The first human gene therapy experiment begun in September 1990 used a retroviral vector containing the human adenosine deaminase (ADA) cDNA to transduce mature peripheral blood lymphocytes from patients with ADA deficiency, an inherited disorder of immunity. Two patients who had been treated with intramuscular injections of pegylated bovine ADA (PEG-ADA) for 2 to 4 years were enrolled in this trial and each received a total of approximately 1011 cells in 11 or 12 infusions over a period of about 2 years. No adverse events were observed. During and after treatment, the patients continued to receive PEG-ADA, although at a reduced dose. Ten years after the last cell infusion, approximately 20% of the first patient's lymphocytes still carry and express the retroviral gene, indicating that the effects of gene transfer can be remarkably long lasting. On the contrary, the persistence of gene-marked cells is very low (< 0.1%), and no expression of the transgene is detectable in lymphocytes from the second patient who developed persisting antibodies to components of the gene transfer system. Data collected from these original patients have provided novel information about the longevity of T lymphocytes in humans and persistence of gene expression in vivo from vectors driven by the Moloney murine leukemia virus long-terminal repeat (LTR) promoter. This long-term follow-up has also provided unique evidence supporting the safety of retroviral-mediated gene transfer and illustrates clear examples of both the potential and the pitfalls of gene therapy in humans.
Introduction
Adenosine deaminase (ADA, EC3.5.4.4) is an enzyme in the purine salvage pathway that catalyzes the deamination of adenosine and deoxyadenosine to inosine and deoxyinosine, respectively. Mutations of the ADA gene can result in profound enzyme deficiency and severe combined immunodeficiency (SCID) with virtual lack of T- and B-cell function.1,2 HLA-identical bone marrow transplantation (BMT) is the therapy of choice; however only a minority of patients has access to a fully matched donor. Alternative options are haploidentical BMT3,4 and enzyme replacement with polyethylene glycol-conjugated bovine ADA (PEG-ADA).5
In the mid 1980s gene transfer was proposed as a therapy for ADA deficiency. The simple regulation of the ADAgene6 and the expected selective advantage of gene-corrected cells suggested ADA deficiency as an ideal initial candidate disease to test gene therapy. Because of the difficulty of transducing hematopoietic stem cells, and the observation that BMT patients with only donor T-lymphocyte engraftment had good immunologic reconstitution, it was postulated that genetic correction of autologous T cells could be clinically beneficial.7 Testing of the hypothesis was feasible because patients on PEG-ADA therapy had developed circulating peripheral T lymphocytes that could be targeted with gene transfer.
The first phase 1 gene therapy trial began September 14, 1990, with infusion of a 4-year-old girl with ADA deficiency with autologous T cells transduced ex vivo with the retroviral vector LASN containing the cDNA for human ADA.8 In January 1991, identical treatment was begun for a 9-year-old girl. Patient histories,ADA mutations, and details of the protocol have been described previously.9,10 Between 1990 and 1992, approximately 1011 total lymphocytes were administered over 11 or 12 infusions to each patient. Twenty percent of the circulating T cells from patient 1 contain the ADA vector 10 years after treatment began, whereas less than 0.1% of T cells from patient 2 have detectable vector. Both patients continue to receive weekly injections of PEG-ADA although at a reduced dose (7-13 U/kg per week). We present the results of immunologic and molecular studies carried out over a decade documenting the safety of retroviral gene transfer and long-term persistence of vector-containing, polyclonal T cells. In addition we offer an hypothesis to explain the disparity in the ultimate responses of these 2 patients.
Patients and methods
Patients
The patients involved in this study have been described previously.9 The clinical research protocol and procedures involving human subjects were approved by the National Cancer Institute (NCI); National Heart, Lung, and Blood Institute (NHLBI); and National Human Genome Research Institute (NHGRI) Institutional Review Boards (IRBs). Informed consent was provided according to the Declaration of Helsinki. Patient blood samples were obtained before infusion of gene-corrected cells during the treatment phase and at various time points after treatment.
ADA enzyme assay
ADA assays were performed as described11 with some modifications. Briefly, peripheral blood mononuclear cells (PBMCs) were obtained by Ficoll separation and counted, and 5 × 105cells were lysed in 100 μL M-Per (Pierce, Rockford, IL) at room temperature or by repeated freezing/thawing in 100 mM Tris (tris(hydroxymethyl)aminomethane), pH 7.4. After centrifugation, 10 μL 1 mM 14C-adenosine (Amersham, Piscataway, NJ) was added to 10 μL cell lysate, either neat or at a 1:5 dilution and incubated for 3 to 20 minutes at 37°C. The reaction was stopped at 95°C for 5 minutes, and samples were transferred to ice. The reaction mixture (4-10 μL) was spotted onto thin-layer chromatography (TLC) plates (Kodak, Newark, NJ) previously loaded with 3 μL 0.1 M standards of unlabeled adenosine, hypoxanthine, and inosine (all from Sigma, St Louis, MO) and dried. Separation was obtained by placing TLC plates in a solution of 0.1 M sodium biphosphate, pH 6.8 (10 parts), 6 parts saturated ammonium sulfate, and 0.2 parts n-propyl alcohol for 60 to 120 minutes. Plates were dried and exposed to a phosphoimager screen for 18 to 36 hours. Pixels were counted with a FujiFilm BAS 1500 phosphoimager (Stamford, CT). ADA units were determined as nanomolar adenosine deaminated per minute per 108 cells.
Real-time PCR analysis
DNA was isolated using the PureGene kit (Gentra, Minneapolis, MN). A total of 500 ng of each DNA sample was added to a mixture of 10 μM neomycin (neo) primers (forward primer, TTGTCAAGACCGACCTGTCC; reverse primer, TTCGCTTGGTGGTCGAATG) and 5 μM LASN probe (CAGCTGTGCTCGACGTTGTCACTGAA) with Taqman 2× polymerase chain reaction (PCR) Master Mix. Samples were run in triplicate. Serial dilutions of DNA obtained from a cell line containing a single copy of the LASN vector were used to establish a standard curve. Reactions were run in an ABI Prism 7700,12 and analysis of the percentage of LASN+ cells was performed with Sequence Detection Systems 1.6.3 (PE Applied Biosystems, Foster City, CA).
Proliferation assay
PBMCs (2 × 105/well) were stimulated in triplicate with phytohemagglutinin (PHA; 10 μg/mL; Wellcome Diagnostics, Research Triangle, NC) for 3 days, tetanus toxoid (3 LF/mL; Connaught Laboratories, Toronto, ON, Canada), and diphtheria toxoid (25 LF/mL; Commonwealth of Massachusetts, Boston) for 7 days. Plates were then pulsed for 4 to 5 hours with 0.037 MBq (1 μCi)/well of 3H-thymidine (New England Nuclear, Boston, MA), harvested with a Tomtec plate washer, and counted on a Betaplate counter (Wallac, Gaithersburg, MD). Stimulation index was calculated as net counts per minute of the stimulated samples/counts per minute of media control.
Fluorescence-activated cell sorter (FACS) analysis
PBMCs (5 × 105) were stained with Vβ monoclonal antibodies (Immunotech/Coulter, Miami, FL), followed by fluorescein isothiocyanate (FITC)–labeled goat antimouse (BioSource, Vacaville, CA), blocked with mouse immunoglobulin G1 (IgG1) kappa (Sigma, St Louis, MO), stained with phycoerythrin (PE)–labeled CD3 antibody (Immunotech/Coulter) and either PECy5-labeled CD4 or CD8 antibody (BD Biosciences, San Jose, CA). Cells were collected (105/tube) on a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ), analyzed using CellQuest software (BD Biosciences), and compared with normal range values provided by Immunotech/Coulter.
Complementarity-determining region-3 (CDR3) size distribution analysis of T-cell receptor (TcR) Vβ14
Total RNA was isolated from PBMCs of patient 1 and a healthy control subject using the PureScript Isolation kit (Gentra, Minneapolis, MN). RNA (5 μg) was used to generate the first cDNA strand that was amplified using a Vβ14-specific primer in conjunction with a 6-fluoroscein phosphoramidite-labeled Cβ primer and analyzed as described.13
Single cell clones
PHA (5 μg/mL) or OKT3 (100 ng/mL) activated patient lymphocytes were cloned by limiting dilution and transformed with human T-cell leukemia virus type I (HTLV-I)–producing cells irradiated at 12 500 rad as described.14
Southern blot analysis
DNA was isolated using the PureGene kit. A total of 10 μg of each DNA sample was digested with EcoRI, BglII,HindIII, or SstI (Life Technologies, Rockville, MD). DNA digests were electrophoresed in 1% agarose gels and transferred to Hybond-N+ membranes (Amersham). Hybridization was performed with 32P-neo probe,10 and bands were visualized by autoradiography.
Immunoprecipitation assay
Ouchterlony plates were poured with a 0.6% agarose layer (SeaKem; FMC Bioproducts, Rockland, ME) in barbitol buffer (Sigma). Antigens (neat or diluted as indicated) were placed in the outer wells, and whole patient serum (10 μL) was placed in the center well. After radial diffusion, antigen/antibody complex precipitin bands were visualized using reflected light.
Western blot
Analysis of patients' sera for the presence of anti-Moloney murine leukemia virus (MMLV) p30 was performed as described.15 Briefly, concentrated stock of murine retrovirus was denatured and separated on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel followed by blotting to a nitrocellulose membrane and staining with patient serum. Monkey sera of known reactivity were used as positive and negative controls. Each serum sample was run with a molecular weight marker lane to the left.
Results
ADA levels in PBMCs reflect the percentage of gene-corrected cells
Using real-time PCR technology, we performed a series of retrospective studies on archived cryopreserved PBMCs and quantitated the levels of gene transfer achieved in the cell infusions and patients' circulating lymphocytes. The difference in numbers of gene-corrected cells received by the 2 patients was remarkable: infusions given to patient 1 ranged from 18.2% to 53.3% LASN positive (LASN+), whereas infusions given to patient 2 ranged from 0.9% to 3.1% LASN+ cells (Table1).
ADA enzyme activity and percentage of cells containing LASN in patient infusions
| Infusion . | Protocol day . | ADA, nM/min/108cells . | LASN+ cells, % . | Total cells infused × 108 . | LASN+ cells infused × 107 . |
|---|---|---|---|---|---|
| Patient 1 | |||||
| 1 | 0 | 2.7 | — | 14 | — |
| 2 | 31 | 3.6 | — | 100 | — |
| 3 | 77 | 7.9 | — | 120 | — |
| 4 | 126 | — | — | 6 | — |
| 5 | 153 | 5.3 | 17.8 | 172 | 306 |
| 6 | 202 | 11.8 | — | 180 | — |
| 7 | 262 | 10.5 | 18.2 | 120 | 220 |
| 8 | 314 | 12.4 | 21.2 | 180 | 380 |
| 9* | 511 | 8.2 | 53.3 | 129 | 690 |
| 10* | 601 | 20.8 | 46.8 | 64 | 300 |
| 11* | 707 | 12.4 | 27.4 | 200 | 548 |
| Patient 2 | |||||
| 1 | 0 | 3.9 | 1.51 | 13 | 1.96 |
| 2 | 27 | 2.1 | 1.53 | 150 | 23 |
| 3 | 81 | 4.2 | 2.55 | 153 | 39 |
| 4 | 127 | 3.5 | 1.59 | 54 | 8.6 |
| 5* | 186 | 3.9 | 2.75 | 36.4 | 10 |
| 6* | 242 | 4 | 3 | 24 | 7.25 |
| 7* | 274 | 4.6 | 3.1 | 32 | 9.9 |
| 8* | 300 | 2.5 | 2.46 | 80.3 | 19.8 |
| 9* | 343 | 4.9 | — | 74.5 | — |
| 10* | 398 | 3.3 | 2.02 | 120 | 24 |
| 11* | 448 | 2.8 | 0.87 | 200 | 17.4 |
| 12* | 511 | 4 | 0.96 | 161 | 15.5 |
| Infusion . | Protocol day . | ADA, nM/min/108cells . | LASN+ cells, % . | Total cells infused × 108 . | LASN+ cells infused × 107 . |
|---|---|---|---|---|---|
| Patient 1 | |||||
| 1 | 0 | 2.7 | — | 14 | — |
| 2 | 31 | 3.6 | — | 100 | — |
| 3 | 77 | 7.9 | — | 120 | — |
| 4 | 126 | — | — | 6 | — |
| 5 | 153 | 5.3 | 17.8 | 172 | 306 |
| 6 | 202 | 11.8 | — | 180 | — |
| 7 | 262 | 10.5 | 18.2 | 120 | 220 |
| 8 | 314 | 12.4 | 21.2 | 180 | 380 |
| 9* | 511 | 8.2 | 53.3 | 129 | 690 |
| 10* | 601 | 20.8 | 46.8 | 64 | 300 |
| 11* | 707 | 12.4 | 27.4 | 200 | 548 |
| Patient 2 | |||||
| 1 | 0 | 3.9 | 1.51 | 13 | 1.96 |
| 2 | 27 | 2.1 | 1.53 | 150 | 23 |
| 3 | 81 | 4.2 | 2.55 | 153 | 39 |
| 4 | 127 | 3.5 | 1.59 | 54 | 8.6 |
| 5* | 186 | 3.9 | 2.75 | 36.4 | 10 |
| 6* | 242 | 4 | 3 | 24 | 7.25 |
| 7* | 274 | 4.6 | 3.1 | 32 | 9.9 |
| 8* | 300 | 2.5 | 2.46 | 80.3 | 19.8 |
| 9* | 343 | 4.9 | — | 74.5 | — |
| 10* | 398 | 3.3 | 2.02 | 120 | 24 |
| 11* | 448 | 2.8 | 0.87 | 200 | 17.4 |
| 12* | 511 | 4 | 0.96 | 161 | 15.5 |
— indicates not available for analysis.
CD8-depleted cultures.
Accordingly, patient 1 received more gene-marked cells in the 7th infusion (2.2 × 109) than patient 2 received in toto from the 11 (of 12 total) infusions that were tested (1.76 × 109). Thus, patient 1 received more than 2 logs more vector-marked T cells during the course of the entire protocol than did patient 2.
ADA enzyme activity (negligible to undetectable before treatment) was analyzed to assess transgene expression. ADA activity in the infusions received by patient 1 ranged from 2.7 to 20.8 units (1 unit = 1 nmol/min/108 cells), whereas infusions given to patient 2 had ADA levels from 2.1 to 4.9 units (Table 1).
This finding suggests that the higher ADA values and percentage of gene-marked cells found in the circulation of patient 1 over time reflect in part the larger numbers of gene-marked cells received (Figure 1A). Three and a half years after the last infusion, the gene-marked cells constituted more than 40% of her circulating lymphocytes. Sixteen months after treatment began, ADA levels had risen to a level approximately one fifth of normal and have fluctuated between one third to one fifth of normal over the past 5 years.
ADA enzyme activity and vector detection in PBMCs of gene therapy patients.
(A-B) Units of cytoplasmic ADA enzyme activity (▪) and percentage of LASN vector–positive cells (○) detectable in patient circulating lymphocytes obtained prior to infusion during the treatment phase and at various time points after cell infusions ended. T-cell infusions for each patient are indicated by the inverted blue triangles. The infusion of CD34+ cells to patient 2 is indicated as an upright triangle.
ADA enzyme activity and vector detection in PBMCs of gene therapy patients.
(A-B) Units of cytoplasmic ADA enzyme activity (▪) and percentage of LASN vector–positive cells (○) detectable in patient circulating lymphocytes obtained prior to infusion during the treatment phase and at various time points after cell infusions ended. T-cell infusions for each patient are indicated by the inverted blue triangles. The infusion of CD34+ cells to patient 2 is indicated as an upright triangle.
The lower numbers of gene-corrected cells received by patient 2 are reflected in the lower numbers of gene-marked cells in the blood and lower ADA values achieved (Figure 1B). Patient 2's ADA levels did increase during the time of cell infusions. On protocol day 90, after the 3rd infusion, and on protocol day 291, after the 7th infusion, approximately 2.5% of patient 2's circulating lymphocytes contained vector sequences; however, this dropped to less than 1% by protocol day 333 before the 9th infusion. An attempt at stem cell gene therapy with 7.4 × 107 autologous granulocyte colony-stimulating factor (G-CSF)–mobilized peripheral CD34+ cells transduced with the G1NaSvAd.24 vector16 was unsuccessful. This treatment produced a transient rise in the ADA levels of circulating PBMCs, but ADA levels subsequently fell and have remained below 2 units for the past 9 years (Figure 1B), whereas the percentage of detectable gene-corrected cells has been less than 0.1%. No G1NaSvAd.24 vector sequences were detectable in patient 2's PBMCs at any of the time points tested.
T-cell numbers and function after gene therapy
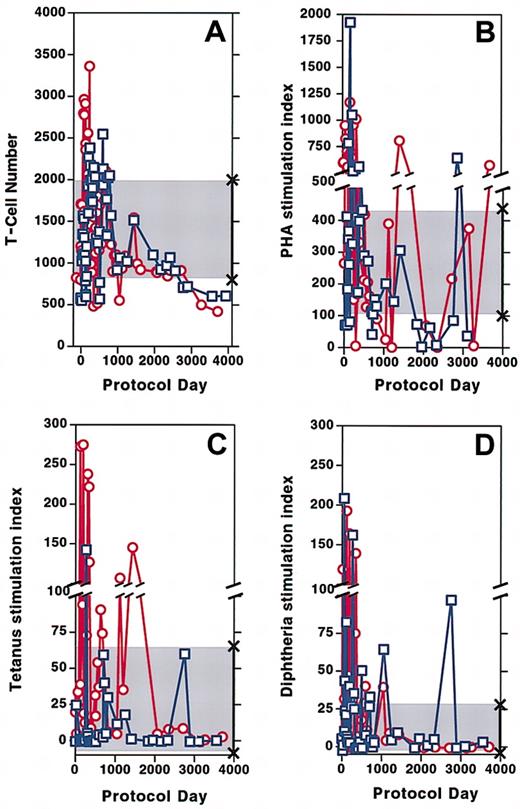
Following initiation of gene therapy, the total number of CD3+ T cells circulating in both patients increased to normal values. After cell infusions stopped, T-cell numbers remained within the normal range for approximately 5 years even as the dose of PEG-ADA was decreasing by approximately 30% in patient 1 and approximately 40% in patient 2. During the past 4 years, both patients' T-cell counts have declined below the normal range. Patient 1 has approximately 610 × 106 circulating CD3+ T cells/L, whereas patient 2 has approximately 420 × 106 CD3+ T cells/L (normal range, 832-2028 × 106/L) (Figure2A). Over the 12 years of the protocol, proliferative responses of patient lymphocytes to PHA have fluctuated above or below the range for healthy control subjects for both patients (Figure 2B). Response to the recall antigens tetanus (Figure 2C) and diphtheria (Figure 2D) were vigorous for several years following cell infusions and have diminished over the past 3 years, although they remain within the lower range for healthy control subjects. Isohemagglutinin titers that had increased to a high of 1:256 in both patients 3 to 4 months after cell infusions began9are now at titers of 1:32 in both subjects. Delayed type hypersensitivity skin tests that had become vigorously positive soon after treatment9 have been very weak or absent for the past 4 years.
Circulating T-cell numbers and patient proliferation responses to mitogen and antigen over time.
(A) CD3+ T cells in the circulation of patient 1 (■) and patient 2 (○). (B-D) Stimulation index (SI) defined as counts per minute in wells treated with mitogen or antigen divided by counts per minute of media alone. SI of patient 1 (■) and patient 2 (○) to PHA (B), tetanus (C), and diphtheria (D). The shaded area marked by × symbols indicates the normal range of CD3+ T cells, mitogen, and antigens along the y-axis.
Circulating T-cell numbers and patient proliferation responses to mitogen and antigen over time.
(A) CD3+ T cells in the circulation of patient 1 (■) and patient 2 (○). (B-D) Stimulation index (SI) defined as counts per minute in wells treated with mitogen or antigen divided by counts per minute of media alone. SI of patient 1 (■) and patient 2 (○) to PHA (B), tetanus (C), and diphtheria (D). The shaded area marked by × symbols indicates the normal range of CD3+ T cells, mitogen, and antigens along the y-axis.
Expansion of T lymphocytes expressing the T-cell receptor (TcR) Vβ14 family in patient 1
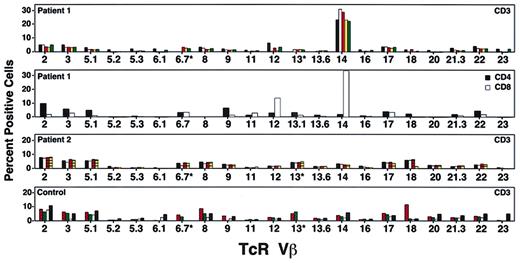
To assess the diversity of the T-cell repertoire following treatment, we tested peripheral blood lymphocytes from both patients using monoclonal antibodies to human TcR Vβ families. Five years after the last cell infusion, patient 1 had cells expressing most of the TcR Vβ families, with an expansion in the percentage of CD3+ T cells that stained positive for TcR Vβ14 (Figure3). Subset analysis revealed that 35% of CD8 T cells and 2% of CD4 T cells expressed Vβ14. A pretreatment sample contained 8% Vβ14+ T cells, suggesting that this population had expanded over time. Oligoclonal expansion of CD8+ T cells has been reported in the elderly17 and in normal individuals.18 19However, because monoclonal cell expansion could also be the result of insertional mutagenesis, or a growth advantage because of genetic correction, further studies were initiated.
Studies of patient T-cell receptor (TcR) Vβ repertoire.
Percentage of T cells in each TcR Vβ family at various times during the study. The top panel shows patient 1 at protocol days 1795 (black bars), 2295 (white bars), 2733 (orange bars), 3063 (yellow bars), and 3559 (green bars). The second panel shows patient 1's cells divided into CD4 and CD8 cells on protocol day 2973; TcR Vβ14 was found predominantly in the CD8+ T-cell subset. The third panel shows patient 2's T cells tested on protocol days 2049 (black boxes), 2070 (white bars), 2718 (orange lined bars), and 3149 (yellow lined bars). No single TcR Vβ family was predominant at any of these time points. The bottom panel shows T cells from healthy control subjects showing range of TcR Vβ families detectable by FACS. Normal 1 (orange dotted bars), Normal 2 (green dotted bars), Immunotech normal low (white bars), and Immunotech normal high (black bars).
Studies of patient T-cell receptor (TcR) Vβ repertoire.
Percentage of T cells in each TcR Vβ family at various times during the study. The top panel shows patient 1 at protocol days 1795 (black bars), 2295 (white bars), 2733 (orange bars), 3063 (yellow bars), and 3559 (green bars). The second panel shows patient 1's cells divided into CD4 and CD8 cells on protocol day 2973; TcR Vβ14 was found predominantly in the CD8+ T-cell subset. The third panel shows patient 2's T cells tested on protocol days 2049 (black boxes), 2070 (white bars), 2718 (orange lined bars), and 3149 (yellow lined bars). No single TcR Vβ family was predominant at any of these time points. The bottom panel shows T cells from healthy control subjects showing range of TcR Vβ families detectable by FACS. Normal 1 (orange dotted bars), Normal 2 (green dotted bars), Immunotech normal low (white bars), and Immunotech normal high (black bars).
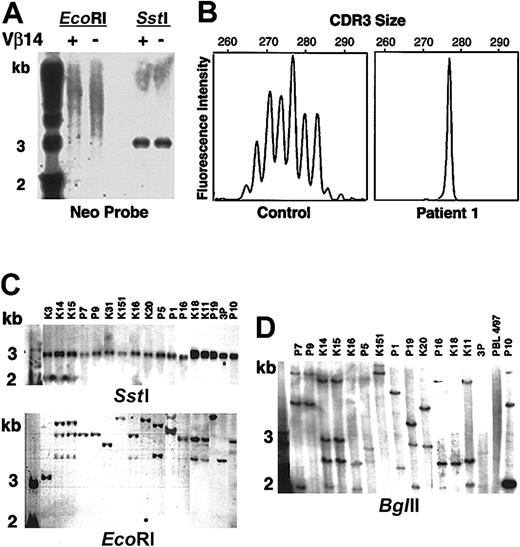
Cells were separated into TcR Vβ14+ and Vβ14− populations and analyzed by Southern blot after digestion with EcoRI that cuts only once within the LASN vector. The results showed diffuse positivity (“smear”), indicating that both cell fractions contained multiple LASN integrants (Figure4A). Digestion with SstI that excises the full-length vector confirmed that there were no gross vector rearrangements (Figure 4A).
Molecular analysis of patient 1's T-lymphocyte complexity.
(A) Patient 1's PBLs were separated into TcR Vβ14-positive and -negative fractions, cut with restriction enzymesSstI (double cutter) and EcoRI (single cutter). SstI digests demonstrate that the vector DNA has not been rearranged. The smear in the lanes cut withEcoRI indicates that there have been multiple LASN integrations. The first lane of panels A, C, and D contains molecular weight markers. (B) Spectratyping of TcR CDR3 of Vβ14 in PBMCs from a healthy control subject and patient 1. (C) Southern blot analysis of single cell clones. SstI andEcoRI digests. (D) BglII (single cutter) digests.
Molecular analysis of patient 1's T-lymphocyte complexity.
(A) Patient 1's PBLs were separated into TcR Vβ14-positive and -negative fractions, cut with restriction enzymesSstI (double cutter) and EcoRI (single cutter). SstI digests demonstrate that the vector DNA has not been rearranged. The smear in the lanes cut withEcoRI indicates that there have been multiple LASN integrations. The first lane of panels A, C, and D contains molecular weight markers. (B) Spectratyping of TcR CDR3 of Vβ14 in PBMCs from a healthy control subject and patient 1. (C) Southern blot analysis of single cell clones. SstI andEcoRI digests. (D) BglII (single cutter) digests.
Spectratyping20 of the TcR β chain complementarity-determining region 3 (CDR3) of the Vβ14+ cells from a healthy control subject revealed the expected Gaussian curve with 6 peaks or more (Figure 4B). Similar analysis of the Vβ14+ population of patient 1 showed a skewed distribution of CDR3 sizes with one prominent peak at 277 nt and a smaller peak of 271 nt, indicating that the patient's Vβ14+ T cells consisted of an oligoclonal population (Figure 4B).
Single T-cell clones were obtained by limiting dilution from HTLV-1–immortalized peripheral blood lymphocytes (PBLs) at various time points (approximately 3.5, 4, 5, and 6.4 years after the last cell infusion). Of the clones shown in Figure 4, 5 were CD4+, TcR Vβ14− and 12 were CD8+, TcR Vβ14+. Digestion of the clones' DNA with SstI, followed by Southern blot analysis, demonstrated the presence of full-length vector in all clones (Figure4C). DNA from the clones was also digested with EcoRI (Figure 4C), BglII (Figure 4D), and HindIII followed by Southern blot. Multiple clones appeared to share similar integration sites using EcoRI (Figure 4C); however, digestion with BglII (Figure 4D) and HindIII (data not shown) confirmed that there were multiple and different LASN integration sites among different Vβ14+ clones. The presence of multiple integrants was also confirmed by inverse PCR21 on clones obtained 3.5 and 4 years after the last cell infusion (data not shown). Together, these results indicated that patient 1's Vβ14+ T cells were multiclonal with multiple LASN integrants. The TcR Vβ14+ population has remained stable and multiclonal for vector insertion over the past 6 years.
Patient 2 had a normal TcR repertoire distribution, with occasional absence of TcR Vβ 6.1 and 23, a finding also observed in PBMCs from some healthy control subjects (Figure 3).
Immune response to fetal calf serum (FCS)
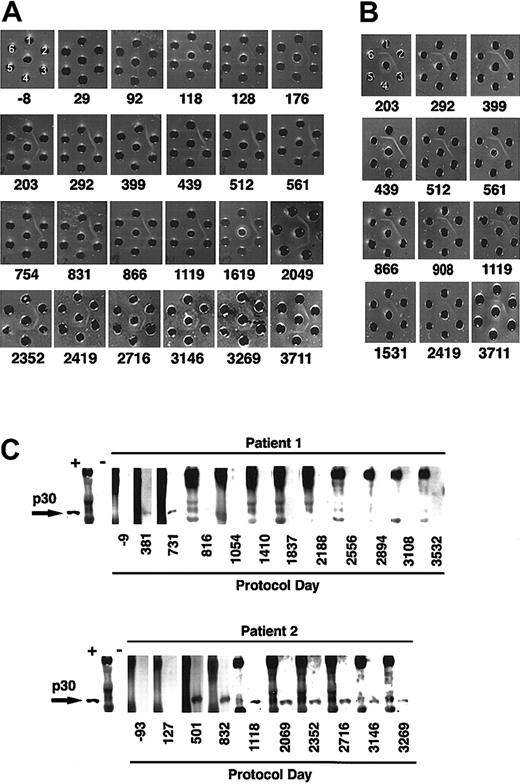
The rapid disappearance of LASN+ cells after each infusion observed in patient 2 (Figure 1B) raised suspicions of an immune-mediated elimination. Therefore, stored serial serum samples from both patients were analyzed for the presence of precipitating antibodies to components of the gene transfer system using the double diffusion Ouchterlony technique.22 No antibodies against FCS were detectable in the sera obtained from patient 1, nor in the sera of 50 healthy blood donors. Similarly, no precipitating antibodies to FCS were detectable in sera collected prior to gene therapy from patient 2, nor after the first and second cell infusion. However, serum collected from patient 2 about 1 month after the third infusion demonstrated clear precipitating antibodies to FCS (Figure5A). By that time, patient 2 had received a total of 31.6 × 109 cultured lymphocytes, of which approximately 6.4 × 108 contained the LASN vector. The level of antibodies increased with subsequent cell infusions. Precipitating antibodies to FCS remained present in patient 2's serum 8 years after the last cell infusion (Figure 5A).
Immune response to bovine protein and retroviral envelope.
(A) Ouchterlony gel of sera from patient 2 at various protocol days. Patient sera (center well); FCS, neat (well no. 1), diluted 1:4 (well no. 2), and 1:8 (well no. 3); bovine serum albumin neat (well no. 4), diluted 1:4 (well no. 5), and 1:8 (well no. 6). (B) Ouchterlony gel of sera from patient 2 (center well), bovine lipoprotein (well no. 1), FCS (well no. 2), lipoprotein-deficient bovine sera (well no. 3), bovine α-2 macroglobulin (well no. 4), bovine ceruloplasmin (well no. 5), Cohn fractions II/III containing primarily β and γ globulins (well no. 6). (C) Western blot assay of patients' sera obtained at various time points to detect antiretroviral antibodies. Each blot begins with a positive monkey serum control (+), followed by molecular weight markers (M) and then a control negative monkey serum (−).
Immune response to bovine protein and retroviral envelope.
(A) Ouchterlony gel of sera from patient 2 at various protocol days. Patient sera (center well); FCS, neat (well no. 1), diluted 1:4 (well no. 2), and 1:8 (well no. 3); bovine serum albumin neat (well no. 4), diluted 1:4 (well no. 5), and 1:8 (well no. 6). (B) Ouchterlony gel of sera from patient 2 (center well), bovine lipoprotein (well no. 1), FCS (well no. 2), lipoprotein-deficient bovine sera (well no. 3), bovine α-2 macroglobulin (well no. 4), bovine ceruloplasmin (well no. 5), Cohn fractions II/III containing primarily β and γ globulins (well no. 6). (C) Western blot assay of patients' sera obtained at various time points to detect antiretroviral antibodies. Each blot begins with a positive monkey serum control (+), followed by molecular weight markers (M) and then a control negative monkey serum (−).
The primary bovine component in FCS recognized by precipitating antibodies from patient 2 was lipoprotein (Figure 5B). Other bovine serum components tested (Cohn fractions II and III, lipoprotein-deficient bovine serum, ceruplasmin, α2-macroglobulin, siderophilin, transferrin, apo-lipoprotein, fetuin, bovine ADA, PEG-ADA, bovine IgM, and bovine IgG) were not recognized by the precipitating antibodies (data not shown).
A third ADA patient treated in Japan using the same transduction protocol23 also developed precipitating antibodies against FCS after the fifth infusion of in vitro–cultured cells (L.M.M. and Y. Sakyiama, unpublished observations, 1998).
Immune response to retroviral vector
In light of the immune response to FCS, we used Western blot analysis15,24 to study patient serum samples for the presence of antibodies to the Moloney murine leukemia virus p30 antigen contained in the envelope of the LASN vector. A study of 53 healthy human sera demonstrated no immune reactivity to MMLV p30. Neither patient exhibited immune reactivity to MMLV p30 prior to treatment. However, antibodies to MMLV p30 were detected in samples obtained from both patients following the cell infusions (Figure 5C). One year after the last cell infusion, patient 1 had lost reactivity to MMLV p30. In contrast, antibodies reactive with MMLV p30 persist in patient 2's serum 8 years after the final infusion of transduced cells. The presence of replication competent retrovirus was excluded by PCR and coculture assays.25
Discussion
Our results demonstrate that autologous T cells from ADA-deficient patients, stimulated in vitro with OKT3 and IL-2, and transduced with first-generation retroviral vectors, can be safely infused into patients and that the transduced cells survive and express ADA enzyme activity in vivo for more than 10 years. Although the number of circulating T cells has declined over the past 3 years, both patients have remained healthy and free from serious infections on reduced doses of PEG-ADA. In a retrospective analysis of archived cells, we found that the average ADA enzyme activity per cell infused to patient 1 was 3 times that of patient 2, and the percentage of lymphocytes containing the LASN vector given to patient 1 was more than 15 times that given to patient 2. PBMCs obtained from patient 1 have maintained 16 to 22 units of ADA activity over the past 5 years (approximately one fourth the enzyme activity of healthy individuals), which is significantly higher than before gene therapy, whereas PBMCs from patient 2 now have less than 2 units of ADA activity, similar to pre–gene therapy levels. These differences of transduction efficiency and gene expression have been consistent throughout this trial.
LASN vector–positive cells have persisted in both ADA-deficient patients for more than 10 years since their last cell infusion. Patient 1 has shown levels of LASN+ cells ranging from 20% to 40% of her circulating lymphocytes, whereas the highest level of circulating transduced cells detectable in patient 2 was approximately 2.5% on protocol day 90, about a week after her third cell infusion, and protocol day 291, 2 weeks after the seventh infusion. Subsequently, the level of circulating vector-containing cells in this patient fell and remained at less than 1%, despite 5 additional T-cell infusions.
This striking difference between patients appears to be the result of several factors, including differential transduction efficiency and cell expansion capability in vitro, as well as the immune response that developed in patient 2 against the retroviral envelope and the lipoprotein components of the FCS used in culturing the cells. Although both patients produced antibodies to MMLV p30 during the period of cell infusions, only patient 2 demonstrated persistence of these antibodies after the cell infusions were completed. The original immune defect in patient 2 was milder,26 and this could explain the generation and persistence of antibodies to FCS and p30. The immune response to foreign proteins observed in both these ADA-deficient patents fits with previous reports of immune responses directed at genetically modified cells, including the generation of antibodies25,27 and specific T-cell cytotoxicity in immunocompromised subjects,28 29 and stresses the importance of rendering gene-modified cells less immunogenic for successful future gene therapy in ADA-deficiency and for treatment in other disorders involving immunocompetent patients.
Five years after gene therapy ended, approximately 30% of the circulating T lymphocytes from patient 1 were CD8+ and Vβ14+. Although spectratyping of the TcR Vβ14 family demonstrated a predominant CDR3 fragment, Vβ14+ T-cell clones contained different and multiple vector integration sites. These findings are not consistent with selective expansion of a single T-cell clone mediated by a particular integration event or a particularly high expression of the ADA transgene. Because the TcR repertoire is oligoclonal in ADA-deficient patients30 and because the original transduction conditions favored the expansion of CD8+ cells in vitro,9 it is reasonable to theorize that a population of CD8+/Vβ14+ T cells was prominently present among patient 1's cells before gene therapy. These cells may have been repeatedly sampled, further expanded, and transduced during the periodic in vitro culture and gene transfer procedures.
The long-term clinical benefit of this first gene therapy trial is difficult to evaluate. Although clearly helped by at least 2 years of PEG-ADA treatment, both patients had persistent immunodeficiency at the time that the trial was initiated. Significant improvement of a series of immunologic parameters was demonstrated following treatment,9 although it is less clear how much of that improvement to attribute to each component of the treatment that they were receiving (infusion of in vitro–expanded and activated T cells, gene correction, and continued PEG-ADA treatment). Objective measures of immune function have gradually diminished in vigor over the past few years in both patients, which fits with the prediction that genetic correction of mature lymphocytes will have a finite effect if not administered periodically.
Nevertheless, several unique conclusions can be drawn from this study. First and foremost, our clinical experience demonstrates the safety of administering genetically modified autologous T cells after in vitro culture and transduction with first-generation retroviral vectors. A second important finding is the previously unanticipated demonstration that human T cells have a life span in the peripheral circulation that can be more than 12 years. In addition, it is clear that transgenes driven by the Moloney retroviral promoter can continue to be expressed in T cells for more than a decade, thus demonstrating resistance to gene silencing in vivo. Finally, we learned that immune responses, even in immunodeficient patients, can confound the outcome of gene therapy experiments in unpredictable ways.
It was our original intention to stop PEG-ADA treatment of these patients after observing the quality and duration of their response to the ADA gene treatment for 2 to 3 years. PEG-ADA was continued at the original dose the children were taking at the start of the protocol but without any adjustment for growth. Concurrent studies in ADA-deficient patients treated with allogeneic BMT without myeloablation had shown that engraftment of donor T cells resulted in adequate immune reconstitution but did not provide enough ADA activity to eliminate the excessive body burden of toxic adenosine metabolites.2 Therefore, we assumed that in the absence of PEG-ADA the gene-corrected cells would not have produced enough ADA to allow recruitment and survival of the ADA-deficient T lymphocytes.
Because our gene therapy treatment depended on the culture expansion of already differentiated and presumably antigen-committed peripheral T cells, the immune repertoire of corrected cells could only reflect the repertoire found in the circulation at the time the blood was taken for culture expansion. Therefore, if PEG-ADA were withdrawn, the patient's residual immune repertoire would be fixed to that already present, with little chance to develop new specificities to new environmental pathogens. For these reasons, we elected to follow a conservative approach and continue to administer low doses of PEG-ADA. It had always been our opinion that the ultimate treatment for this disease would be hematopoietic stem cell gene therapy and that we could retreat the initial patients when that treatment became available. Although the immune response mounted by patient 2 to some of the gene therapy components could be of some concern, elimination of FCS from the culture system and the use of vectors packaged in different envelopes should reduce the likelihood of immune reactions against future treatments. Recent results reported by Aiuti et al31 have shown that gene therapy in the absence of concomitant PEG-ADA treatment, with the addition of mild myeloablation prior to infusion of gene-corrected cells, can successfully treat ADA deficiency and pose the basis for future applications of gene transfer into the hematopoietic stem cells as therapy for this disease.
We thank B. Martin, M. Warner, T. Li, F. May, C. Everett, V. Fellowes, K. Snitzer, A. Telchin, L. Top, D. Gladden, B. Sink, the staff of the Dowling Clinic and the Cell Processing Section, Department of Transfusion Medicine, Clinical Center, NIH for superb nursing care and technical assistance. Dr M. S. Hershfield monitored the plasma levels and metabolites of ADA for which we are grateful. We also thank our earlier collaborators, Drs K. W. Culver, A. D. Miller, M. Clerici, G. Shearer, L. Chang, Y. Chiang, P. Tolstoshev, J. J. Greenblatt, H. Klein, M. Berger, C. A. Mullen, R. A. Morgan, and W. F. Anderson for their contributions that led the way to these studies. An additional special thank you to the patients and their families.
Prepublished online as Blood First Edition Paper, November 27, 2002; DOI 10.1182/blood-2002-09-2800.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Linda Mesler Muul, NIAMS, NIH, Bldg 10, Rm 9N252, Bethesda, MD 20892; e-mail:muul1@mail.nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal