The potential prognostic value of quantitative real-time reverse transcription–polymerase chain reaction (RT-PCR [qrtPCR]) measurements of PML-RARα mRNA in acute promyelocytic leukemia was retrospectively assessed before treatment and at 3 posttreatment intervals in 123 patients on intergroup protocol 0129. The primary measure was the PML-RARαGAPDH normalized quotient (NQ), that is, PML-RARα mRNA copies divided by glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) mRNA copies. Only samples with more than 2.5 × 105 copies of the housekeeping gene GAPDH mRNA (detection sensitivity exceeding 104) were considered NQ evaluable. With RNA from low-density selected cells, paired peripheral blood (PB) and bone marrow samples (n = 140) had comparable NQs (P < .001). Before treatment, high NQ was associated with short-form PML-RARα (P < .001), but not with white blood cell count or clinical outcome. Following treatment, NQ was lower in all-trans retinoic acid–induced complete remission (CR) than chemotherapy-induced CR (P = .018) and at first test after consolidation chemotherapy (P = .037). After consolidation chemotherapy, patients with NQ exceeding 10−5 had 4.1-fold increased relapse risk (P = .008); however, 73% of patients who experienced relapse had NQ lower than 10−5. In the follow-up period (FUP), any NQ exceeding 10−5 and 10−6 had 17.5-fold and 7.6-fold increased relapse risk, respectively (P < .001), while no gradation of relapse risk (approximately 18%) could be identified at NQ lower than 10−6, including NQ−. These results indicate that qrtPCR monitoring of PML-RARα NQ can identify patients at high risk of relapse and suggest that clinically practical PB NQ monitoring at more frequent FUP intervals may improve predictive accuracy for relapse or continuing CR in patients with persistent, fluctuating minimal residual disease levels.
Introduction
Recent improvements in the treatment of acute promyelocytic leukemia (APL), which accounts for about 10% of acute myeloid leukemias, have increased the long-term remission and apparent cure rate to approximately 70%.1-6 The primary factor in this improvement has been the addition of all-trans retinoic acid (ATRA) to anthracycline-containing chemotherapy, with the best results reported in clinical trials in which ATRA is given concurrently with chemotherapy during the remission-induction phase.2,4-6 Additionally, 2 trials have provided evidence that, after various types of consolidation therapy, ATRA maintenance therapy has a positive effect in securing long-term remissions.1 4 Nevertheless, disease relapse has been a major contributor to treatment failure and death.
A powerful tool that has been used to attempt to prospectively identify patients at high risk of experiencing relapse is reverse transcription–polymerase chain reaction (RT-PCR) monitoring of mRNA encoding the APL-specific fusion protein PML-RARα in order to detect minimal residual disease (MRD) (reviewed in Diverio et al11). Early studies, using RT-PCR assays with a sensitivity of 103 to 104, found that patients who were persistently PCR+ after completing induction and consolidation therapy with ATRA plus chemotherapy regimens had a high incidence of relapse within a few months (approximately 75%).7-10 Conversely, following consolidation chemotherapy, patients who were PCR− experienced relapse much less frequently, with incidences of 17% and 27% in 2 large clinical trial studies.5,11 However, because fewer than 10% of patients in these studies were PCR+ after consolidation, the total number of relapses were greater in the PCR− patients. These considerations pointed out the importance of continued PCR monitoring in the postconsolidation follow-up period (FUP). In one of these studies, only 8 (6%) of 142 cases with 2 or more PCR− tests during the FUP subsequently experienced relapse, while 20 of 21 cases that converted to PCR+ experienced relapse (median follow-up, 18 months).11 These results were recently confirmed in another study with long-term follow-up (median, 63 months) in which only 3 (7%) of 41 cases with 2 or more PCR− tests experienced relapse, while 4 of 4 cases with 2 or more PCR+assays experienced relapse.12 The therapeutic relevance of these findings was further supported by a pilot study in which the administration of additional treatment after detection of PML-RARα by PCR produced superior clinical outcome compared with a historical control group in which salvage therapy was administered only after hematologic relapse.13 The concept of basing salvage therapy on the detection of persistent or recurrent MRD has been incorporated in the principal Italian protocol for de novo APL,13 and the same criteria have been proposed for the evaluation of novel therapeutic approaches with the goal of increasing therapeutic index.14
In the current study, we used quantitative real-time RT-PCR (qrtPCR) to study samples from intergroup protocol 0129 (INT0129), in which patients were initially randomized to induction therapy with either ATRA or chemotherapy (daunorubicin and cytarabine [DA]) and, following 2 courses of DA consolidation therapy, were rerandomized to either ATRA maintenance for 1 year or no further treatment (termed “observation”).1 In preclinical experiments using cells and RNA from the APL cell lines NB4 and UF-1 that express the long (L) and short (S) forms of PML-RARα, respectively,15,16 we established assay conditions and evaluation criteria that enabled us to confidently measure PML-RARα mRNA expression levels over a 6 log-linear range, excluding from analysis all samples with a detection sensitivity of less than 104.17 This methodology has the potential of extending the effectiveness of MRD monitoring in APL to be higher than previously applied, nonquantitative manual RT-PCR (mrtPCR) procedures by providing a more precise estimate of relapse risk from defined MRD levels detected, on average, with greater than 10-fold higher sensitivity. In addition to identifying a threshold for high relapse risk, it might identify a low-level MRD threshold below which the risk of relapse is minimal, as suggested by previous findings of persistent low levels of PML-RARα transcripts in long-term complete remission (CR) cases of APL using high-sensitivity mrtPCR.18 Also, quantification of PML-RARα mRNA levels prior to and following initial therapy might be of prognostic value. The results of this investigation indicate that this methodology indeed has the capacity to identify cases at defined high risk of relapse following the completion of consolidation chemotherapy. They also indicate, however, that other potential prognostic applications are limited by an apparent lack of a tight relationship between PML-RARα mRNA levels and clinical outcome. In some instances, it may be possible to overcome these limitations by performing more frequent assays, possibly abetted by our additional finding that qrtPCR assays using RNA extracted from peripheral blood cells are nearly as effective as those from less conveniently obtained bone marrow.
Patients, materials, and methods
Patients and samples
Patient materials used for this study were obtained from 123 patients (aged 15 years or older) registered to protocol INT0129 by the Eastern Cooperative Oncology Group (ECOG), the Cancer and Leukemia Group B (CALGB), and the Southwest Oncology Group (SWOG) under a uniform consent form approved by the institutional review boards of all participating institutions. This patient subgroup was derived from a previously reported group of 203 patients determined to have either the S form or L form of PML-RARα, a requirement for RT-PCR monitoring.19 Selection for the study group was based solely on the availability of more than 1 postinduction treatment sample. Of the 123 pretreatment cases, 116 achieved CR and 111 completed consolidation therapy. Of the latter group, 97 cases were randomized to either ATRA maintenance (47 cases; 24 and 23 cases treated with ATRA or DA induction, respectively) or observation (50 cases; 30 and 20 cases treated with ATRA or DA induction, respectively). Fourteen additional patients who received no postconsolidation treatment owing either to assignment or to default were monitored in the FUP and, for the current analyses, were combined with the observation group (64 total no-treatment cases). Of these cases, 9 had been induced with ATRA, 4 with DA, and 1 with ATRA (failure) followed by DA induction therapy.
Collection of bone marrow (BM) and peripheral blood (PB) samples was not mandated by INT0129. Nevertheless, 822 samples from the 123-patient study group (median, 6 samples per patient; range, 2-27 samples) were retrospectively available from recommended collection intervals for RT-PCR analysis. The analyses in this report used 593 samples from 4 strictly defined monitoring intervals: (1) before any antileukemic therapy (pretreatment); (2) after achieving clinical and hematological remission on either ATRA or DA induction therapy but before the administration of consolidation chemotherapy (at CR); (3) after completion of 2 courses of consolidation chemotherapy but before the second protocol randomization (after consolidation chemotherapy); and (4) during the follow-up period after the second protocol randomization (FUP). The checkpoint used after consolidation chemotherapy was the first evaluable sample after completion of consolidation therapy. The FUP samples were irregularly collected at recommended 6-month intervals for the first 2 years and at 12-month intervals for up to 5 years, including samples up to the time of clinical relapse or to the last official entry in the central protocol database of continuing CR (CCR).
Sample processing and RT-PCR procedures
Low-density mononuclear cells (LDMNCs) (density, 1.077 g/mL or lower) were prepared from BM and PB19 and were either immediately extracted in guanidinium buffer or viably conserved. Total RNA was isolated from fresh LDMNCs (5 to 20 × 106cells) by a modification of the guanidine-HCl extraction/cesium chloride density-gradient procedure, by a modification of the acid phenol extraction method,19 or from thawed, fresh-frozen LDMNCs by means of 1 of 2 proprietary column affinity procedures (RNAqueous kits [Ambion, Austin, TX] or RNeasy kits [Qiagen, Valencia, CA]). All RNAs prepared by initial guanidinium extraction were repurified by means of an RNeasy kit (Qiagen) to provide uniform material for cDNA synthesis reactions.17 Reverse transcriptase reactions, using antisense gene-specific primers for first-strand cDNA synthesis, and PCR amplification were performed at a single site (by W.B. at Applied Biosystems [ABI], Foster City, CA), as previously described.17
The semiautomated collection of qrtPCR amplification data by TaqMan technology, using the ABI PRISM 7700 DNA Sequence Detection System was as previously reported.17,20 Briefly, PML-RARα and glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) transcript copy numbers were determined by the first PCR cycle during real-time analysis in which the initiation of exponential cDNA template amplification exceeds background by 10-fold (the threshold cycle [CT]) and by reference to plasmid DNA standard curves, in which a single template copy is detectable at a CT of 40.20 In this study, we considered as positive any sample with an average CT for PML-RARα lower than 40, even if 1 or 2 of the triplicate values was 40. Although such low-level activity could represent nonspecific background, we did not observe this in a large series of negative controls.17 Thus, these low-level values are likely to reflect the stochastic nature of PCR assays near the limit of detection,17,21,22 and they were regarded as quantifiable positives. Reported PML-RARα and GAPDH copy numbers were the geometric average of triplicate (infrequently, duplicate) determinations derived from independent PCRs performed from the same cDNA. PML-RARα values are reported as the normalized quotient (NQ), derived by dividing the PML-RARα copy number by the GAPDH copy number. The use of GAPDH for intersample normalization was based on a preclinical study in which we found GAPDH mRNA to be equally expressed, with little variation in LDMNCs from normal BM and PB.17
Quantitative real-time RT-PCR sample assessment
In the preclinical study, we determined that a GAPDH copy number of 2.5 × 105 corresponds to a detection sensitivity of 1 leukemic cell in 104 LDMNCs.17 In this report, samples with fewer than 2.5 × 105 GAPDH copies, reflecting low RNA quantity and/or poor efficiency of cDNA synthesis, were considered nonevaluable and excluded from analysis. By this criteria, 92 (16%) of 593 samples from the 4 monitoring intervals were nonevaluable. Although a higher fraction of PB than BM samples were nonevaluable (21% versus 13%; P = .009), the fractions of PB and BM samples with GAPDH copy numbers in lower (greater than 2.5 × 105 to 2.5 × 106) and upper (greater than 2.5 × 106) evaluable sensitivity ranges were equal (P = .777). Thus, a total of 501 evaluable samples, 328 BM, 171 PB, and 2 BM + PB were available for assessment at the 4 specified monitoring intervals.
Statistical methods
The distribution of GAPDH ranges between BM and PB was compared by Fisher exact test.23 Agreement between NQs from simultaneous BM and PB samples and the association between NQs before treatment and at CR were assessed by Spearman rank-order correlation.24 Difference in the distribution of NQs between 2 patient groups was evaluated by the Wilcoxon rank-sum test.24 Disease-free survival (DFS) for the analysis after consolidation therapy is defined as the time from achievement of CR per protocol INT0129 to relapse, death, or last follow-up, while DFS for the FUP analysis is from randomization for the maintenance phase or end of consolidation therapy among the nonrandomized patients. Proportional hazards regression of DFS was used to analyze the predictive value of NQ cutoff levels while adjusting for treatment, PML-RARα type, and white blood cell (WBC) count at presentation; NQ during FUP was analyzed as a binary time-varying covariate according to cutoff level.23 DFS rates were estimated by the method of Kaplan and Meier.25 All values for P are based on a 2-sided hypothesis owing to the exploratory nature of the statistical tests and, thus, should not be interpreted by strict conventional criteria.
Results
Patient characteristics
Four important characteristics (age, sex distribution, WBC count, and DFS follow-up) of the 123-patient study group were similar to those of 344 total adult protocol patients (Table1).1,26 The proportion of study group patients who received ATRA as induction or maintenance therapy deviated from the equal randomization in INT0129; however, the impact of ATRA on DFS was adjusted by multivariate modeling. Also, the fraction of patients in CCR was higher in the study group than in total adult patients, 66 (54%) of 123 versus 129 (38%) of 344. Accordingly, we also compared the distribution of characteristics of patients in the 2 cohorts who achieved CR, but found no differences. Finally, there was no difference in the distribution of L- or S-form PML-RARα types from 203 previously reported adult INT0129 patients,19 further indicating the representative nature of the study group.
Characteristics of 123 patients monitored with serial PML-RARα NQ determinations compared with 344 overall adult patients on protocol INT0129
| Characteristic . | Study group . | Total adults* . |
|---|---|---|
| Age, y, median (range) | 41 (16-77) | 41 (15-81) |
| Sex, M/F, % | 55/45 | 53/47 |
| Pretreatment WBC count, cells/μL, median (range) | 1700 (300-550 000) | 2100 (200-550 000) |
| DFS follow-up | ||
| Months, median no. | 69 | 66 |
| Patients in CCR, no. | 66 | 129 |
| Induction Rx, ATRA/DA, no. patients | 68/55 | 171/173 |
| Postconsolidation Rx, ATRA/no-ATRA, no. patients | 47/64† | 93/130‡ |
| PML-RARα type, L-form/S-form, % | 61/38 | 60/401-153 |
| Characteristic . | Study group . | Total adults* . |
|---|---|---|
| Age, y, median (range) | 41 (16-77) | 41 (15-81) |
| Sex, M/F, % | 55/45 | 53/47 |
| Pretreatment WBC count, cells/μL, median (range) | 1700 (300-550 000) | 2100 (200-550 000) |
| DFS follow-up | ||
| Months, median no. | 69 | 66 |
| Patients in CCR, no. | 66 | 129 |
| Induction Rx, ATRA/DA, no. patients | 68/55 | 171/173 |
| Postconsolidation Rx, ATRA/no-ATRA, no. patients | 47/64† | 93/130‡ |
| PML-RARα type, L-form/S-form, % | 61/38 | 60/401-153 |
Rx indicates treatment.
ATRA patients were randomized to ATRA maintenance therapy for 1 year per protocol; the no-ATRA group did not receive any further therapy, whether by official protocol randomization (50 cases) or for other reasons (14 cases).
These values deviate from the anticipated 50:50 protocol distribution, because they have been corrected for patients who did not register for maintenance randomization and, thus, did not receive further treatment in the postconsolidation period and for patients who were randomized to maintenance treatment (7 to ATRA, 3 to observation) without confirmed CR.
Based on 203 clinically eligible adult cases positive for the L- or S-form of PML-RARα from Gallagher et al.19
Comparison of samples from bone marrow versus peripheral blood and from 4 monitoring intervals
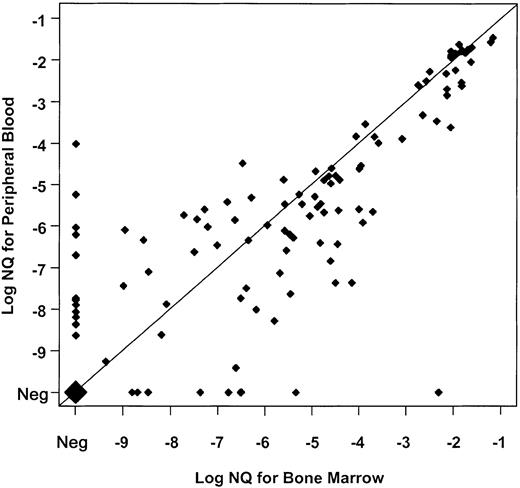
The comparable distribution of GAPDH copy numbers in PB and BM between the lower and upper evaluable sensitivity ranges (“Patients, materials, and methods”) suggested that PB might provide similar sensitivity to BM for detecting PML-RARα transcripts. To evaluate this further, we compared PML-RARα NQs for 140 points at which evaluable RNA samples were available from both BM and PB, and a strong correlation of NQs between these simultaneous pairs was found (Spearman rank-order coefficient = 0.84, P < .001) (Figure1). Moreover, a good correlation was found between NQs of 102 sample pairs at posttreatment time points with MRD (Spearman = 0.67, P < .001). Overall, PB was only slightly less effective than BM for monitoring PML-RARα NQs, and on the basis of this assessment, we substituted NQ data from evaluable PB samples for which a BM sample was not available or not evaluable.
Paired analysis of PML-RARα NQs from 140 simultaneously obtained, evaluable BM and PB samples from INT0129 patients.
Values obtained by dividing PML-RARα copy number by GAPDH copy number for BM (abscissa) and PB (ordinate) are co-plotted on a log10 scale marked by negative exponents. Values below 10−10 were derived from samples in which PML-RARα was undetectable. To be evaluable, both BM and PB samples had to have a GAPDH copy number exceeding 2.5 × 105. Concordance of BM and PB values is indicated by coincident values on the diagonal, and the magnitude of discordance is indicated by the distance from the diagonal. The large diamond represents 27 BM-PB pairs in which evaluable samples from each were NQ−. Thirty-eight NQ+ pairs were also concordant on the diagonal, while 35 pairs disagreed by 1 log (21 with PB lower than BM; 14 with BM lower than PB); 14 pairs by 2 logs (8 with PB lower than BM; 6 with BM lower than PB); and 4 pairs by 3 logs (4 with PB lower than BM). In 9 comparisons, BM was positive and PB negative, while in 13 comparisons PB was positive and BM negative. The overall discordance was 42 pairs with BM greater than PB, and 33 with PB greater than BM. Neg indicates negative.
Paired analysis of PML-RARα NQs from 140 simultaneously obtained, evaluable BM and PB samples from INT0129 patients.
Values obtained by dividing PML-RARα copy number by GAPDH copy number for BM (abscissa) and PB (ordinate) are co-plotted on a log10 scale marked by negative exponents. Values below 10−10 were derived from samples in which PML-RARα was undetectable. To be evaluable, both BM and PB samples had to have a GAPDH copy number exceeding 2.5 × 105. Concordance of BM and PB values is indicated by coincident values on the diagonal, and the magnitude of discordance is indicated by the distance from the diagonal. The large diamond represents 27 BM-PB pairs in which evaluable samples from each were NQ−. Thirty-eight NQ+ pairs were also concordant on the diagonal, while 35 pairs disagreed by 1 log (21 with PB lower than BM; 14 with BM lower than PB); 14 pairs by 2 logs (8 with PB lower than BM; 6 with BM lower than PB); and 4 pairs by 3 logs (4 with PB lower than BM). In 9 comparisons, BM was positive and PB negative, while in 13 comparisons PB was positive and BM negative. The overall discordance was 42 pairs with BM greater than PB, and 33 with PB greater than BM. Neg indicates negative.
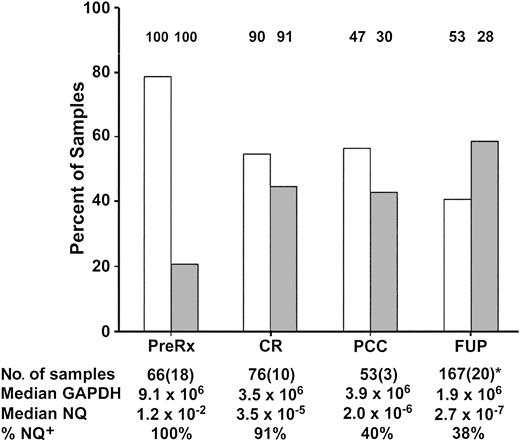
Figure 2 details the number and source of samples used for analysis, which varied depending on availability at the 4 monitoring intervals. Figure 2 also illustrates that the assay sensitivity was variable: maximal for pretreatment and least for FUP samples (median, 9.1 × 106 and 1.9 × 106 GAPDH copies, respectively). This had an impact on the ability to detect low levels of MRD in the interval after consolidation therapy and the FUP interval, since the fraction of PML-RARα+ samples was reduced in the lower- compared with the upper-sensitivity range: after consolidation chemotherapy, 30% versus 47%; FUP, 28% versus 53%. The lower assay sensitivity during the FUP interval was not due to the higher fraction of substitute PB samples (20 of 167, 12%) compared with the interval after consolidation therapy (3 of 53, 6%), since the distribution of GAPDH and of NQs between PB and BM samples in the FUP was equivalent (data not shown).
Distribution of evaluable low- and high-sensitivity range samples and of PML-RARα+ samples at 4 INT0129 monitoring-interval checkpoints.
The columns represent the percentage of evaluable samples (ordinate) in the higher-sensitivity range (GAPDH exceeding 2.5 × 106copies; ■) or lower-sensitivity range (GAPDH exceeding 2.5 × 105-2.5 × 106; ░). The percentage of cases that were PML-RARα+ (NQ+) in each sensitivity range at the 4 checkpoints is indicated above each column, while the overall percentage of NQ+ cases is summarized in the last row at the bottom of the Figure. The first row at the bottom of the Figure presents the total number of samples at each checkpoint interval, with the number of PB samples substituted for BM indicated in parentheses. The median number of GAPDH copies and the median NQ for NQ+ cases at each checkpoint interval are indicated in the second and third rows at the bottom of the Figure, respectively. *During the FUP, the median number of samples per patient was 2 (range, 1-8).
Distribution of evaluable low- and high-sensitivity range samples and of PML-RARα+ samples at 4 INT0129 monitoring-interval checkpoints.
The columns represent the percentage of evaluable samples (ordinate) in the higher-sensitivity range (GAPDH exceeding 2.5 × 106copies; ■) or lower-sensitivity range (GAPDH exceeding 2.5 × 105-2.5 × 106; ░). The percentage of cases that were PML-RARα+ (NQ+) in each sensitivity range at the 4 checkpoints is indicated above each column, while the overall percentage of NQ+ cases is summarized in the last row at the bottom of the Figure. The first row at the bottom of the Figure presents the total number of samples at each checkpoint interval, with the number of PB samples substituted for BM indicated in parentheses. The median number of GAPDH copies and the median NQ for NQ+ cases at each checkpoint interval are indicated in the second and third rows at the bottom of the Figure, respectively. *During the FUP, the median number of samples per patient was 2 (range, 1-8).
Evaluation of PML-RARα NQ for association with pretreatment APL characteristics
We evaluated NQs for an association with 2 pretreatment determinations that have been extensively studied for an association with treatment outcome: PML-RARα mRNA type and presenting WBC count.19 We found a highly significant association between cases with S-form PML-RARα and higher NQs (Table2). At CR, however, no difference was observed (P = .457), and in fact, a greater proportion of S-form cases converted to PML-RARα−: 5 (17%) of 29 S-form cases versus 2 (4%) of 47 L-form cases. Similarly, after consolidation therapy, there was no difference in the distribution of S- and L-form cases (P = .659) in terms of PML-RARα− cases, 9 (56%) of 16 and 22 (61%) of 36, respectively.
PML-RARα NQ tested for association with PML-RARα type and white blood cell count in pretreatment samples
| PML-RARα NQ* . | PML-RARα type† . | WBC count‡ . | ||
|---|---|---|---|---|
| L-form . | S-form . | 2000/μL or less . | Greater than 2000/μL . | |
| 10−2-10−1 | 11 | 26 | 18 | 19 |
| 10−3-10−2 | 25 | 2 | 16 | 11 |
| 10−4-10−3 | 1 | 1 | 1 | 1 |
| Total | 37 | 29 | 35 | 31 |
| PML-RARα NQ* . | PML-RARα type† . | WBC count‡ . | ||
|---|---|---|---|---|
| L-form . | S-form . | 2000/μL or less . | Greater than 2000/μL . | |
| 10−2-10−1 | 11 | 26 | 18 | 19 |
| 10−3-10−2 | 25 | 2 | 16 | 11 |
| 10−4-10−3 | 1 | 1 | 1 | 1 |
| Total | 37 | 29 | 35 | 31 |
P was assessed by the 2-sided Wilcoxon rank-sum test.
There was no difference in the GAPDH copy numbers or NQs between the BM and PB samples used for this analysis (P = .643 and P = .320, respectively).
P = < .001. The significantly higher PML-RARα NQs of S-form versus L-form cases were not due to lower GAPDH copy numbers: S-form median 9.3 × 106 (range, 2.9 × 105 to 2.0 × 108); L-form median 8.9 × 106 (range, 3.7 × 105 to 1.3 × 108).
P = .808.
In contrast, we found no significant association between pretreatment WBC count and NQ (P = .808) (Table 2). For this analysis, we asked if there was a difference in NQs for patients with WBC counts greater than versus less than 2 ×109/L (less than 2000/μL), because this cutoff approximates the median WBC count (1.7 ×109/L [1700/μL]) for the current study and because a concurrent re-evaluation of the INT0129 trial indicates that patients with greater than 2 ×109/L (greater than 2000/μL) have significantly reduced DFS.26 Independent analyses of evaluable samples from BM (n = 48) or from PB (n = 31) also showed no association of NQs with WBC count (P = .851, BM; P = .294, PB).
Evaluation of PML-RARα NQ for relation to induction-therapy response
Of the 123-patient cohort, there were no early deaths, and only 7 patients failed to achieve CR. Pretreatment NQ determinations were available on 5 of the latter cases (NQ exceeding 0.01, 4 cases; NQ less than 0.01, 1 case), too few to analyze. Alternatively, we asked whether NQ at CR correlated with pretreatment NQ, but found no significant association (n = 42; P = .814). Nor was there a difference in the time required to achieve CR between the higher and lower pretreatment NQ groups at the 0.01 cutoff (median times, 45 and 47 days; respectively; P = .586). Additionally, we found no association of pretreatment NQ with 2 complications that occur relatively frequently during the remission induction period in APL, bleeding/coagulation disorders (P = .163), or, in ATRA-treated patients, the ATRA syndrome (P = .619).27-30 Most important, we found no association between any NQ level, including PML-RARα−versus PML-RARα+, and relapse risk in 61 patients who were evaluated before treatment and subsequently achieved CR or who, in 76 cases, were evaluated at CR (data not shown).
Another question was whether there was a difference in the clearance of PML-RARα+ cells on the ATRA- or DA-induction arms of protocol INT0129. Analysis of 76 CR samples demonstrated significantly lower NQs for ATRA-treated compared with DA-treated patients (P = .018; 2-sided Wilcoxon rank-sum test) (Table3). This included 6 (16%) ATRA-treated cases versus 1 (3%) DA-treated case with undetectable PML-RARα. The association of lower NQs with ATRA versus DA induction therapy was also seen after consolidation therapy (P = .037), at which time 20 (71%) of 28 ATRA-treated versus 12 (48%) of 25 DA-treated patients were PML-RARα−.
Distribution of PML-RARα NQs after remission induction with chemotherapy or ATRA at clinical remission and after consolidation chemotherapy
| PML-RARα NQ . | Clinical remission3-150 . | After consolidation chemotherapy3-151 . | ||
|---|---|---|---|---|
| CTH . | ATRA . | CTH . | ATRA . | |
| 10−2-10−1 | 3 | 1 | 0 | 0 |
| 10−3-10−2 | 3 | 3 | 1 | 0 |
| 10−4-10−3 | 11 | 2 | 1 | 1 |
| 10−5-10−4 | 9 | 12 | 4 | 2 |
| 10−6-10−5 | 4 | 8 | 1 | 1 |
| 10−7-10−6 | 4 | 5 | 1 | 1 |
| 10−8-10−7 | 1 | 1 | 0 | 0 |
| 10−9-10−8 | 2 | 0 | 5 | 1 |
| Less than 109 | 0 | 0 | 0 | 2 |
| Negative | 13-152 | 63-152 | 12 | 20 |
| Totals | 38 | 38 | 25 | 28 |
| PML-RARα NQ . | Clinical remission3-150 . | After consolidation chemotherapy3-151 . | ||
|---|---|---|---|---|
| CTH . | ATRA . | CTH . | ATRA . | |
| 10−2-10−1 | 3 | 1 | 0 | 0 |
| 10−3-10−2 | 3 | 3 | 1 | 0 |
| 10−4-10−3 | 11 | 2 | 1 | 1 |
| 10−5-10−4 | 9 | 12 | 4 | 2 |
| 10−6-10−5 | 4 | 8 | 1 | 1 |
| 10−7-10−6 | 4 | 5 | 1 | 1 |
| 10−8-10−7 | 1 | 1 | 0 | 0 |
| 10−9-10−8 | 2 | 0 | 5 | 1 |
| Less than 109 | 0 | 0 | 0 | 2 |
| Negative | 13-152 | 63-152 | 12 | 20 |
| Totals | 38 | 38 | 25 | 28 |
P assessed by 2-sided Wilcoxon rank sum test; similar results are obtained when cases are stratified by PML-RARα type or pretreatment WBC count. CTH indicates chemotherapy.
P = .018.
P = .037.
The GAPDH quantity of the 7 PML-RARα− cases was in the same range as the overall study (median, 2.7 × 106; range, 4.4 × 105 to 1.1 × 107).
Evaluation of PML-RARα NQ after consolidation for relation to disease-free survival
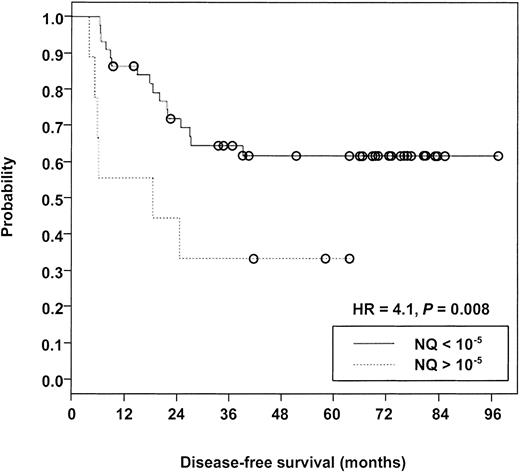
Evaluable samples were available from 53 patients after consolidation therapy, 22 (42%) of whom subsequently experienced relapse over a median follow-up of 69 months. Measurable NQs from these patients were distributed over a greater than 6-log range (lower than 10−9-10−3). Table4 presents the data related to relapse risk at 3 different NQ cutoff levels, comparing cases with NQs above the cutoff (poor risk) with those below the cutoff (good risk). The maximum difference between poor-risk and good-risk cases was observed at the 10−5 cutoff level (hazard ratio [HR] = 4.1; P = .008) (Figure3). A similar trend was noted at the 10−7/−8 cutoff (HR = 2.4;P = .074), but there was no significant risk of relapse in a comparison of PML-RARα− versus PML-RARα+cases (HR = 1.7; P = .244). Notably, 11 (50%) of 22 relapse cases were PML-RARα− at the checkpoint after consolidation therapy (Table 4).
Hazard ratio and 1- to 3-year DFS analysis at 3 PML-RARα NQ cutoff levels of samples after consolidation therapy in 53 INT0129 cases
| NQ cutoff . | Relapses/patients . | HR4-150,4-151 . | P4-151 . | DFS, % . | ||
|---|---|---|---|---|---|---|
| 1 y . | 2 y . | 3 y . | ||||
| Greater than 10−5 | 6/9 | 4.1 | .008 | 56 | 44 | 33 |
| Less than 10−5 | 16/44 | 86 | 72 | 65 | ||
| Greater than 10−7/10−8 | 8/13 | 2.4 | .074 | 62 | 54 | 46 |
| Less than 10−7/10−8 | 14/40 | 88 | 72 | 63 | ||
| Greater than 10−10 | 11/21 | 1.7 | .244 | 67 | 62 | 52 |
| Negative | 11/32‡ | 91 | 70 | 64 | ||
| NQ cutoff . | Relapses/patients . | HR4-150,4-151 . | P4-151 . | DFS, % . | ||
|---|---|---|---|---|---|---|
| 1 y . | 2 y . | 3 y . | ||||
| Greater than 10−5 | 6/9 | 4.1 | .008 | 56 | 44 | 33 |
| Less than 10−5 | 16/44 | 86 | 72 | 65 | ||
| Greater than 10−7/10−8 | 8/13 | 2.4 | .074 | 62 | 54 | 46 |
| Less than 10−7/10−8 | 14/40 | 88 | 72 | 63 | ||
| Greater than 10−10 | 11/21 | 1.7 | .244 | 67 | 62 | 52 |
| Negative | 11/32‡ | 91 | 70 | 64 | ||
Cases with NQs higher than the cutoff level are defined as poor-risk cases as compared with good-risk cases, those with NQs below the cutoff level. Percentages indicate DFS of the good-risk versus bad-risk categories at each level estimated for up to 3 years of follow-up. (Figure 3 shows plot of < 10−5versus > 10−5 cutoff level.) HR indicates hazard ratio.
The hazard ratio indicates the relative risk of relapse in poor-risk versus good-risk cases calculated at 3 selected cutoff levels.
The HRs and P's are adjusted for induction treatment, PML-RARα type, and pretreatment WBC count at 2000/μL cutoff.
The median GAPDH copy number for PML-RARα− relapse cases was 4.3 × 106, higher than the median of 3.9 × 106 for all cases after consolidation therapy.
Kaplan-Meier analysis comparing disease-free survival of patients with PML-RARα NQ exceeding 10−5 versus patients with NQ below 10−5 after completion of protocol INT0129 consolidation therapy.
Open circles indicate censored cases. Median follow-up was 69 months measured from the achievement of complete remission.
Kaplan-Meier analysis comparing disease-free survival of patients with PML-RARα NQ exceeding 10−5 versus patients with NQ below 10−5 after completion of protocol INT0129 consolidation therapy.
Open circles indicate censored cases. Median follow-up was 69 months measured from the achievement of complete remission.
Evaluation of PML-RARα NQ in the follow-up period
The subgroup of patients at the checkpoint after consolidation therapy with high relapse risk (NQ exceeding 10−5) constituted only a minor fraction of evaluable patients (9 of 53; 17%) and included only about one quarter of cases destined to experience relapse (6 of 22; 27%). Thus, an important question was whether a criterion could be established to identify an additional subgroup at increased risk of relapse in the FUP. For this purpose, 167 evaluable samples (147 BM plus 20 PB) were available from 70 patients who had a least 1 sample in the FUP (Figure 2). Each patient was initially assigned to the good-risk group, but was reassigned to the poor-risk category at the actual time when the NQ exceeded the specific cutoff for risk determination. Thus, a patient whose NQ exceeded 10−6 in a particular FUP sample was considered good risk before but became poor risk with respect to this cutoff level at the sample date and remained poor risk even if lower NQs were recorded at subsequent qrtPCR checkpoints. Table 5illustrates the analysis of poor risk versus good risk for the possibility of relapse at the 3 most informative NQ cutoff levels. The analysis was structured in this manner because the data suggested that there was a substantial difference between the risk of relapse at the 10−5 (HR = 17.5) and the 10−6 (HR = 7.6) cutoff levels, while the HR values at cutoff levels below 10−6 (eg, HR = 5.8 for NQ+ versus NQ−) appeared to be almost entirely driven by the high relapse rate in cases with NQs exceeding 10−6. As can be easily calculated, of 45 cases with NQs below 10−6, relapse occurred in 4 (19%) of 21 cases, with NQs between greater than 10−11 and lower than 10−6, and in 4 (17%) of 24 NQ− cases. These data suggest that a relatively low and constant risk of relapse is present at NQ below 10−6, but that there is a progressively higher risk at values above this cutoff.
Hazard ratio and relapse incidence at 3 PML-RARα NQ levels in follow-up period (FUP) samples from 70 INT0129 cases after completion of consolidation chemotherapy
| NQ cutoff . | Relapses/patients . | HR5-150,5-151 . | P5-151 . |
|---|---|---|---|
| Greater than 10−5 | 10/13 | 17.5 | < .001 |
| Less than 10−5 | 13/57 | ||
| Greater than 10−6 | 15/25 | 7.6 | < .001 |
| Less than 10−6 | 8/45 | ||
| Greater than 10−11 | 19/46 | 5.8 | .002 |
| Negative | 4/24 |
| NQ cutoff . | Relapses/patients . | HR5-150,5-151 . | P5-151 . |
|---|---|---|---|
| Greater than 10−5 | 10/13 | 17.5 | < .001 |
| Less than 10−5 | 13/57 | ||
| Greater than 10−6 | 15/25 | 7.6 | < .001 |
| Less than 10−6 | 8/45 | ||
| Greater than 10−11 | 19/46 | 5.8 | .002 |
| Negative | 4/24 |
Data are based on the analysis of at least one evaluable FUP sample with the patient entering the poor-risk category, as defined in Table 4, at the time point that the NQ rises above the indicated cutoff level. Abbreviations are explained in Table 4.
The hazard ratio indicates the relative risk of relapse in poor-risk versus good-risk cases calculated at 3 selected cutoff levels.
Adjusted for the maintenance treatment, PML-RARα type, and pretreatment WBC count.
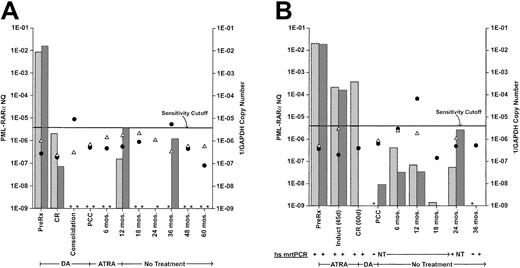
Insufficiencies in the number and regularity of samples did not permit analysis for a possible relationship of PML-RARα NQs to clinical outcome in successive samples. Further, serially monitored NQs in individual cases showed marked heterogeneity of pattern. In only a minority of FUP cases was CCR associated with persistent NQ-negativity or was relapse presaged by a distinctive ascending pattern at the 6-month or longer protocol checkpoint intervals. Most cases showed a descent to low or undetectable NQs immediately after consolidation therapy, frequently followed, in cases with 2 or more samples, by an erratic pattern of various levels of positivity interspersed with negative points. Two examples of such cases that persisted in CCR, despite intermittent NQ-positivity, are illustrated in Figure 4. The transient NQ increases at 12 and 36 months in patient 1 (Figure 4A) or at 6, 12, and 24 months in patient 2 (Figure 4B) are indistinguishable from similar elevations in other cases that were succeeded by relapse before the next 6-month checkpoint (eg, Figure 4; Slack et al17). The data in Figure 4 also illustrate the comparability of PB- and BM-monitoring values and the qualitative confirmation of low-level qrtPCR+ NQs by high-sensitivity mrtPCR detection (Figure4B).
Clinical course and serial monitoring of PML-RARα NQs derived from bone marrow and/or peripheral blood by real-time quantitative RT-PCR in 2 patients treated on protocol INT0129.
(A) Patient 1 received induction treatment with daunorubicin plus cytarabine (DA) and, after 2 courses of DA consolidation chemotherapy, received all-trans retinoic acid (ATRA) maintenance therapy for 1 year. (B) Patient 2 received remission induction treatment with ATRA and, after 2 courses of DA consolidation chemotherapy, received no further treatment. Both patients are in continuing clinical remission after longer than 5 years follow-up. The monitoring intervals designated on the abscissa are representational and not time-linear. PML-RARα NQs, indicated on the left ordinate, were determined by dividing PML-RARα copy number by GAPDH copy number. Bone marrow NQs, stippled columns; peripheral blood NQs, hatched columns. Assay sensitivity is represented by the reciprocal of the GAPDH copy number (1 divided by the GAPDH copy number), indicated on the right ordinate. The sensitivity cutoff at 1/2.5 × 105 GAPDH copies corresponds to a sensitivity detection level of 104.17 Thus, the further below the cutoff line, the more sensitive the assay, while values above the line were considered nonevaluable for use in cohort analyses, whether PML-RARα NQ− or NQ+, although the calculated PML-RARα NQs for positive samples are included in this chart. Reciprocal GAPDH values: bone marrow, closed circles; peripheral blood, open triangles. The presence of these symbols indicates that the sample was assayed; if the corresponding PML-RARα NQ column is absent and marked with an asterisk, it means that the sample was NQ−. Several bone marrow and peripheral blood samples for patient 2 were assayed by a high-sensitivity (hs) mrtPCR assay,17 as well as by real-time qrtPCR, and ± results are shown below the graph. NT, not tested.
Clinical course and serial monitoring of PML-RARα NQs derived from bone marrow and/or peripheral blood by real-time quantitative RT-PCR in 2 patients treated on protocol INT0129.
(A) Patient 1 received induction treatment with daunorubicin plus cytarabine (DA) and, after 2 courses of DA consolidation chemotherapy, received all-trans retinoic acid (ATRA) maintenance therapy for 1 year. (B) Patient 2 received remission induction treatment with ATRA and, after 2 courses of DA consolidation chemotherapy, received no further treatment. Both patients are in continuing clinical remission after longer than 5 years follow-up. The monitoring intervals designated on the abscissa are representational and not time-linear. PML-RARα NQs, indicated on the left ordinate, were determined by dividing PML-RARα copy number by GAPDH copy number. Bone marrow NQs, stippled columns; peripheral blood NQs, hatched columns. Assay sensitivity is represented by the reciprocal of the GAPDH copy number (1 divided by the GAPDH copy number), indicated on the right ordinate. The sensitivity cutoff at 1/2.5 × 105 GAPDH copies corresponds to a sensitivity detection level of 104.17 Thus, the further below the cutoff line, the more sensitive the assay, while values above the line were considered nonevaluable for use in cohort analyses, whether PML-RARα NQ− or NQ+, although the calculated PML-RARα NQs for positive samples are included in this chart. Reciprocal GAPDH values: bone marrow, closed circles; peripheral blood, open triangles. The presence of these symbols indicates that the sample was assayed; if the corresponding PML-RARα NQ column is absent and marked with an asterisk, it means that the sample was NQ−. Several bone marrow and peripheral blood samples for patient 2 were assayed by a high-sensitivity (hs) mrtPCR assay,17 as well as by real-time qrtPCR, and ± results are shown below the graph. NT, not tested.
Discussion
In the current study, we investigated the application of high-sensitivity qrtPCR in a representative subgroup of APL patients from the phase 3 trial INT0129, which is the first application of this methodology to a large clinical trial study of APL. The data permit an assessment of the potential prognostic value of qrtPCR at 4 phases of disease relative to protocol therapy: pretreatment; at CR; after the completion of 2 cycles of consolidation chemotherapy but before the second protocol randomization (after consolidation therapy); and during the postconsolidation FUP (median follow-up, 69 months).
The study accomplished the major objective of identifying high-risk thresholds for disease relapse in the postconsolidation period. At the checkpoint after consolidation therapy, it was determined that the subgroup with a PML-RARα NQ exceeding 10−5 had a 4.1-fold increased risk of relapse, which occurred in 6 (67%) of 9 patients in this category (Table 4; Figure 3). However, these 6 high-risk patients represented only 27% of total relapse cases, which emphasized the importance of continued monitoring in the FUP to attempt to identify most relapse cases present after consolidation therapy in the good-risk subgroup with NQ below 10−5. Partly related to sample limitations, this analysis was based on the highest single NQ achieved by each patient at any time during the FUP. Using time-varying covariate analysis,23 31 we found that relapse risk was increased 17.5-fold or 7.6-fold for poor-risk cases at NQ cutoff levels of 10−5 or 10−6, respectively (Table 5). These cutoffs were associated with relapse rates of 77% for NQ exceeding 10−5 and of 42% in the NQ 10−6 to 10−5 range, and they identified 43% (NQ exceeding 10−5) and 65% (NQ exceeding 10−6) of total relapse cases. These results strongly suggest that patients with NQ exceeding 10−5 after the completion of consolidation therapy should be considered for further therapy and that patients with NQ 10−6 to 10−5 should be monitored frequently for a possible increase into the very high-risk range of greater than 10−5.
A second important finding in the postconsolidation phase was that there was no apparent stratification of relapse risk at PML-RARα NQs below the higher-risk cutoffs. At the checkpoint after consolidation therapy, relapse occurred in 16 (36%) of 44 patients with NQ below 10−5 and in 11 (34%) of 32 NQ− patients (Table 4). Similarly, during the FUP, relapse occurred in 4 (19%) of 21 patients in the NQ 10−10 to 10−6 range and in 4 (17%) of 24 NQ− patients (Table 5). Thus, our PML-RARα NQ evaluation appears to define a single threshold in the NQ 10−6 to 10−5 range, above which there is high risk, and below which there is lower but still significant risk of relapse. Interestingly, serial monitoring of individual patients demonstrated that there are frequently marked and sporadic quantitative variations in NQs within the lower-risk range (Figure 4). At the relatively infrequent sampling intervals used in this study (6 to 12 months), it was not possible to discern patterns of NQs that distinguished patients who would continue in CR (Figure 4) from those who would subsequently experience relapse (Figure 4; Slack et al17). These observations are consistent with reports that clinical relapse can occur rather suddenly, within 3 to 6 months, from low MRD levels, including several reports after negative mrtPCR assays5,11,12,32 and 7 patients in this study with negative or very low NQs (below 10−8). These considerations argue for more frequent MRD monitoring for 1 to 2 years after finishing consolidation therapy when relapse risk is highest. Such monitoring of MRD “kinetics” at short intervals may be able to detect an upward trend toward the high-risk MRD level, as has been successfully used, for example, in predicting hematological relapse in chronic myeloid leukemia.31 Our documentation that monitoring blood is nearly as effective as monitoring BM could greatly facilitate in reducing the MRD-monitoring interval to shorter than 3 months, although this requires more systematic confirmation with simultaneous blood and marrow testing.
Our observation of frequent sporadic changes in persistent low-level PML-RARα mRNA levels during continuing CR also has more basic implications related to the process of leukemia progression. In addition to confirming that a persistent low level of PML-RARα+ cells is compatible with prolonged DFS,18 it suggests that the interplay between residual APL disease and the host is a dynamic process. In combination with the observation that clinical relapse not infrequently occurs rapidly from low MRD levels, it suggests that relapse occurs from a subpopulation of APL cells that undergoes further change(s) leading to escape from host control mechanisms. Consistent with this hypothesis, we have found several INT0129 patients in whom mutations in the PML-RARα gene emerged only late at the time of clinical relapse.33 Presumably, clonal escape due to this or other molecular mechanisms would be reflected in the quantitative rate of clonal emergence, and thus, another benefit of qrtPCR monitoring may be to learn more about molecular biological changes that perturb the leukemic cell-host control balance leading to relapse.
In contrast to results in the postconsolidation period, we did not find any indication that qrtPCR had prognostic value at earlier time points in the disease course. At CR, there was no association between PML-RARα NQ level and DFS (P = .814), and 3 of 7 NQ− patients subsequently experienced relapse. Interestingly, the distribution of NQs at CR was significantly lower in patients treated with ATRA versus chemotherapy for induction (P = .018) (Table 4). These findings were unexpected since previous reports with mrtPCR reported a greater frequency of PML-RARα− assays after chemotherapy versus ATRA induction therapy.2,8,9 The reason for this difference is not clear, since INT0129 used a standard ATRA dose (45 mg/m2/d) and schedule and since it was not related to a difference in the timing of CR and CR sampling (median, 47 and 48 days, respectively), as identified in a recent report.12 The greater reduction of NQ by ATRA was also seen after consolidation, when 71% ATRA-treated versus 48% chemotherapy-treated patients were PML-RARα− (Table 4). These observations suggest that the rate of disease clearance may be an additional quantitative parameter derived from qrtPCR studies that can be used to assess prognosis and/or the comparative effectiveness of different induction therapies.
We also found no evidence that the pretreatment level of PML-RARα NQ can add to accepted clinical risk factors in APL, particularly the presenting WBC count.2,5,19,26,34,35 We found no association between pretreatment NQ and WBC count or clinical outcome, nor was PML-RARα NQ a predictor for development of bleeding/coagulation problems and/or the ATRA syndrome during the induction period. We did find a strong association between high pretreatment NQ and S-form versus L-form PML-RARα (P < .001), but this association did not persist after achieving CR or following consolidation treatment, a finding in agreement with previous conclusions that PML-RARα type does not significantly affect the outcome of combined ATRA/chemotherapy regimens.2,5,19,34 35 A caveat to the conclusion that pretreatment NQ has no prognostic value relates to our use of RNA extracted from Ficoll-selected LDMNCs for performing the assays. Indeed, the lack of association between NQ using RNA from PB samples and WBC count seems surprising, considering the marked intercase heterogeneity of APL cell penetration into the vascular compartment at disease presentation. Thus, we cannot exclude the possibility that NQ determinations from unfractionated PB leukocytes could have prognostic significance, although it is unclear if this would add to the value of WBC count per se.
In summary, this initial monitoring study of APL using qrtPCR indicates that a significant relationship between the level of PML-RARα mRNA and clinical outcome was limited to the postconsolidation phase. These findings are qualitatively similar to those previously reported using conventional, nonquantitative mrtPCR.5,11 12 By both methods, the criteria for high risk after consolidation therapy (NQ exceeding 10−5 by qrtPCR; PCR+ by mrtPCR) fail to detect most subsequent relapse cases, but during the FUP (NQ exceeding 10−5 by qrtPCR; 2 successive PCR+ assays by mrtPCR) are highly predictive of relapse. Because of limitations of sample size and of differences in treatment regimens and sampling schedules, it is not possible to formally compare the results presented here with those reported in previous studies using mrtPCR methods. QrtPCR has the theoretical advantage of providing a precise assessment of the effectiveness of each test specimen assay. Since, as shown in this study, the assay sensitivity of evaluable samples can vary more than 40-fold (GAPDH greater than 2.5 × 105 to greater than 107), it seems reasonable to assume that results from mrtPCR methods are subject to similar variations that might affect individual assay results, that is, PCR+ versus PCR−, especially near the sensitivity limit of the mrtPCR assay. Increasing the sensitivity of mrtPCR to attempt to identify patients at a lower risk of relapse would probably diminish the clinical predictive value of this procedure, since, as we determined by qrtPCR, low levels of PML-RARα mRNA are not tightly associated with relapse risk. We conclude that further studies using common samples to directly compare the effectiveness of the 2 methodologies are required to determine if the theoretical advantage of qrtPCR is of sufficient practical importance to justify the additional expense of this high-technology procedure for clinical monitoring of MRD in APL.
We express our gratitude to the physicians, nurses, data managers, and patients from ECOG-, SWOG- and CALGB-affiliated institutions for their cooperation in providing clinical specimens and information for our investigations. We express our special thanks to Dr David Harrington for his encouragement leading to the completion of this investigation and to Dr Jerry Radich for critical reading of the manuscript.
Prepublished online as Blood First Edition Paper, December 5, 2002; DOI 10.1182/blood-2002-05-1357.
Supported by grants CA56771, CA21115, and CA31946 from the National Institutes of Health. This study is based on an intergroup clinical trial (INT0129) involving participation of the Eastern Cooperative Oncology Group (ECOG; Robert L. Comis, MD, Group Chair); the Cancer and Leukemia Group B (CALGB; Richard L. Schilsky, MD, Group Chair); and the Southwest Oncology Group (SWOG; Charles A. Coltman Jr, MD, Group Chair).
Two of the authors (W.B. and K.J.L.) are employed by a company (Applied Biosystems, Inc, Foster City, CA) whose product was studied in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Robert E. Gallagher, Department of Oncology, Montefiore Medical Center, Rm 601, Hofheimer Bldg, 111 East 210th St, Bronx, NY 10467; e-mail:rgallagh@aecom.yu.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal