reported in the murine system and is believed to be due to a microsatellite insertion in Stat2.37 Type I IFN induces the formation of both homodimers (STAT1,38STAT3,39 and STAT5) and heterodimers (STAT1/STAT238 and STAT1/STAT340). These STAT dimers are translocated to the nucleus where they bind type I IFN-stimulated response elements leading to gene transcription. Downstream STAT1/STAT2 heterodimers bind p48 to form the mature IFN-stimulated gene factor 3 (ISGF3) complex, a p38 mitogen-activated protein kinase (MAPK)–dependent process.41,42 During signaling for growth inhibition, IFN-α activates the Rac1/p38 MAPK pathway,43 while recombinant huIFN-α2 has been shown to activate JNK-1 and p38 MAPK in apoptotic cell death.44 Alternatively, IFN-α activation of p42 MAPK is essential for cell proliferation.45
Previously, J2E cells stimulated with mixed murine IFN-α (muIFN-α), IFN-α1, and IFN-α4 differed in their antiproliferative capacity,46 while levels of Epo-induced differentiation were maintained.47 In this report, we investigated the differential antileukemic properties of a panel of 7 type I IFN subtypes (-α1, -α2, -α4, -α5, -α6, -α9, and -β) using delivery of an expression vector encoding the IFN subtype gene. Individual IFN subtypes were studied in vitro for effect on erythroid cell proliferation, differentiation, viability, and apoptosis. Members of the STAT and MAPK signaling families in J2E cells were examined for differential activation/phosphorylation in response to individual IFN subtypes. Finally, the efficacy of IFN naked DNA treatment was assessed in the development of J2E cell-induced erythroleukemia in vivo.
Materials and methods
Cell culture
J2E cells2 were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (GIBCO BRL, Mount Waverly, Victoria, Australia) at 37°C in a humidified atmosphere of 5% CO2. Cells were stimulated with IFN (1-1000 IU/mL) determined by antiviral bioassay, as indicated. For signaling and viability experiments, cells were washed 3 times to be free of fetal calf serum (FCS) and incubated 5 hours at 37°C with 5% CO2 prior to stimulation with interferon. Cell viability was determined by 0.4% eosin exclusion, while hemoglobin production was detected by benzidine staining.48
Measurement of apoptosis
Apoptosis was measured by TdT-mediated dUTP nick end labeling (TUNEL) assay (Apoptosis Detection Kit; TA200; R&D Systems, Minneapolis, MN) and by DNA integrity.49TUNEL assays were performed according to the manufacturer's instructions, staining with anti-BrdU antibody followed by streptavidin horseradish peroxidase and diaminobenzidine (DAB) substrate development. Cells were counterstained with hematoxylin, mounted, and viewed with light microscopy. DNA integrity was determined by extraction of DNA by the proteinase K method.50 DNA concentration was determined by spectrophotometry at 260 nm before electrophoresis using a 1% agarose gel.
Protein analyses
Proteins were extracted in lysis buffer (150 mM NaCl2, 20 mM Tris, pH 7.5, 1% Triton X-100 [vol/vol], 40 mM Na4P2O7, 1 mM Na3VO4, 50 mM NaF, 5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/mL aprotinin, 10 μg/mL leupeptin, and 1 mM benzamidine) and samples (100 μg) immunoblotted with antiphospho-STAT1, -STAT3, -STAT5, and -STAT6 antibodies (phospho-STAT antibody sampler, no. 9914; New England Biolabs, Beverly, MA) or antiphospho-JNK, -p38, -p42, or -p44 MAPK antibodies (phospho-MAPK antibody sampler, no. 9910; New England Biolabs). Proteins (2 mg/mL) were immunoprecipitated with anti-STAT2 and -STAT4 antibodies (sc-476, sc-478; Santa Cruz Biotechnology, Santa Cruz, CA) and immunoblotted with antiphosphotyrosine IgG (no. 05-321, clone 4G10; Upstate Biotechnology, Lake Placid, NY). Antibody binding was determined by enhanced chemiluminescence (ECL; Pharmacia Biotech, Buckinghamshire, United Kingdom) and visualized by autoradiography. Levels of p42 MAPK (sc-154; Santa Cruz Biotechnology) were used as a loading control in each experiment.
Northern analysis of double-stranded (ds)RNA-activated protein kinase (PKR) and 2′5′ oligoadenylate synthetase (2′5′OAS)
Total RNA from J2E cells was extracted using TRIZOL (GIBCO BRL), in accordance with the manufacturer's instructions, and RNA samples (20 μg) were electrophoresed on formaldehyde-agarose gels prior to transfer to nylon membranes (MSI, Westborough, MA) in 10 × SSC (1.5 M sodium chloride, 150 nM trisodium citrate, pH 7.0). Full-length murine cDNA (100 ng) for PKR (1.4 kilobase [kb]) and 2′5′OAS (1.8 kb), obtained from plasmids provided by B. Williams (Cleveland Clinic Foundation, Cleveland, OH), or glyceraldehyde phosphate dehydrogenase (GAPDH) was 32P-labeled using a Gigaprime labeling kit (Geneworks, Adelaide, Australia). Membranes were hybridized to α-32P-labeled probes at 65°C in 6 × sodium chloride tri-sodium (SSC); 10 nM sodium phosphate buffer, pH 7.0; 1 mM ethylenediaminetetraacetic acid; 1 × Denhardt; 5% dextran sulfate (wt/vol); 0.1% sodium dodecyl sulfate (SDS); and 100 μg/mL herring sperm DNA for 18 hours. Following hybridization, membranes were washed to 0.2 × SSC/0.1% SDS (wt/vol) and visualized by autoradiography.
Preparation of interferons
IFN DNA constructs comprising IFN-A1, -A2, -A4, -A5, -A6, -A9, and -B genes cloned into the mammalian expression vector pkCMVint.polylinker (VICAL, San Diego, CA) have been described elsewhere.51 Plasmid DNA was prepared from Terrific Broth culture of transformed Escherichia coli (DH5α; Life Technologies, Grand Island, NY) using standard extraction procedures with LiCl precipitation. Concentration and purity of plasmid DNA was determined by spectrophotometric analysis at 260 nm, with a 260:280 nm ratio of 1.6 or higher.
IFN protein was prepared from transfected COS-7 cells expressing the individual IFN DNA constructs. In brief, COS-7 cells were calcium phosphate-transfected with 20 μg plasmid DNA.52 Cells were cultured in 5% FCS or serum-free conditions for 24 or 48 hours. Supernatants were harvested, pH 2 acid–treated to remove acid-labile IFNs, and IFN content determined by titration against the international standard IFN-α/β (Lee Biomolecular, San Diego, CA; 1000 IU/mL) using encephalomyocarditis virus–infected L929 cells in a bioassay as previously described.53 All COS-7 cells transfected with the IFN-A/B transgenes showed secretion of various amounts of IFN-α (pg/mL) by enzyme-linked immunosorbent assay (ELISA) using an antimurine IFN-α antibody kit (PBL Biomedical Laboratories, Piscataway, NJ). Though cells transfected with the blank vector or IFN-βtransgene secreted nominal amounts of IFN-α. The 6 alpha IFN subtypes investigated in this study were equated to pg/1000 IU based on these assays (IFN-α1, 2583 pg; IFN-α2, 1549 pg; IFN-α4, 1294 pg; IFN-α5, 3111 pg; IFN-α6, 371pg; and IFN-α9, 94 pg).
Naked DNA treatment
Specific pathogen-free, 4-week-old homozygousnu/nu BALB/c mice (Animal Resources Centre, Murdoch, Western Australia) were inoculated bilaterally in each tibialis anterior (TA) muscle with 100 μg plasmid DNA (vehicle or encoding type I IFN subtype) in a 25 μL volume, as previously described.51,53 Circulating expression levels of IFN transgenes in vivo have previously been found to be similar, with an average of 20 IU/mL sera.49
Erythroleukemia
At 14 days following DNA treatment, mice were inoculated with 1 × 106 J2E cells by the intravenous route as described previously,3 and culled as they became moribund. Hematocrit readings were taken via tail-vein bleeds prior to culling, and spleen and liver weights were determined at autopsy.
Statistical analyses
Significant differences between means were determined using a 2-tailed Student t test (P ≤ .05). Statistical significance between cumulative survival rates of animals was determined by the Cox proportional hazards regression model.
Results
Antiproliferative effects of IFN stimulation of J2E cells
Type I IFNs were initially examined for their antiproliferative effect on J2E cell growth via IFN dose-response analysis (1-1000 IU/mL) during 48 hours of cell culture. All IFN subtypes examined (IFN-α1, -α2, -α4, -α5, -α6, -α9, and -β) reduced J2E cell number (Figure 1) with IFN-α1, -α4, -α5, and -α6 acting in a dose-dependent manner. Interestingly, suppression of J2E cell growth required a higher dose of IFN-α2 and -β (100-1000 IU/mL) while very low concentrations of IFN-α9 (1 IU/mL) significantly reduced cell number after 48 hours of culture.
Type I IFN dose-response analysis of J2E cell growth.
J2E cells were seeded at 5 × 104 cells/mL and stimulated with IFN-α1, -α2, -α4, -α5, -α6, -α9, or -β (1-1000 IU/mL) as indicated. Cell number was determined by eosin exclusion at 48 hours of culture (statistical significance compared with unstimulated J2E cells, *P ≤ .05; **P ≤ .01; mean ± SE, n = 3).
Type I IFN dose-response analysis of J2E cell growth.
J2E cells were seeded at 5 × 104 cells/mL and stimulated with IFN-α1, -α2, -α4, -α5, -α6, -α9, or -β (1-1000 IU/mL) as indicated. Cell number was determined by eosin exclusion at 48 hours of culture (statistical significance compared with unstimulated J2E cells, *P ≤ .05; **P ≤ .01; mean ± SE, n = 3).
Individual IFN subtype efficacies of J2E cell differentiation
J2E cells respond to Epo stimulation by synthesizing hemoglobin. Therefore we next determined the effect of IFN stimulation, in the absence of Epo, on differentiation of J2E cells (Figure2). IFN-α4 was the only IFN subtype that significantly enhanced J2E cell differentiation at 100 IU/mL, while stimulation at 1000 IU/mL induced significant increases in differentiation of J2E cells with all the IFNs subtypes (up to 5.5-fold with IFN-α1) except IFN-α9 and -β. The maturation of J2E cells in the presence of IFN was therefore dependent on the IFN subtype chosen.
Effect of type I IFNs on J2E erythroid cell differentiation.
J2E cells were seeded at 5 × 104 cells/mL and stimulated with IFN subtypes at (A) 100 and (B) 1000 IU/mL. Hemoglobin production was measured by benzidine staining at 48 hours of culture. (statistical significance compared with unstimulated J2E cells, *P ≤ .05; **P ≤ .01; mean ± SE, n = 3).
Effect of type I IFNs on J2E erythroid cell differentiation.
J2E cells were seeded at 5 × 104 cells/mL and stimulated with IFN subtypes at (A) 100 and (B) 1000 IU/mL. Hemoglobin production was measured by benzidine staining at 48 hours of culture. (statistical significance compared with unstimulated J2E cells, *P ≤ .05; **P ≤ .01; mean ± SE, n = 3).
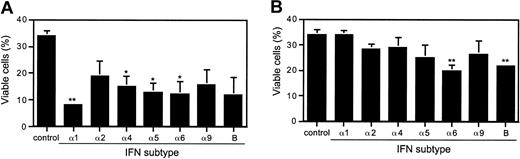
Differential IFN subtype efficacies of J2E cell viability and apoptosis
Since type I IFNs are known inducers of apoptotic cell death, we therefore investigated the effect of the IFN subtypes on J2E cell viability and apoptosis. Cells stimulated with 100 IU/mL of IFN-α1, -α4, -α5, and -α6 all showed a significant reduction in J2E cell viability by 75.6%, 55.3%, 61.8%, and 63.8%, respectively (Figure 3). In direct contrast, however, when using higher concentrations of IFN (1000 IU/mL), only IFN-α6 and -β significantly reduced J2E cell viability. We speculate that the reduced level of cell death at high IFN concentration may be due to the necessity for maintaining circulating erythrocyte numbers during distress or infection.
Effect of type I IFNs on J2E erythroid cell viability.
J2E cells were washed free of serum, seeded at 3 × 105cells/mL, and stimulated with IFN subtypes at (A) 100 or (B) 1000 IU/mL. Percentage viable cells was determined by eosin exclusion at 48 hours of culture (statistical significance compared wtih unstimulated J2E cells, *P ≤ .05; **P ≤ .01; mean ± SE, n = 3).
Effect of type I IFNs on J2E erythroid cell viability.
J2E cells were washed free of serum, seeded at 3 × 105cells/mL, and stimulated with IFN subtypes at (A) 100 or (B) 1000 IU/mL. Percentage viable cells was determined by eosin exclusion at 48 hours of culture (statistical significance compared wtih unstimulated J2E cells, *P ≤ .05; **P ≤ .01; mean ± SE, n = 3).
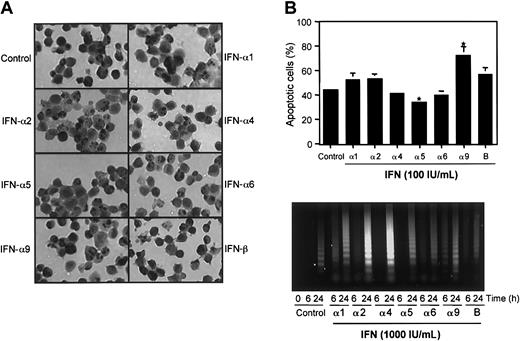
Next, IFN-induced apoptosis was examined in J2E cells by TUNEL assay following stimulation with 100 IU/mL IFN (Figure 4A-B). A significant enhancement in the rate of apoptosis was observed for IFN-α9 (71.3%) with a moderate increase detected for IFN-β (57.6%) in contrast to control, unstimulated cultures (44.7%). Notably IFN-α5 significantly reduced apoptotic cell death to 34.3%, a 10.4% reduction compared with control cells. These results indicate differential apoptotic potential of individual IFN subtypes in J2E cells.
Apoptosis was also measured by DNA degradation in response to J2E cell stimulation with 1000 IU/mL of the IFN subtypes (Figure 4C). Interestingly, DNA fragmentation at 6 hours after IFN stimulation was minimal for all subtypes examined (IFN-α5 at 6 hours, not done), with the exception of IFN-α9. At 24 hours, stimulation with IFN-α2 and -α4 (1000 IU/mL) increased while IFN-β reduced DNA fragmentation. These data indicate that while lower doses of IFN-α9 or IFN-α5 may enhance or reduce apoptosis, it appears that high concentrations of IFN potentiate the onset of apoptosis of J2E cells cultured in the absence of serum.
Effect of type I IFNs subtypes on J2E cell apoptosis.
(A) Apoptosis of J2E cells in response to stimulation with IFN subtypes (100 IU/mL), in the absence of serum, at 24 hours by TUNEL assay. Apoptotic cell bodies stained with diaminobenzidine (DAB, black) and cells counterstained with hematoxylin (gray). Original magnification, × 40. (B) The percentage of positive staining apoptotic cells was determined by light microscopy. (Statistical significance compared with unstimulated J2E cells, *P ≤ .05, mean ± SE, n = 3.) (C) At 6 and 24 hours after stimulation with 1000 IU/mL IFN, in the absence of serum, DNA fragmentation (≤ 6 kb) was determined in J2E cells. Control cultures were unstimulated.
Effect of type I IFNs subtypes on J2E cell apoptosis.
(A) Apoptosis of J2E cells in response to stimulation with IFN subtypes (100 IU/mL), in the absence of serum, at 24 hours by TUNEL assay. Apoptotic cell bodies stained with diaminobenzidine (DAB, black) and cells counterstained with hematoxylin (gray). Original magnification, × 40. (B) The percentage of positive staining apoptotic cells was determined by light microscopy. (Statistical significance compared with unstimulated J2E cells, *P ≤ .05, mean ± SE, n = 3.) (C) At 6 and 24 hours after stimulation with 1000 IU/mL IFN, in the absence of serum, DNA fragmentation (≤ 6 kb) was determined in J2E cells. Control cultures were unstimulated.
IFN subtypes activate different members of the STAT family
Having established different IFN effects on J2E cell proliferation, differentiation, viability, and apoptosis, we next examined signaling pathways that may be involved in this process. First, we analyzed the activation of STAT molecules in response to stimulation with the IFN subtypes. Phosphorylation of STAT molecules in response to IFN (100 IU/mL) was examined using antiphospho-STAT antibodies. Tyrosine phosphorylation of STAT1 was induced in response to IFN-α1, -α2, -α4, and -α5, while tyrosine phosphorylation of STAT3 was induced in response to IFN-α1, with partial activation in response to IFN-α4, -α5, and -β at 15 minutes after stimulation (Figure 5A). Tyrosine phosphorylation of STAT5a, STAT5b, or STAT6 (data not shown) and serine phosphorylation of STAT3 was not detected at this low concentration of IFN (Figure 5A).
Phosphorylation of STAT and MAPK family members in response to IFN stimulation of J2E cells.
Cells were serum starved for 5 hours prior to stimulation with IFN subtypes (100-1000 IU/mL). Total cell protein (100 μg) was extracted at the indicated times and immunoblotted with (A) antiphospho-STAT1 (STAT1-Y) and antiphospho-STAT3 (STAT3-Y, STAT3-S) antibodies; (B) antiphospho-STAT1 (STAT1-Y), antiphospho-STAT3 (STAT3-Y, STAT3-S), antiphospho-STAT5 (STAT5a-Y), and antiphospho-STAT6 (STAT5b-Y) antibodies; and (C) antiphospho-p38, antiphospho-p42, and antiphospho-p44 MAPK antibodies. Immunoblot with anti-p42 MAPK antibodies indicates protein loading.Y- denotes tyrosine; S-, serine.
Phosphorylation of STAT and MAPK family members in response to IFN stimulation of J2E cells.
Cells were serum starved for 5 hours prior to stimulation with IFN subtypes (100-1000 IU/mL). Total cell protein (100 μg) was extracted at the indicated times and immunoblotted with (A) antiphospho-STAT1 (STAT1-Y) and antiphospho-STAT3 (STAT3-Y, STAT3-S) antibodies; (B) antiphospho-STAT1 (STAT1-Y), antiphospho-STAT3 (STAT3-Y, STAT3-S), antiphospho-STAT5 (STAT5a-Y), and antiphospho-STAT6 (STAT5b-Y) antibodies; and (C) antiphospho-p38, antiphospho-p42, and antiphospho-p44 MAPK antibodies. Immunoblot with anti-p42 MAPK antibodies indicates protein loading.Y- denotes tyrosine; S-, serine.
Stimulation of J2E cells with IFN at 1000 IU/mL was next examined for phosphorylation of STAT molecules (Figure 5B). Tyrosine phosphorylation of STAT1 was induced in J2E cells in response to all 7 IFN subtypes. IFN-α1 and -α4 (1000 IU/mL) induced tyrosine phosphorylation of STAT5a and STAT5b. Tyrosine phosphorylation of STAT3 was not detected with IFN stimulation at 1000 IU/mL, however, serine phosphorylation of STAT3 was detected in response to all subtypes with an elevated level of phosphorylation for IFN-α9 and IFN-β (Figure 5B). STAT2 was tyrosine phosphorylated in response to all subtypes at both IFN concentrations (100 and 1000 IU/mL), while phosphorylation of STAT4 was not detected (data not shown).
In summary, stimulation of J2E cells with the IFN subtypes led to differential phosphorylation/activation of STAT molecules that altered with the concentration of stimulating cytokine. While STAT1 and STAT2 were phosphorylated at high and low concentrations for all IFN subtypes examined, STAT3 tyrosine and serine phosphorylation was subtype- and concentration-dependent. Tyrosine phosphorylation has been found to be important for dimerization of STAT proteins, whereas serine phosphorylation is required for maximum transcriptional activation mediated by some STAT proteins, including STAT3.54 The specific activation of STAT5a and STAT5b with IFN subtypes at 1000 IU/mL was an unexpected finding and interestingly correlated with increased hemoglobin-positive staining in J2E cells stimulated with IFN. Indeed, STAT5b activation has been shown to be critical for differentiation of erythroid cells.55 56
Differential activation of MAPK family members by IFN-α and -β subtypes
As stimulation of J2E cells with low concentrations of IFN (100 IU/mL) activated a limited panel of STAT molecules, we therefore examined the phosphorylation of MAPK family members in response to IFN stimulation. Phosphorylation of JNK was detected upon stimulation with IFN-β only and not with any of the other IFN-α subtypes (data not shown). However, p38 MAPK was activated with all of the IFN-α subtypes but not with IFN-β, while p42 and p44 MAPK were activated by all IFN-α and IFN-β subtypes (Figure 5C). Thus the MAPK family members displayed distinct activation patterns compared with the STAT molecules.
Type I IFNs activate ISGs
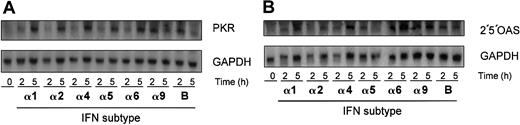
Type I IFNs are known to activate interferon-stimulated genes (ISGs), therefore we examined the expression of PKR and 2′5′OAS. Stimulation of J2E cells with all IFN subtypes led to the activation of PKR by 5 hours, with a higher expression of PKR detected at 2 hours for IFN-α9 and IFN-β subtypes (Figure6A). Similarly, all IFN subtypes activated 2′5′OAS over 2 to 5 hours (Figure 6B). These data indicate that IFN (100 IU/mL) effectively induced gene expression of PKR and 2′5′OAS in the J2E cell system.
Type I IFNs stimulate expression of antiviral ISGs in J2E cells.
J2E cells were seeded at 1 × 105 cells/mL, stimulated with type I IFNs (100 IU/mL), and RNA extracted at indicated time points. RNA (20 μg) was electrophoresed on formaldehyde gels, Northern blotted, and hybridized with (A) PKR or (B) 2′5′OAS. GAPDH loading control as indicated.
Type I IFNs stimulate expression of antiviral ISGs in J2E cells.
J2E cells were seeded at 1 × 105 cells/mL, stimulated with type I IFNs (100 IU/mL), and RNA extracted at indicated time points. RNA (20 μg) was electrophoresed on formaldehyde gels, Northern blotted, and hybridized with (A) PKR or (B) 2′5′OAS. GAPDH loading control as indicated.
IFN therapy for J2E cell-induced erythroleukemia
J2E cells induce a rapid fatal erythroleukemia, following intravenous injection into mice, which is characterized by severe anemia and hepatosplenomegaly.3 Previously, it has been shown that altering signaling molecules can impede the progression of erythroleukemia induced by J2E cells.6,57 Therefore, since the IFN subtypes varied in their antigrowth, differentiation, cell viability, apoptotic response, and induced altered signaling pathways in J2E cells, we investigated their therapeutic potential for J2E cell-induced erythroleukemia. Mice were injected with IFN plasmid DNA into the TA muscle 2 weeks prior to intravenous injection of J2E cells, thus allowing expression of the transgene in vivo.51Animals were culled as they became moribund and the degrees of anemia, hepatosplenomegaly, and tumor burdens were monitored.
All IFN-α subtypes delayed the onset of mortality compared with the death rate of mice inoculated with the control vector (vehicle). Notably, IFN-A1, IFN-A6, and IFN-A9 delayed the onset of mortality by 12 days compared with the vehicle-treated mice, which commenced dying at day 35 (P ≤ .1, Figure7). In addition, survival rates of mice following DNA treatment with IFN-A6 and IFN-A9were 60% (P ≤ .1) and 40% (P ≤ .1), respectively, compared with 0% survival of control mice. In contrast,IFN-A2 increased the rate of death induced by J2E cells with all mice killed by day 49. IFN-B treatment, however, neither delayed onset of mortality nor increased survival rate compared with treatment with the vehicle. Thus, specific IFN-α subtypes were effective in extending the time to disease onset and life span of leukemia-cell recipients.
Effect of IFN transgene expression on the onset of J2E cell-induced erythroleukemia.
Mice were vaccinated bilaterally with 100 μg IFN DNA in the TA muscle and 14 days later injected with 1 × 106 J2E cells via the tail vein. The cumulative survival was calculated as the mice became moribund (n = 5-7). Vehicle-treated mice were injected with vector DNA without IFN gene cassette.
Effect of IFN transgene expression on the onset of J2E cell-induced erythroleukemia.
Mice were vaccinated bilaterally with 100 μg IFN DNA in the TA muscle and 14 days later injected with 1 × 106 J2E cells via the tail vein. The cumulative survival was calculated as the mice became moribund (n = 5-7). Vehicle-treated mice were injected with vector DNA without IFN gene cassette.
Reduced hematocrit readings indicating anemia and the presence of hepatosplenomegaly are indicators of erythroleukemia development. Therefore, hematocrit readings, and liver and spleen weights for normal, control, and IFN-treated mice were monitored (Table1). The vehicle-treated group developed typical J2E-induced erythroleukemia with characteristic hepatosplenomegaly. In the moribund mice, hepatomegaly was apparent forIFN-A1 (P ≤ .05), IFN-A4(P ≤ .05), and IFN-B (P ≤ .01) DNA treatment while splenomegaly was observed for IFN-A1(P ≤ .01), IFN-A2 (P ≤ .05),IFN-A6 (P ≤ .05), IFN-A9(P ≤ .05), and IFN-B (P ≤ .01) DNA treatment. However, healthy mice sampled at day 83 (end of experiment) had normal spleen weights for IFN-A6 andIFN-A9 DNA treatment groups, 0.16 g ± 0.05 g and 0.34 g ± 0.07 g, respectively. As expected, hematocrit readings were reduced at time of death for vehicle-, IFN-A2, IFN-A4, IFN-A5, and IFN-B DNA-treated mice compared with normal mice. Interestingly, hematocrit readings for DNA treatment with effective subtypes IFN-A1, IFN-A6, andIFN-A9 were not significantly altered from control animals. Indeed, hematocrit readings for survivors of IFN-A6 andIFN-A9 DNA treatment groups at day 83 were normal, at 56.6% ± 1.3 and 48.9% ± 1.1, respectively.
Liver, spleen, and hematocrit readings near death in J2Eand IFN-treated mice
| Treatment group . | Liver, g . | Spleen, g . | Hematocrit, % . |
|---|---|---|---|
| Control | 1.30 ± 0.02 | 0.135 ± 0.007 | 52.5 ± 0.8 |
| Vehicle | 1.90 ± 0.12* | 0.636 ± 0.155* | 39.5 ± 2.6† |
| A1 | 1.79 ± 0.04* | 0.543 ± 0.072† | 44.9 ± 2.9 |
| A2 | 1.60 ± 0.24 | 0.993 ± 0.222* | 36.7 ± 2.1† |
| A4 | 2.27 ± 0.14* | 0.666 ± 0.229 | 44.1 ± 1.5* |
| A5 | 2.59 ± 0.58 | 1.130 ± 0.336 | 36.1 ± 2.7* |
| A6 | 1.99 ± 0.25 | 0.902 ± 0.232* | 42.0 ± 5.8 |
| A9 | 1.58 ± 0.10 | 0.616 ± 0.232* | 42.8 ± 2.8 |
| B | 1.83 ± 0.09† | 0.392 ± 0.066† | 44.1 ± 2.5* |
| Treatment group . | Liver, g . | Spleen, g . | Hematocrit, % . |
|---|---|---|---|
| Control | 1.30 ± 0.02 | 0.135 ± 0.007 | 52.5 ± 0.8 |
| Vehicle | 1.90 ± 0.12* | 0.636 ± 0.155* | 39.5 ± 2.6† |
| A1 | 1.79 ± 0.04* | 0.543 ± 0.072† | 44.9 ± 2.9 |
| A2 | 1.60 ± 0.24 | 0.993 ± 0.222* | 36.7 ± 2.1† |
| A4 | 2.27 ± 0.14* | 0.666 ± 0.229 | 44.1 ± 1.5* |
| A5 | 2.59 ± 0.58 | 1.130 ± 0.336 | 36.1 ± 2.7* |
| A6 | 1.99 ± 0.25 | 0.902 ± 0.232* | 42.0 ± 5.8 |
| A9 | 1.58 ± 0.10 | 0.616 ± 0.232* | 42.8 ± 2.8 |
| B | 1.83 ± 0.09† | 0.392 ± 0.066† | 44.1 ± 2.5* |
As mice became moribund, blood was harvested by tail bleed; spleen and liver were removed immediately following death and weighed. Control mice received no treatment and were killed after the last mouse became moribund.
Values represent mean ± standard error.
Significance compared with normal mice, P < .05
P ≤.01
Numerous tumors were observed in the lymph nodes ofvehicle-, IFN-A1–, IFN-A2–, and IFN-B–treated mice (Table 2). IFN-A9treament was not associated with tumor involvement of lymph nodes. Furthermore, neither IFN-A6- nor IFN-A9DNA-treated mice showed evidence of multiple tumors that was observed in the vehicle-treated mice. Interestingly, introduction ofIFN-B also induced cachexia in 2 mice. These data therefore indicate a possible influence of IFN subtypes on tumor homing in the mice.
Tumor localization in J2E- and IFN-treated moribund mice
| Treatment group . | Cachexia, n . | Axillary, n . | Inguinal, n . | Cervical, n . | Total, n/N . |
|---|---|---|---|---|---|
| Normal | 0 | 0 | 0 | 0 | 0/5 |
| Vehicle | 0 | (1) | 0 | (1) | 1/5 |
| A1 | 0 | 0 | (3) | (3) | 3/6 |
| A2 | 0 | 0 | 0 | 3 | 3/4 |
| A4 | 0 | 1 | 0 | 0 | 1/5 |
| A5 | 0 | 0 | 1 | 0 | 1/4 |
| A6 | 0 | 0 | 1 | 0 | 1/5 |
| A9 | 0 | 0 | 0 | 0 | 0/6 |
| B | 2 | 0 | 1(1) | 1(1) | 5/7 |
| Treatment group . | Cachexia, n . | Axillary, n . | Inguinal, n . | Cervical, n . | Total, n/N . |
|---|---|---|---|---|---|
| Normal | 0 | 0 | 0 | 0 | 0/5 |
| Vehicle | 0 | (1) | 0 | (1) | 1/5 |
| A1 | 0 | 0 | (3) | (3) | 3/6 |
| A2 | 0 | 0 | 0 | 3 | 3/4 |
| A4 | 0 | 1 | 0 | 0 | 1/5 |
| A5 | 0 | 0 | 1 | 0 | 1/4 |
| A6 | 0 | 0 | 1 | 0 | 1/5 |
| A9 | 0 | 0 | 0 | 0 | 0/6 |
| B | 2 | 0 | 1(1) | 1(1) | 5/7 |
As mice became moribund they were killed and autopsied.
Numbers in parentheses indicate mice with multiple tumors.
Discussion
The recent establishment, in this laboratory, of a large panel of murine IFN-α subtypes and IFN-β for DNA gene therapy51and IFN protein production51,58 has provided an avenue through which to explore the anticancer effects of this multigene cytokine family. The divergent antiviral effects of the IFN subtypes are evident against numerous viruses including murine cytomegalovirus,51,58 herpes simplex virus type 1 (HSV-1),59 and HSV-2.60 Furthermore, covaccination of IFN subtypes with viral vaccines produces differential protective effects on viral infection.51 To our knowledge this is the first investigation into the antileukemic effects of such a broad panel of IFN subtypes on the development of erythroleukemia.
The antiproliferative effects of type I IFNs have been established in numerous and diverse cancers and cell lineages.15,61Specifically in erythroid cells, constitutive expression of consensus type I IFN (IFN-con1) has been shown to revert the malignant phenotype in vitro as indicated by growth inhibition in culture.62Previous reports in erythroid cells have shown antiproliferative effects with mixed IFN-α, IFN-α1, and IFN-α4.46,47We report here, the antiproliferative effects of 7 type I IFNs on erythroid J2E cells. In this study, all IFN subtypes examined induced an antiproliferative response in J2E cells, with cell numbers reduced by 51.3% to 67.9%, and IFN stimulation at 100 IU/mL, which was further reduced by 66.1% to 89.4% using 1000 IU/mL. Significantly, IFN-α9 was able to effectively reduce cell proliferation at 1 IU/mL. Previously, J2E cell growth inhibition has been observed at 10 IU/mL when stimulated with mixed muIFN-α. Studies in other erythroid systems have indicated the effectiveness of as little as 0.01 IU/mL recombinant IFN-α2a in suppressing erythroblast numbers, while alternative IFNs, including recombinant IFN-α2b, hybrid IFN-α1-8, and IFN-con1, were not effective at this dose.63
J2E cells respond to the hormone Epo with increased differentiation, with a proportion of cells enucleating to form mature reticulocytes.64 Prior findings have indicated that mixed muIFN-α (100-300 IU/mL) stimulation of J2E cells did not intervene with Epo-induced J2E cell differentiation.47 However, in this study, in the absence of Epo, IFN-α4 (100 IU/mL) was able to significantly increase the percentage of hemoglobin-positive J2E cells, while higher doses of IFN-α1 and -α4 (1000 IU/mL) induced 5-fold and 4-fold enhancement of differentiation, respectively. Interestingly, decreased cell proliferation has been shown to potentiate Epo-induced differentiation.4,65 In the absence of Epo, chemical induction of J2E cells with sodium butyrate severely restricts proliferation while stimulating hemoglobin production in up to 100% of cells.66 Data presented here indicate that stimulation of J2E cells with specific IFN subtypes elevates the number of resting hemoglobin-positive cells. However, decreased rate of cell growth alone was not sufficient to induce cell differentiation, indicating that other intracellular mechanisms are at hand in this process.
In addition to differentiation and proliferation, J2E cells respond to Epo with a third biologic function, namely enhanced viability.5 Recent reports have indicated that type II IFN (IFN-γ) promotes enhanced viability in peripheral blood cells and stimulates the survival of erythroid colony-forming cells.67 We therefore investigated the differential effects of type I IFN on the viability of J2E erythroid cells in vitro. Low concentrations (100 IU/mL) of IFN-α1, -α4, -α5, and -α6 reduced J2E cell viability, while stimulation with higher doses of IFN (1000 IU/mL) did not reduce J2E cell viability with the exception of IFN-α6 and -β treatment. In response to localized infection, IFN is secreted to stimulate a danger signal and subsequent initiation of an innate immune response.68 Elevated doses of IFN may be indicative of systemic infection, a situation in which decreased circulating erythrocytes would be detrimental to the individual. Indeed, therapy with high- dose IFN-α has proved effective in normalizing hemoglobin levels in anemic patients.14
The IFNs are inducers of apoptosis in malignancies including herpesvirus-associated lymphomas, acute promyelocytic leukemia, non–small cell lung cancer, nonmelanoma skin cancer, and glioma.20 Early findings of growth inhibition in erythroid progenitor cells implicated an apoptotic mechanism via DNA fragmentation.69 At low IFN concentrations (100 IU/mL), apoptosis in the J2E cell was enhanced by IFN-α9, and partially but significantly reduced with IFN-α5 treatment. At higher concentrations of IFN (1000 IU/mL), all IFN-α subtypes induced DNA fragmentation. IFN-β however, had minimal apoptotic activity on J2E cells. Similarly, other researchers have shown decreased rates of hematopoietic cell proliferation in the presence or absence of apoptosis.70 71
Stimulation of J2E cells with the type I IFNs led to differential effects at the cellular level, presumably via stimulation of various signaling cascades following activation of the IFN-α receptor (IFNAR). Surprisingly, the majority of signaling studies have used a limited range of type I IFN subtypes. Divergence between IFN-α and IFN-β signaling can be observed at the levels of Jak/STATs,72,73 Ras/Raf/MAPK,45,74,75IRS-1,76,77 and CrkL phosphorylation.73 Most studies in humans have concentrated on signal events stimulated by either unspecified IFN-α IFN-α mixtures,76,78recombinant IFN-α2,32,41-43,73,79IFN-α2c,78 IFN-α A/D,76 IFN-β, or IFN-β-1b.32,42,73-75,77-80 However, Hilkens et al81 have recently examined recombinant human IFN subtypes -α1, -α2, and -α21 for differential STAT1 activation and found that IFN-α1 was weaker in tyrosine phosphorylation of STAT1 than the other IFN subtypes. In the murine system, IFN signaling has been examined in response to IFN-α,37IFN-α/β mix,80 and IFN-β.9 Given that purified IFN-α mixture is predominantly IFN-α2,15 most of these studies compare only IFN-α2 and IFN-β subtypes.
IFN-α (500 IU/mL) in the human lymphoblastoid cell line Daudi activates STAT1, STAT2, STAT3, STAT5, and STAT6, and Fasler-Kan et al35 propose that cell type-specific activation of STATs could be responsible for cell type-specific responses to IFN-α.35 In the J2E erythroid system, while all IFN subtypes activated STAT2 regardless of IFN concentration, activation of other STAT family members was more specific to subtype and dosage. Low concentrations of IFN-α1, -α2, -α4, and -α5 phosphorylated STAT1, while high concentrations of all IFN subtypes induced STAT1 tyrosine phosphorylation and STAT3 serine phosphorylation. Further, low levels of IFN-α1 activated STAT3 tyrosine phosphorylation (partial activation by IFN-α4, -α5, and -β; 100 IU/mL), while high levels of IFN-α1 and -α4 activated STAT5a and STAT5b. The biologic relevance between serine and tyrosine phosphorylation of STAT3 via IFN signaling in J2E cells is undefined and warrants further investigation. Type I IFNs primarily bind the IFNAR2 subunit, which is thought to confer differential subtype recognition, followed by association with IFNAR1 that results in a high-affinity receptor complex.82-84 Data presented here comply with differential activation of the IFNAR complex with individual IFN subtypes; moreover, these receptor complexes led to the specific activation of downstream STAT molecules.
Many signaling cascades culminate in activation of members of the MAPK family. IFN-α has been shown to activate p38 MAPK43 and p42 MAPK,45 while IFN-β activates Raf-1 and p42 MAPK.74 All 6 IFN-α subtypes stimulated phosphorylation/activation of p38 MAPK, while only IFN-β activated JNK phosphorylation. Both IFN-α and -β induced p42 MAPK and p44 MAPK phosphorylation. Other studies have shown that activation of MAPK family members is necessary for STAT activation.43 Data presented here indicate phosphorylation of p42 MAPK and p44 MAPK; however relevance to STAT activation is an area for further investigation.
PKR has antiviral85 as well as antitumor activities86,87 and is firmly established as an IFN-inducible gene.88 PKR modulates cytokine signaling and transcriptional activation via NF-κB,89STAT,90 and c-Myc91 and is implicated in regulation of cell growth and apoptosis. Similarly, 2′5′OAS, another IFN-induced protein, is implicated in suppression of cell growth.92 All 7 type I IFN subtypes examined in the present study induced the expression of both PKR and 2′5′OAS, indicating a potential mechanism for the suppression of cell growth in the J2E erythroid system.
Gene therapy has been highlighted recently as an alternative form of antiviral and anticancer treatment. IFN DNA therapy has clearly demonstrated that type I IFNs differ in their antiviral capacity and modulation of the immune response.51,57-59,93 Other investigators have found that type I IFN is a clinical target with promising antileukemic potential. In cancer treatment, IFN-α electroporation gene therapy results in tumor regression.94 Splenic CTL activity against a murine colorectal adenocarcinoma cell line, MC38, was markedly enhanced with murine IFN-α2 transduction of such tumor cells.95 Mice with Friend virus–induced leukemia and treated with poly I:C, a strong inducer of type I IFNs, were shown to have higher numbers of NK cells, no evidence of erythroleukemic tumor cells, and prolonged survival.96 Furthermore, athymic nude mice treated with neutralizing antibodies to IFN-α/β have marked enhancement in growth of 6 different subcutaneous tumors.13,97 In this study, J2E cell–induced erythroleukemia responded differentially to type I IFN gene therapy, with IFN-A6 and IFN-A9most effective in prolonging animal survival (P ≤ .1) despite similar low levels of all IFN DNA transgenes expressed in vivo (10-40 IU/mL).51,58 Thus IFN subtype transgene expression in mice is biologically effective using the naked DNA delivery system. Indeed research by Piehler et al98suggested that “for maintaining transcription of IFN-responsive genes over a longer time period, low but continuous signaling through the IFN receptor is essential.”98(p40425) Since the subtypes of IFN-α have multiple properties, the antileukemic mechanisms for IFN-α6 and IFN-α9 may be pleiotropic and include both direct antitumor effects and indirect immunomodulatory effects (eg, NK cell activation and MHC class I expression). Interestingly at a low concentration in vitro, IFN-α6 was most potent at decreasing J2E cell viability, while IFN-α9 was superior at decreasing proliferation and enhancing apoptosis. Both of these IFN subtypes were shown to exert their effects accompanied by activation of PKR and 2′5′OAS in vitro, which are involved in down-regulation of expression of growth factors leading to antiproliferation and/or apoptosis. Alternatively, we have previously shown the immunomodulatory capacity of IFN-A6 andIFN-A9 against viral infection.51 It may be that effective subtypes stimulate the innate immune response and subsequent clearance of invasive erythroleukemic cells.
This is the first time that the individual type I IFN subtypes have been found to display differential antileukemic mechanisms. A summary of both the in vitro and in vivo antileukemic effects of the individual IFN subtypes used at low doses is shown in Table3. Taken together, the results indicate that IFN subtypes differentially regulate the onset of erythroleukemia, with significantly delayed onset and increased survival possibly via reduction in cell viability with IFN-A6, and enhanced antiproliferative and apoptotic effects with IFN-A9 DNA treatment. The in vivo data show that IFN-A6 andIFN-A9 treatment may inhibit the development of anemia and increase survival time. Interestingly, the results observed with IFN-α6 and IFN-α9 are similar to the effects we previously observed when the activity of the Epo receptor and its associated tyrosine kinase Lyn were altered in J2E cells.6 57 The consequence of manipulating these signaling molecules was an increased survival time in mice given J2E-induced erythroleukemia. In summary, our data indicate differential capacity of type I IFNs in immunomodulation of cellular responses such as proliferation, differentiation, viability, and apoptosis. Furthermore, the type I IFN subtypes, shown here to activate different subsets of STAT molecules, may allude to their specific recognition of the IFNAR. Finally, we demonstrate the efficacy of IFN gene therapy in the treatment of erythroleukemia, highlighting the differential capacity of IFN subtypes and the need for best subtype choice in disease treatment.
Summary of antileukemic properties for IFN subtypes
| IFN subtype . | A1 . | A2 . | A4 . | A5 . | A6 . | A9 . | B . |
|---|---|---|---|---|---|---|---|
| Antiproliferative | + | + | + | + | + | ++ | + |
| Differentiation | − | + | ++ | + | + | − | − |
| Decreased viability | ++ | + | ++ | ++ | ++ | + | + |
| Apoptosis | − | − | − | = | − | ++ | − |
| Delayed onset of erythroleukemia | ++ | + | + | + | ++ | ++ | − |
| Increased survival rate from erythroleukemia | + | − | + | + | ++ | ++ | − |
| IFN subtype . | A1 . | A2 . | A4 . | A5 . | A6 . | A9 . | B . |
|---|---|---|---|---|---|---|---|
| Antiproliferative | + | + | + | + | + | ++ | + |
| Differentiation | − | + | ++ | + | + | − | − |
| Decreased viability | ++ | + | ++ | ++ | ++ | + | + |
| Apoptosis | − | − | − | = | − | ++ | − |
| Delayed onset of erythroleukemia | ++ | + | + | + | ++ | ++ | − |
| Increased survival rate from erythroleukemia | + | − | + | + | ++ | ++ | − |
++ indicates significant increase; +, increase; −, no change; and =, significant decrease.
The authors would like to thank S. Meakins and Dr L. Manning for excellent technical assistance. We thank VICAL for generously providing us with the vector pkCMVint.polylinker. We thank Associate Prof I. Robertson for skilled assistance in statistical analyses.
Prepublished online as Blood First Edition Paper, November 21, 2002; DOI 10.1182/blood-2002-05-1521.
Supported by National Health and Medical Research Council, Australia (project grant no. 990393), the Cancer Foundation of Western Australia, and the Medical Research Foundation of Royal Perth Hospital.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Cassandra James, Division of Veterinary and Biomedical Sciences, Murdoch University, South St, Murdoch 6150, Western Australia, Australia; e-mail:casjames@central.murdoch.edu.au.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal