The (11;19)(q23;p13.1) translocation in acute leukemia results in the formation of an MLL-ELL fusion protein. ELL is an RNA polymerase II elongation factor that interacts with the recently identified EAF1 protein. To characterize the normal functions of ELL and its aberrant activities when fused to MLL, we isolated a second protein that interacts with ELL named EAF2 for ELLAssociated Factor 2. EAF2 is highly homologous to EAF1, with 58% identity and 74% amino acid conservation. Using specific antibodies generated to EAF2, we coimmunoprecipitated ELL and EAF2 from multiple cell lines. Confocal microscopy revealed that endogenous EAF2 and ELL colocalized in a nuclear speckled pattern. Database comparisons with EAF2 identified a region with a high content of serine, aspartic acid, and glutamic acid residues that is conserved with EAF1 and exhibited amino acid similarity with several translocation partner proteins of MLL, including AF4 and ENL. We found that EAF2 and EAF1 both contain transcriptional activation domains within this region. Using retroviral bone marrow transduction, we observed that a heterologous fusion of EAF2 to MLL immortalized hematopoietic progenitor cells. In contrast to EAF1, EAF2 does not bind to the carboxy-terminus of ELL. We identified a protein-protein interaction domain within the amino-terminus of ELL that binds to both EAF1 and EAF2. This amino-terminal interaction domain is disrupted in the formation of the MLL-ELL fusion protein. Thus, MLL-ELL retains an interaction domain for EAF1 but not for EAF2. Taken together, these data suggest that MLL-ELL may disrupt the normal protein-protein interactions of ELL.
Introduction
The ELL gene was first identified as a fusion partner gene of MLL in the (11;19)(q23;p13.1) translocation, a recurring chromosomal aberration in acute myeloid leukemia.1 Subsequent studies revealed that ELL has the capacity to function as an RNA polymerase II (Pol II) transcriptional elongation factor.2 As a result of the (11;19)(q23;p13.1) translocation, an MLL-ELL chimeric protein is formed that contains the amino-terminal region of MLL, including its AT hooks, methyltransferase domain, and repression domain, fused to amino acids 46 to 621 of ELL, including its elongation domain, a lysine-rich region, and a protein-protein interaction domain. ELL2 was identified by sequence homology to ELL and has been shown to exhibit similar activities to ELL in transcriptional elongation assays.3However, ELL2 has not been observed in association with chromosome translocations in leukemia or in other malignancies. In addition to ELL and ELL2, several other factors have been identified that regulate the elongation phase of mRNA synthesis. The function of one group of these factors, which includes Elongin, TFIIF, ELL, and ELL2, is to facilitate the processivity of transcription by suppressing transient pausing by Pol II.4
More than 30 different recurring cytogenetic aberrations that affect the MLL gene at 11q23 have been described.5,6The critical feature of these chromosomal rearrangements is the generation of a chimeric transcript consisting of 5′ MLL and 3′ sequences of the gene on the partner chromosome. Although no consistent homologies or motifs among the partner gene sequences have been identified that might explain how their fusion to MLLresults in leukemia, certain groups of partner proteins have similar features. These include ENL and AF9, which are serine- and proline-rich and share extensive amino acid homology.7,8 AF4, AF5q31, and LAF4 are highly homologous proteins that also are rich in serines and prolines and exhibit limited homology with ENL and AF9.9-11 Although these proteins contain transcriptional activation domains with similar properties in reporter gene assays, the functions of AF4, LAF4, AF5q31, ENL, and AF9 are not yet known.12 13
Using ELL as the bait in a yeast 2-hybrid screen, we recently isolated a novel protein that we named EAF1 for ELLAssociated Factor 1.14We found that endogenous ELL and EAF1 coimmunoprecipitated as a complex in multiple cell types. We identified an EAF1 interaction domain within the carboxy-terminus of ELL that mapped to amino acids 508-621. EAF1 contains a region that is rich in serine, aspartic acid, and glutamic acid residues that exhibits limited amino acid similarity with the transactivation domains of AF4, LAF4, and AF5q31 proteins that fuse to MLL in 11q23 chromosome translocations. We identified a similar transactivation domain within this region of EAF1. By confocal microscopy, ELL and EAF1 colocalize in a distinct speckled pattern within nuclei.
Recently, we showed that retroviral infection of murine hematopoietic progenitor cells with MLL-ELL followed by transplantation into lethally irradiated littermates resulted in the development of acute myeloid leukemia.15 Similarly, murine hematopoietic progenitor cells transduced with MLL-ELL became immortalized in vitro. Using a hematopoietic progenitor cell immortalization assay, we demonstrated that the amino-terminal elongation domain of ELL is dispensable but that a carboxy-terminal domain that retains the potential to interact with EAF1 is necessary and sufficient for the immortalization of hematopoietic progenitor cells.16 To address whether the EAF1 interaction domain was the critical functional contribution of ELL to the MLL-ELL fusion protein, we examined the transforming properties of a heterologous MLL-EAF1 fusion. Although EAF1 has not been identified as an MLL partner protein, this chimeric fusion recapitulated the phenotype observed with MLL-ELL. MLL-EAF1 demonstrated the capacity to immortalize primary hematopoietic cells in vitro, and mice receiving transplants of cells transduced with MLL-EAF1 developed acute myeloid leukemia. Taken together, these data suggest that the protein-protein interactions of MLL partner genes may have important functional contributions to leukemias that result from 11q23 translocations.
To further characterize the normal functions of ELL and to analyze its aberrant functions when fused to MLL, we have now identified and characterized the EAF2 gene, which encodes a protein that is highly homologous to EAF1. EAF2 exhibits similar functional properties to EAF1; both proteins contain a transcriptional activation domain and exhibit the capacity to immortalize hematopoietic cells when fused to MLL. However, EAF2 exhibits an alternative pattern of binding to ELL, and the MLL-ELL fusion protein retains an interaction domain for EAF1 but not for EAF2.
Materials and methods
Yeast 2-hybrid interaction assay
The full-length open reading frame of ELL was cloned in the pAS2-1 vector and used as the bait to screen for interacting proteins in a yeast 2-hybrid screen. Using the lithium acetate method, yeast strain Y190 was sequentially transformed with pAS2-1–ELL and then subsequently with a human bone marrow cDNA library (Clontech, Palo Alto, CA) fused to the GAL4 transactivation domain in the pGAD10 vector.17Approximately 5 × 106 independent clones were screened. To exclude false-positives, cDNA clones isolated from the library screening were retransformed in yeast strains Y190 and CG1945 along with positive and negative controls and assessed for β-galactosidase activity and for growth on media lacking histidine in the presence of 30 mM 3-amino-1,2,4-triazole (3-AT). The bait plasmid, pAS2-1–ELL, was used as a positive control. As negative controls, we used the pAS2-1 vector alone, and pAS2 fused tolamin C, p53, CD30, andSNF1.
Nucleotide sequencing
The positive cDNA clones were sequenced on both strands using cycle sequencing with ABI BigDye Terminators (PE Applied Biosystems, Foster City, CA). Database searches were conducted with the basic local alignment search tool (BLAST) search algorithm and with the DeCypher II Similarity Search System.18 To determine the exon-intron boundaries of EAF2, its cDNA sequence was aligned with the Ensembl genome database atwww.ensembl.org. An expressed sequence tag (EST) clone (zt03e05.r1) that matched the sequence isolated from the library screen was obtained from American Type Culture Collection (Manassas, VA) and sequenced. The nucleotide sequence of theEAF2 cDNA has been deposited in GenBank under accession number AF517829.
Northern blot analysis
Multiple tissue Northern blots (Clontech) containing approximately 2 μg per lane purified poly(A) + RNA from different human tissues were hybridized for 1 hour with a α-32[P] dCTP-labeled EAF2 cDNA probe prepared using random primers (New England Biolabs, Beverly, MA). Filters were washed with 2 × SSC (salt sodium citrate) and 0.1% sodium dodecyl sulfate (SDS) for 40 minutes with agitation at room temperature, and with 0.1 × SSC and 0.1% SDS for 40 minutes at 50°C, and then autoradiographed. To normalize for the relative amount of RNA in each lane, the blots were stripped and then reprobed with a human β-actin cDNA probe.
Production of monoclonal antibodies to EAF2
To produce a histidine-tagged EAF2 protein in bacteria, the full-length open reading frame of EAF2 was cloned into the pET-19b expression vector (Novagen, Madison, WI) and transformed in theE coli strain BL21(DE3). The histidine-tagged EAF2 fusion protein was purified on a nickel column and eluted in 1 M imidazole, 500 mM NaCl, 20 mM Tris [tris(hydroxymethyl)aminomethane]-HCl, pH 7.9. The eluted EAF2 protein was used to immunize mice for the generation and screening of hybridomas. Monoclonal antibodies to EAF2 were generated using standard methods. We isolated 4 unique hybridomas that produced monoclonal antibodies that recognized EAF2.
Cell culture, transient transfection, and immunoprecipitation
Human 293 and HeLa cells were transiently transfected using 1-2 μg of plasmid DNA and the Effectene transfection reagent (Qiagen, Valencia, CA). Cell pellets were resuspended in 1 mL TEN (40 mM Tris, 1 mM EDTA [ethylenediaminetetraacetic acid], 150 mM NaCl) buffer, centrifuged for 5 minutes at 1200g at 4°C, lysed with 500 μL NETN (100 mM NaCl, 20 mM Tris pH 8.0, 1 mM EDTA), and 0.2% NP-40 containing a cocktail of protease inhibitors (Sigma), incubated on ice for 10 minutes, and centrifuged at 2500gfor 30 minutes at 4°C. To precipitate the complexes, supernatants were precleared with 30 μL of A/G agarose beads (Santa Cruz, Santa Cruz, CA) for 30 minutes and then incubated for 1 hour with the indicated antibody. We then added 30 μL of a 50% slurry of protein A/G agarose beads, incubated overnight at 4°C, washed 5 times at 4°C with lysis buffer, boiled in Laemmli sample buffer, fractionated by SDS–polyacrylamide gel electrophoresis (PAGE), and transferred to nitrocellulose membranes (BioRad, Hercules, CA). Blots were blocked in Tris-buffered saline with 0.05% Tween and 5% nonfat milk followed by incubations with the indicated primary and secondary antibodies in this buffer. For the Western blots, the membranes were incubated with the preimmune and polyclonal anti-ELL antisera at 1:1000, anti–FLAG-M2 at 1:1000, anti-EAF2 (1F11) at 1:10, isotype control at 1:10, anti-GAL4 at 1:500.
Immunofluorescence
HeLa cells were grown for 24 hours on glass coverslips coated with 0.2% gelatin, washed with phosphate-buffered saline (PBS), fixed in 3.7% formaldehyde in PBS at room temperature, and permeabilized with 0.2% Triton X-100. Incubation with the primary and secondary antibodies, quenching, and staining with DAPI (4′,6-diamidino-2-phenlindole) was performed as previously described.14 Fluorescence images were obtained with a Zeiss Axiophot microscope and confocal images were obtained with a Zeiss/NORAN system. For indirect immunofluorescence, affinity-purified polyclonal anti-ELL antiserum was used at a 1:200 dilution, and for the anti-EAF2 monoclonal antibodies: 7A4 at 1:10, 1F11 at 1:20, and 5F6 at 1:10.
CAT assays
293 cells were transfected using Effectene (Qiagen) and 1.5 μg of a GAL4-E1bCAT and E1bCAT or GAL4-tkCAT and tkCAT plasmids, 0.5 μg of a β-gal plasmid, and 2 μg of GAL4-EAF2 or GAL4-EAF1, and GAL4 fused to truncations of EAF2 or EAF1. Total cell lysates were prepared from cells harvested 48 hours after transfection, and expression of each construct was confirmed by Western blot. β-galactosidase values were used to normalize the amount of cell lysate for enzyme-linked immunosorbent assay (ELISA) analysis. Chloramphenicol acetyltransferase (CAT) activity was assayed using an ELISA kit following the manufacturer's recommendations (Boehringer Mannheim, Indianapolis, IN). CAT activity was measured using a microplate reader (Bio-Tek Instruments, Winooski, VT). Transfections and CAT assays were repeated at least 3 times with each set of constructs. Fold activation was expressed as the ratio of the absolute values obtained from samples cotransfected with a reporter plasmid containing GAL4-E1b or tkCAT versus those cotransfected with a reporter plasmid containing E1b or tkCAT (lacking GAL4-DNA binding sites).
Hematopoietic immortalization assay
To obtain normal murine hematopoietic progenitor cells, bone marrow cells were harvested from C57Bl/6 mice 5 days after 5-fluorouracil treatment, incubated with magnetically labeled antibodies to CD5, B220, Cd11b, Gr-1, and TER119, followed by depletion of labeled differentiated cells by passage through a magnetic column (StemCell Technologies, Vancouver, BC, Canada). TheEAF1 and EAF2 fragments were ligated to 5′MLL and cloned in the murine stem cell virus (MSCV) retroviral expression vector. Production of retroviral supernatants in Bosc23 cells was performed as described.15Viral titers were determined by infection of 3T3 cells with Bosc23 retroviral supernatants. For the MSCV vector alone, the viral titers were approximately 5-6 × 105/mL, and for the MLL-EAF2, MLL-EAF1, and MLL-ELL constructs, the viral titers were approximately 2-4 × 105/mL. Infection of lineage-depleted progenitor cells and culture of the transduced progenitors in methylcellulose culture were as previously described.15
Reverse transcriptase–polymerase chain reaction analysis
Cells from primary colonies grown in the presence of G418 were harvested, and total RNA was isolated using RNA STAT-60 (Tel-Test, Friendswood, TX) as recommended by the manufacturer. One microgram of RNA was reversed transcribed into cDNA with enhanced avian myeloblastosis virus (AMV) reverse transcriptase (Sigma). Polymerase chain reaction (PCR) reactions were performed with Taq polymerase using a forward primer from MLL exon 7 and a reverse primer from ELL, EAF1, orEAF2. To exclude a false-positive signal from DNA contamination of the RNA, PCR reactions were also performed on 1 μg aliquots RNA that had not been incubated with reverse transcriptase. As a control for the integrity of the RNA, PCR reactions were also performed using primers from the actin gene.
Results
Isolation of EAF2
To identify proteins that interact with ELL, we performed a yeast 2-hybrid screen using full-length ELL as the bait. We isolated 29 individual clones from this screen, which included 6 clones from the recently described EAF1 gene. We identified 21 identical or overlapping clones from a second novel gene that we namedEAF2 for ELL AssociatedFactor 2. To verify the interaction of EAF2 with ELL and to exclude the possibility that EAF2 might either interact with other proteins in a nonspecific manner or activate theHIS3 or β-gal reporters by itself, we retransformed pGAD10-EAF2 with pAS2-1–ELL and a series of 4 unrelated proteins cloned in the pAS2-1 vector. These assays confirmed the specificity of the interaction of EAF2 with ELL in the yeast 2-hybrid system.
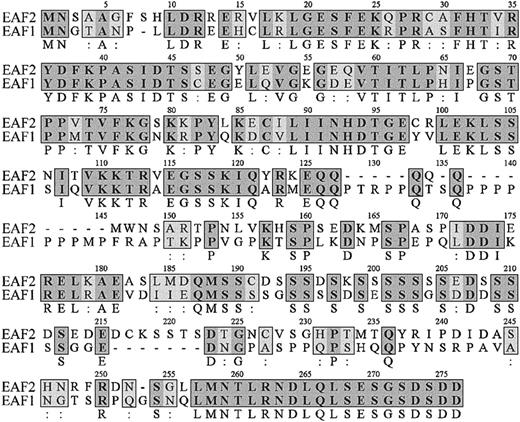
Comparison of EAF2 sequence with GenBank revealed that this gene previously had not been identified. The overlappingEAF2 clones obtained from the library contained a start codon but lacked a termination codon. Searches of the EST database revealed a clone that matched EAF2. Sequence analysis of the 2-hybrid and EST clones enabled us to assemble the full-length open reading frame of EAF2. The EAF2 sequence contains 926 nucleotides with a predicted open reading frame of 780 nucleotides, a 5′ untranslated region of 51 nucleotides and a 3′ untranslated region of 95 nucleotides. Comparison of the predicted amino acid sequences of EAF2 with EAF1 revealed a high level of homology, with 58% identity and 74% amino acid conservation, suggesting that these proteins are members of a novel gene family (Figure1). Moreover, EAF2 andEAF1 demonstrated the highest degree of homology to each other in the GenBank EST database. No other members of this gene family were identified in EST databases. There are several regions of high homology in the predicted amino acid sequences of EAF1 and EAF2, especially at the carboxy-terminal residues, where the last 20 amino acids are identical. To examine the genomic structure ofEAF1 and EAF2, we compared their cDNA sequences to genomic sequence in the Ensembl human genome database and observed that the exon-intron boundaries and reading frames at the exon-intron junctions are highly conserved (Table1).
Amino acid sequence of EAF2.
The amino acid sequence of EAF2 is listed on the top lines with the corresponding sequence of EAF1 below. The numbering of EAF2 residues is shown above each line. Identical amino acids are indicated by bold letters in gray boxes with the residue listed below the box and conserved amino acids by light boxes with 2 dots below the box.
Amino acid sequence of EAF2.
The amino acid sequence of EAF2 is listed on the top lines with the corresponding sequence of EAF1 below. The numbering of EAF2 residues is shown above each line. Identical amino acids are indicated by bold letters in gray boxes with the residue listed below the box and conserved amino acids by light boxes with 2 dots below the box.
Exon-intron boundaries of EAF1 andEAF2
| Exon . | Frame . | Length, bp . | Splice site . |
|---|---|---|---|
| EAF1 | |||
| Exon 1 | — | 257 | CGTTgta--tagATGA |
| Exon 2 | 3 | 95 | CCCTgtg--tagGGAT |
| Exon 3 | 1 | 137 | CAAGgta--cagAGCT |
| Exon 4 | 2 | 191 | AGAGgta--cagAGCT |
| Exon 5 | 3 | 234 | CTCAgta--tagGAAA |
| Exon 6 | 3 | 75 | |
| EAF2 | |||
| Exon 1 | — | 158 | CGCTgtg--tagATGA |
| Exon 2 | 3 | 95 | AGAAgtg--cagGGTT |
| Exon 3 | 1 | 137 | CAAGgta--cagAGTT |
| Exon 4 | 2 | 146 | AGAGgta--cagAACT |
| Exon 5 | 3 | 252 | TTAAgta--tagGAAA |
| Exon 6 | 3 | 138 |
| Exon . | Frame . | Length, bp . | Splice site . |
|---|---|---|---|
| EAF1 | |||
| Exon 1 | — | 257 | CGTTgta--tagATGA |
| Exon 2 | 3 | 95 | CCCTgtg--tagGGAT |
| Exon 3 | 1 | 137 | CAAGgta--cagAGCT |
| Exon 4 | 2 | 191 | AGAGgta--cagAGCT |
| Exon 5 | 3 | 234 | CTCAgta--tagGAAA |
| Exon 6 | 3 | 75 | |
| EAF2 | |||
| Exon 1 | — | 158 | CGCTgtg--tagATGA |
| Exon 2 | 3 | 95 | AGAAgtg--cagGGTT |
| Exon 3 | 1 | 137 | CAAGgta--cagAGTT |
| Exon 4 | 2 | 146 | AGAGgta--cagAACT |
| Exon 5 | 3 | 252 | TTAAgta--tagGAAA |
| Exon 6 | 3 | 138 |
Exon-intron boundaries of EAF1 and EAF2. Exons 2, 3, and 5 encode corresponding amino acids fromEAF1 and EAF2.
— indicates first exon.
The predicted protein of EAF2 contains 260 amino acids and has an estimated pI of 4.84. Similar to EAF1, EAF2 is rich in serine, glutamic acid, and aspartic acid residues. In contrast to EAF1, EAF2 contains a lower percentage of proline residues (4.62% vs 12.69%) and a higher percentage of serines (17.31% vs 14.55%). In light of the amino acid similarity previously observed between EAF1 and AF4 family members, we used the BLASTP and DeCypher II Smith Waterman algorithms to identify a comparable region of EAF2 with limited similarity to AF4 and ENL. This amino acid similarity is highest from amino acids 178 to 201 of EAF2, a domain that is highly enriched in serine, aspartic acid, and glutamic acid residues (Figure 2).
Amino acid similarity of EAF2 to AF4 and ENL.
The region of amino acid similarity identified from the DeCypher II search is rich in serine, aspartic acid, and glutamic acid residues. Identical residues are indicated by black boxes and conserved residues by gray boxes.
Amino acid similarity of EAF2 to AF4 and ENL.
The region of amino acid similarity identified from the DeCypher II search is rich in serine, aspartic acid, and glutamic acid residues. Identical residues are indicated by black boxes and conserved residues by gray boxes.
Northern blot analysis
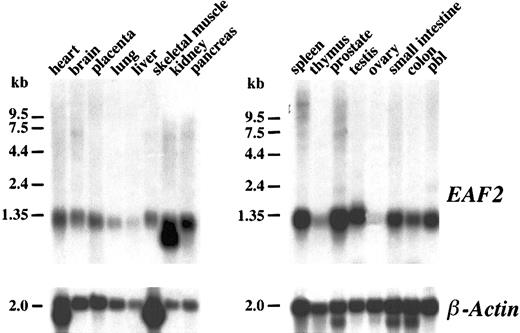
To determine the pattern of expression of EAF2 mRNA in different tissues, a human multiple tissue Northern blot was hybridized with random primed copies of a cDNA fragment containing the open reading frame of EAF2. A single approximately 1.2-kb transcript could be visualized in most tissues (Figure3). However, minimal EAF2expression was observed in the liver, thymus, and ovary. An additional smaller band of approximately 1.0 kb was present in the kidney lane. In multiple tissue Northern blots probed with EAF1, we observed a similar broad pattern of expression. Expression of EAF1also was minimal in the thymus. In contrast to EAF2,EAF1 expression was abundant in the ovary and the liver. Although the size of the predicted proteins of EAF2 and EAF1 are similar (260 vs 268 residues), the size of the EAF1transcript on Northern blots is much greater at approximately 4.5 kb. The large 5′ untranslated sequence of EAF1 accounts for the discrepancy in the size of the transcripts.
Northern blot analysis.
Human multiple tissue Northern blots were hybridized with anEAF2 cDNA probe. EAF2 is broadly expressed in multiple tissues except in liver, thymus, and ovary. The same blot was probed with human β-actin as a control for RNA loading.
Northern blot analysis.
Human multiple tissue Northern blots were hybridized with anEAF2 cDNA probe. EAF2 is broadly expressed in multiple tissues except in liver, thymus, and ovary. The same blot was probed with human β-actin as a control for RNA loading.
EAF2 interacts with ELL and ELL2 in vivo
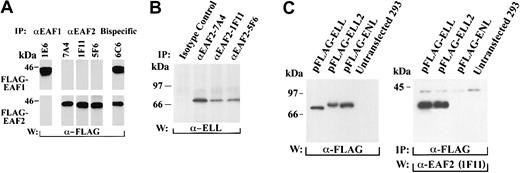
To determine the specificities of the monoclonal antibodies, we transiently transfected 293 cells with FLAG-tagged EAF1 and EAF2, immunoprecipitated with the monoclonal antibodies, and probed with the FLAG antibody. The 7A4, 1F11, and 5F6 antibodies recognized EAF2 but not EAF1 (Figure 4A). The 7A4 antibody recognized the amino-terminal third, 1F11 the middle third, and 5F6 the C-terminal third of EAF2. To determine the specificity of the 1E6 monoclonal antibody that we previously had generated to EAF1, we immunoprecipitated 293 cells transfected with either FLAG-EAF1 or FLAG-EAF2. The 1E6 monoclonal antibody immunoprecipitated EAF1 but not EAF2, confirming its specificity. Clone 6C6 recognized EAF2 and EAF1 equivalently, suggesting that it recognized a highly conserved region in the 2 proteins.
Coimmunoprecipitation of EAF2 and ELL.
(A) Specificity of EAF2 and EAF1 monoclonal antibodies. 293 cells were transfected with FLAG-EAF1 or FLAG-EAF2 constructs. The cell lysates were immunoprecipitated with each of the monoclonal antibodies and blotted with the FLAG antibody. The 1E6 antibody precipitated FLAG-EAF1 but not FLAG-EAF2. In contrast, the 7A4, 1F11, and 5F6 monoclonal antibodies precipitated FLAG-EAF2 but not FLAG-EAF1. The 6C6 monoclonal antibody precipitated FLAG- EAF1 and FLAG-EAF2 equivalently, indicating that this antibody recognizes an epitope that is shared by both proteins. (B) Endogenous EAF2 is in a complex with endogenous ELL in untransfected cells. 293 cell extracts were immunoprecipitated with an isotype-control antibody or with the 3 monoclonal antibodies specific for EAF2. Using an affinity-purified polyclonal ELL antibody, endogenous ELL was detected in the lysates precipitated by the EAF2 antibodies but not in the lysates precipitated by the isotype control. (C) Endogenous EAF2 interacts with transfected ELL2. 293 cells were transfected with FLAG-ELL, FLAG-ELL2, or FLAG-ENL. Expression of these constructs was demonstrated by Western blot analysis of cell lysates with the FLAG antibody (left panel). Cell extracts were immunoprecipitated with the FLAG antibody. Endogenous EAF2 coprecipitated with FLAG-ELL and FLAG-ELL2, as detected by Western blot analysis using the 1F11 anti-EAF2 antibody (right panel). However, EAF2 did not coprecipitate with FLAG-ENL. Endogenous EAF2 migrates at approximately 42 kDa.
Coimmunoprecipitation of EAF2 and ELL.
(A) Specificity of EAF2 and EAF1 monoclonal antibodies. 293 cells were transfected with FLAG-EAF1 or FLAG-EAF2 constructs. The cell lysates were immunoprecipitated with each of the monoclonal antibodies and blotted with the FLAG antibody. The 1E6 antibody precipitated FLAG-EAF1 but not FLAG-EAF2. In contrast, the 7A4, 1F11, and 5F6 monoclonal antibodies precipitated FLAG-EAF2 but not FLAG-EAF1. The 6C6 monoclonal antibody precipitated FLAG- EAF1 and FLAG-EAF2 equivalently, indicating that this antibody recognizes an epitope that is shared by both proteins. (B) Endogenous EAF2 is in a complex with endogenous ELL in untransfected cells. 293 cell extracts were immunoprecipitated with an isotype-control antibody or with the 3 monoclonal antibodies specific for EAF2. Using an affinity-purified polyclonal ELL antibody, endogenous ELL was detected in the lysates precipitated by the EAF2 antibodies but not in the lysates precipitated by the isotype control. (C) Endogenous EAF2 interacts with transfected ELL2. 293 cells were transfected with FLAG-ELL, FLAG-ELL2, or FLAG-ENL. Expression of these constructs was demonstrated by Western blot analysis of cell lysates with the FLAG antibody (left panel). Cell extracts were immunoprecipitated with the FLAG antibody. Endogenous EAF2 coprecipitated with FLAG-ELL and FLAG-ELL2, as detected by Western blot analysis using the 1F11 anti-EAF2 antibody (right panel). However, EAF2 did not coprecipitate with FLAG-ENL. Endogenous EAF2 migrates at approximately 42 kDa.
To exclude an artifact related to transfection and to establish whether endogenous ELL and EAF2 interact in vivo, we examined untransfected 293 and HeLa cells. Cell lysates were immunoprecipitated with the 3 monoclonal antibodies specific for EAF2 or with an isotype control. These immunoprecipitates were then probed with the ELL antiserum. We detected a single band corresponding to endogenous ELL in the cell lysates immunoprecipitated with the EAF2 monoclonals and probed with the ELL antiserum, whereas the control lanes were negative (Figure 4B). Although the ELL antiserum functions in Western blotting and immunofluorescence, it does not immunoprecipitate ELL. To examine whether immunoprecipitation of ELL coprecipitates endogenous EAF2, we transfected 293 cells with FLAG-tagged ELL, immunoprecipitated with the FLAG antibody, and observed a band corresponding to endogenous EAF2 with the 1F11 EAF2 antibody (Figure 4C). Thus, immunoprecipitation of EAF2 coprecipitates ELL, and in the reciprocal experiment, immunoprecipitation of ELL coprecipitates EAF2, confirming that ELL and EAF2 have a direct physical interaction in vivo.
To examine the potential of the endogenous EAF2 protein to interact with ELL2, we transfected 293 cells with FLAG-tagged ELL2, and as a negative control, we used FLAG-ENL. The cell lysates were immunoprecipitated with the FLAG antibody and then probed with the 1F11 EAF2 antibody. We observed that endogenous EAF2 coprecipitated with ELL and ELL2, but not with the ENL control (Figure 4C). Endogenous EAF2 migrated at approximately 42 kDa. Previously, we observed that endogenous EAF1 migrated at approximately 43 kDa.
EAF2 binds to the amino-terminus but not the carboxy-terminus of ELL
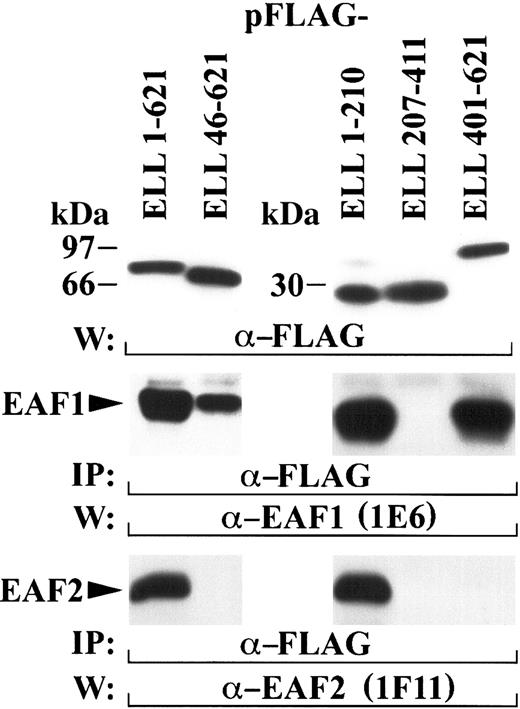
In the (11;19)(q23;p13.1) translocation, amino acids 46-621 of ELL fuse to the amino-terminus of MLL. Previously, we observed that endogenous EAF1 coprecipitated with transiently transfected ELL amino acids 46-621.14 Further mapping revealed that endogenous EAF1 interacts with ELL amino acids 508-621. Fusion of this domain of ELL to MLL was necessary and sufficient for immortalization of hematopoietic cells in vitro and the development of AML in vivo.16 To determine whether the capacity to bind EAF2 was retained by these ELL sequences, we transfected a FLAG-tagged construct containing amino acids 46-621 of ELL. In contrast to EAF1, ELL amino acids 46-621 did not retain the capacity to coprecipitate endogenous EAF2 (Figure 5). To determine the region of ELL that interacts with EAF2, we transfected a series of FLAG-tagged constructs that contained the amino-terminal, middle, and carboxy-terminal regions of ELL. Transfected 293 cells were immunoprecipitated with the FLAG antibody and probed with the EAF2 (1F11) monoclonal. For comparison with the binding of EAF1 to ELL, we also probed with the EAF1 monoclonal. Endogenous EAF2 coprecipitated with the amino-terminal region of ELL (amino acids 1-210), but not with the carboxy-terminus of ELL (amino acids 401-621). Strikingly, endogenous EAF1 also coprecipitated with ELL 1-210, indicating that 2 separable regions of ELL interact with EAF1. However, only the amino-terminus of ELL interacts with EAF2. This amino terminal interaction domain is disrupted in the formation of the MLL-ELL fusion protein, and thus MLL-ELL does not retain the potential to interact with EAF2.
ELL contains 2 separable protein-protein interaction domains.
The amino-terminus of ELL binds to both EAF2 and EAF1, but the carboxy-terminus of ELL binds only to EAF1. 293 cells were transfected with FLAG-tagged constructs containing multiple regions of ELL, immunoprecipitated with the FLAG antibody, and probed with either the 1E6 EAF1 antibody or the 1F11 EAF2 antibody. The top panel shows the expression of the FLAG-tagged ELL constructs detected with the FLAG antibody. Endogenous EAF1 coprecipitated with both the amino-terminus (residues 1-210) and the carboxy-terminus (residues 401-621) of ELL (middle panel). Endogenous EAF2 coprecipitated with amino acids 1-210 of ELL but did not coprecipitate with ELL amino acids 46-621, the region of ELL contributed to the MLL-ELL fusion protein (bottom panel).
ELL contains 2 separable protein-protein interaction domains.
The amino-terminus of ELL binds to both EAF2 and EAF1, but the carboxy-terminus of ELL binds only to EAF1. 293 cells were transfected with FLAG-tagged constructs containing multiple regions of ELL, immunoprecipitated with the FLAG antibody, and probed with either the 1E6 EAF1 antibody or the 1F11 EAF2 antibody. The top panel shows the expression of the FLAG-tagged ELL constructs detected with the FLAG antibody. Endogenous EAF1 coprecipitated with both the amino-terminus (residues 1-210) and the carboxy-terminus (residues 401-621) of ELL (middle panel). Endogenous EAF2 coprecipitated with amino acids 1-210 of ELL but did not coprecipitate with ELL amino acids 46-621, the region of ELL contributed to the MLL-ELL fusion protein (bottom panel).
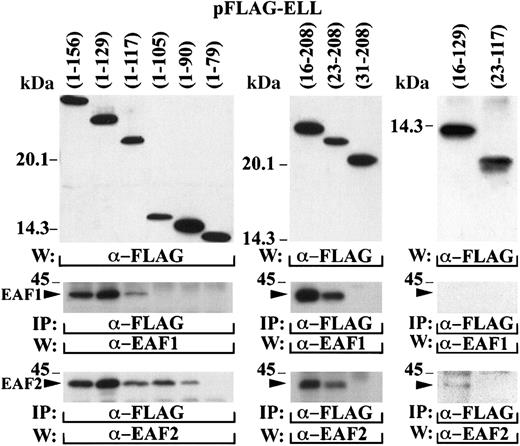
To map the amino-terminal interaction domain more precisely, a series of deletion mutants of ELL were cloned in the pFLAG vector and transiently transfected in 293 cells (Figure6). The cell lysates were immunoprecipitated with the FLAG antibody and probed with the EAF1 and EAF2 monoclonal antibodies. To map the interaction domain, we truncated ELL from both the amino and carboxy-terminal margins of the amino-terminal one third of ELL. Binding to EAF1 was maintained with ELL amino acids 1-117 but not with amino acids 1-105. In contrast, a slightly smaller region at the amino-terminus of ELL (amino acids 1-90) maintained binding to EAF2, indicating a slight heterogeneity in the interaction of EAF2 and EAF1 to this domain of ELL. Binding to both EAF1 and EAF2 was maintained with ELL amino acids 23-208 but not with amino acids 31-108, indicating that the amino-terminal border of the interaction domain was the same for EAF2 and EAF1. We also examined fragments of ELL that truncated both the amino and carboxy-termini of this interaction domain. An ELL fragment that included amino acids 23-117, the minimal boundaries of the interaction domain, did not immunoprecipitate EAF1 or EAF2, indicating that flanking sequence of ELL is important for binding to these proteins. Moreover, the inclusion of additional flanking sequence in the ELL 16-129 construct resulted in weak binding to EAF2 but not to EAF1, confirming that a slightly smaller domain of ELL is sufficient for interaction with EAF2.
Mapping of the amino-terminal interaction domain within ELL.
A series of deletion mutants containing different regions of the amino-terminus of ELL were cloned in the pFLAG vector and transiently transfected in 293 cells. The top panels demonstrate expression of these constructs with the FLAG antibody. The cell lysates were immunoprecipitated with the FLAG antibody and probed with the EAF1 and EAF2 monoclonal antibodies. The bands corresponding to the endogenous EAF1 and EAF2 proteins that coprecipitated with the transfected FLAG-tagged ELL constructs are indicated with an arrowhead.
Mapping of the amino-terminal interaction domain within ELL.
A series of deletion mutants containing different regions of the amino-terminus of ELL were cloned in the pFLAG vector and transiently transfected in 293 cells. The top panels demonstrate expression of these constructs with the FLAG antibody. The cell lysates were immunoprecipitated with the FLAG antibody and probed with the EAF1 and EAF2 monoclonal antibodies. The bands corresponding to the endogenous EAF1 and EAF2 proteins that coprecipitated with the transfected FLAG-tagged ELL constructs are indicated with an arrowhead.
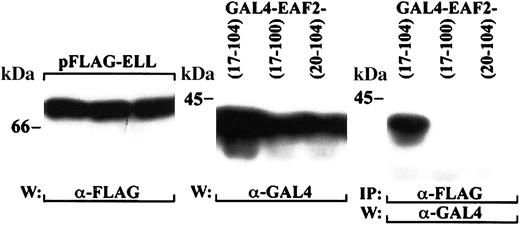
To identify the region of EAF2 required for binding to ELL, we prepared a series of EAF2 truncation mutants in the pGAD10 vector. The pGAD10 EAF2 mutants were cotransfected with pAS2-1-ELL in yeast and assessed for β-galactosidase activity and for growth on histidine dropout media containing 30 mM 3-AT. The smallest fragment that retained binding to ELL in yeast included amino acids 17-104 of EAF2. To determine whether this fragment maintained interaction with ELL in mammalian cells, we cotransfected GAL4-tagged EAF2 17-104 with FLAG-tagged ELL in 293 cells. We also examined 2 constructs that did not interact with ELL in yeast, EAF2 17-100 and EAF2 20-104. As we observed in yeast, EAF2 amino acids 17-104 was sufficient for interaction with ELL in mammalian cells, and further truncation of this region resulted in a loss of binding to ELL (Figure7).
Mapping of the ELL interaction domain within EAF2.
293 cells were cotransfected with GAL4-tagged fragments of EAF2 and FLAG-tagged ELL. The left panel demonstrates expression of full-length ELL with the FLAG antibody for each of the transfections, and the middle panel the expression of the GAL4-EAF2 fragments with the GAL4 antibody. The cell lysates were immunoprecipitated with the FLAG antibody and probed with the GAL4 antibody. EAF2 amino acids 17-104 coprecipitated with ELL, but EAF2 17-100 and 20-104 did not bind to ELL.
Mapping of the ELL interaction domain within EAF2.
293 cells were cotransfected with GAL4-tagged fragments of EAF2 and FLAG-tagged ELL. The left panel demonstrates expression of full-length ELL with the FLAG antibody for each of the transfections, and the middle panel the expression of the GAL4-EAF2 fragments with the GAL4 antibody. The cell lysates were immunoprecipitated with the FLAG antibody and probed with the GAL4 antibody. EAF2 amino acids 17-104 coprecipitated with ELL, but EAF2 17-100 and 20-104 did not bind to ELL.
Colocalization of ELL and EAF2
To determine the subcellular localization of EAF2, we incubated the adherent cell lines HeLa and 293 with the EAF2 antibodies and observed that EAF2 exhibited a speckled nuclear pattern in nondividing cells and a diffuse pattern in dividing cells. Costaining with DAPI revealed that EAF2 localized exclusively to the nucleus. We have previously observed that ELL and EAF1 colocalized in nuclear speckles.14 To determine whether ELL and EAF2 might colocalize, we used each of the 3 EAF2 monoclonal antibodies and examined cells by confocal microscopy. To exclude artifact, we used the fluorochromes Cy2 and Cy5, which have no overlap in spectral emission. Similar to EAF1, we observed that the nuclear speckles of ELL and EAF2 colocalized (Figure 8).
EAF2 colocalizes with ELL in nuclear speckles.
Confocal microscopy with the affinity-purified polyclonal ELL antiserum detected with FITC-labeled goat antirabbit antibodies and with the monoclonal EAF2 antibodies detected with Cy5-labeled goat antimouse antibodies. Merged confocal images of ELL and EAF2 revealed colocalization.
EAF2 colocalizes with ELL in nuclear speckles.
Confocal microscopy with the affinity-purified polyclonal ELL antiserum detected with FITC-labeled goat antirabbit antibodies and with the monoclonal EAF2 antibodies detected with Cy5-labeled goat antimouse antibodies. Merged confocal images of ELL and EAF2 revealed colocalization.
EAF2 contains a transactivation domain in its carboxy terminus
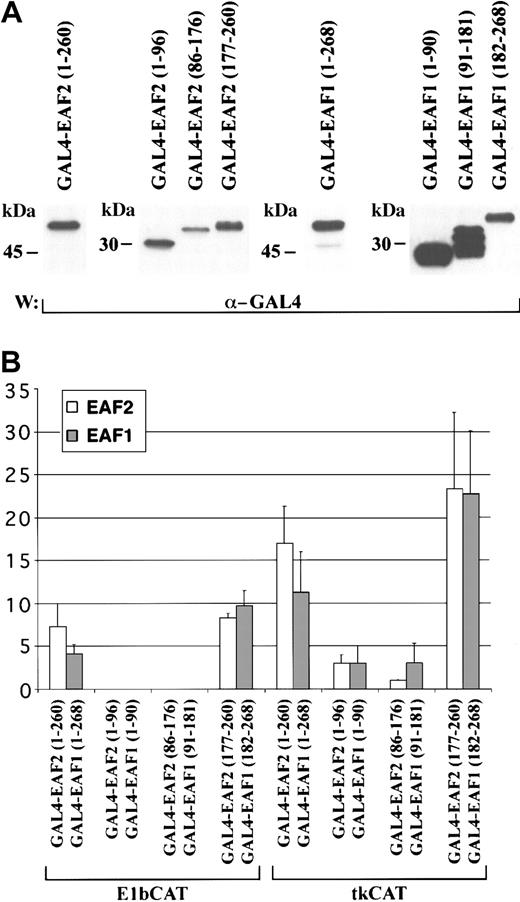
Previously, we observed that EAF1 contains a transactivation domain in the region of its amino acid similarity with AF4 family members.14 To investigate the potential of EAF2 to function as a transcriptional activator, we generated fusions to the GAL4 DNA binding domain with full-length EAF2 and EAF2 deletion mutants. To compare this activity with EAF1, we used comparable deletion mutants of EAF1 in the GAL4 vector. These constructs were then cotransfected into 293 cells with either a GAL4-E1bCAT or a control E1bCAT reporter construct and a β-galactosidase plasmid. In addition, we also assessed the capacity of these constructs to activate transcription using GAL4-tkCAT or a control tkCAT promoter. The E1b construct contains a minimal promoter, and the tk construct exhibits a higher basal activity level. Cell lysates were analyzed in a CAT ELISA assay, normalizing the amount of lysate for each assay by measuring β-galactosidase expression. Expression of each effector construct was confirmed by Western blot analysis (Figure9A). We observed that full-length EAF2 could function as a transactivator. Using the deletion mutants, we mapped this activity to amino acids 177-260 of EAF2 (Figure 9B). This region of EAF2 includes the domain rich in serine, glutamic acid, and aspartic acid residues that exhibits similarity to AF4 and LAF4 (Figure2). This carboxy-terminal region of EAF2 showed the strongest activity, exhibiting a 23.3 ± 8.9-fold increase in activation compared with the control. The carboxy-terminal third of EAF1 (amino acids 182-268) exhibited comparable activity, namely 22.7 ± 7.4-fold activation. The amino-terminal and middle thirds of EAF1 and EAF2 did not exhibit the capacity to activate transcription in this assay. Both EAF1 and EAF2 exhibited a higher overall level of activation with the tk promoter than with the E1b promoter constructs.
EAF2 contains a transactivation domain in its carboxy-terminus.
(A) Western blot of protein extracts from 293 cells transfected with GAL4-EAF2 and GAL4-EAF1 fusion proteins and detected with the GAL4 antibody. (B) Transcriptional activation assay. The indicated fold activation levels are relative to those of the controls. Each bar represents the mean ± standard deviation of at least 3 independent experiments. GAL4-EAF2 constructs are shown in light boxes (■) and the corresponding GAL4-EAF1 constructs in gray boxes (░). The columns represent the following constructs: GAL4-EAF2 1-260 and GAL4-EAF1 1-268, GAL4-EAF2 1-96 and GAL4-EAF1 1-90, GAL4-EAF2 86-176 and GAL4 EAF1 91-181 GAL4-EAF2 177-260, and GAL4-EAF1 182-268. Results with the GAL4-E1bCAT promoter are on the left and the GAL4-tkCAT promoter on the right.
EAF2 contains a transactivation domain in its carboxy-terminus.
(A) Western blot of protein extracts from 293 cells transfected with GAL4-EAF2 and GAL4-EAF1 fusion proteins and detected with the GAL4 antibody. (B) Transcriptional activation assay. The indicated fold activation levels are relative to those of the controls. Each bar represents the mean ± standard deviation of at least 3 independent experiments. GAL4-EAF2 constructs are shown in light boxes (■) and the corresponding GAL4-EAF1 constructs in gray boxes (░). The columns represent the following constructs: GAL4-EAF2 1-260 and GAL4-EAF1 1-268, GAL4-EAF2 1-96 and GAL4-EAF1 1-90, GAL4-EAF2 86-176 and GAL4 EAF1 91-181 GAL4-EAF2 177-260, and GAL4-EAF1 182-268. Results with the GAL4-E1bCAT promoter are on the left and the GAL4-tkCAT promoter on the right.
MLL-EAF2 immortalizes hematopoietic progenitor cells
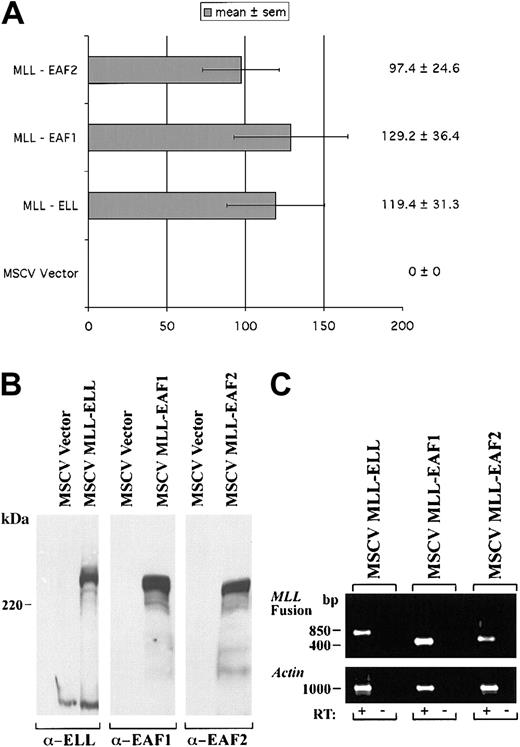
We previously observed that a heterologous fusion of MLL to EAF1 immortalized hematopoietic progenitor cells in serial passages in methylcellulose and induced the development of AML in mice. To assess the potential of EAF2 to exhibit a similar capacity, we prepared a heterologous fusion of EAF2 to MLL, fusing amino acids 1-260 of EAF2 to the amino terminus of MLL in the MSCV vector. Murine hematopoietic progenitor cells were obtained from 5-FU pretreated mice and “spinoculated” with retroviral supernatants containing MLL-ELL, MLL-EAF1, MLL-EAF2, or the MSCV vector alone as a control. The hematopoietic progenitor cells transduced with the MSCV vector alone did not become immortalized. However, we observed a similar number of colonies on tertiary plating with MLL-EAF2, MLL-EAF1, and MLL-ELL, indicating that EAF2 also exhibits the capacity to immortalize hematopoietic cells when fused to MLL (Figure10).
A heterologous MLL-EAF2 fusion immortalizes hematopoietic cells.
The open reading frame of EAF2 was fused to the amino-terminus of MLL in the MSCV vector. High titer retroviruses were generated in Bosc23 cells and used to infect primary murine hematopoietic progenitor cells. For comparison, MLL-ELL, MLL-EAF1, and MSCV vector alone retroviral constructs were also examined. (A) The number of colonies generated in tertiary passage in methylcellulose cultures. (B) Expression of the MLL fusion proteins by Western blot in transiently transfected Bosc23 cells using antibodies to ELL, EAF1, and EAF2. (C) Expression of these constructs by RT-PCR in the transduced hematopoietic cells. Amplification from reverse transcribed cDNA is indicated by a plus symbol (+). To exclude contamination with integrated retroviral genomic DNA, a no reverse transcriptase control is indicated by a minus symbol (−).
A heterologous MLL-EAF2 fusion immortalizes hematopoietic cells.
The open reading frame of EAF2 was fused to the amino-terminus of MLL in the MSCV vector. High titer retroviruses were generated in Bosc23 cells and used to infect primary murine hematopoietic progenitor cells. For comparison, MLL-ELL, MLL-EAF1, and MSCV vector alone retroviral constructs were also examined. (A) The number of colonies generated in tertiary passage in methylcellulose cultures. (B) Expression of the MLL fusion proteins by Western blot in transiently transfected Bosc23 cells using antibodies to ELL, EAF1, and EAF2. (C) Expression of these constructs by RT-PCR in the transduced hematopoietic cells. Amplification from reverse transcribed cDNA is indicated by a plus symbol (+). To exclude contamination with integrated retroviral genomic DNA, a no reverse transcriptase control is indicated by a minus symbol (−).
Discussion
We have isolated and characterized EAF2, a protein highly homologous with EAF1 that also interacts with ELL. The pattern of expression of the 2 genes is similar with the exception of the liver and the ovary, which exhibit minimal expression of EAF2 but not EAF1. Previously, we observed high levels ofELL expression in the liver, suggesting that the potential of ELL to form complexes with EAF1 or EAF2 may vary in different tissues.19 In addition, the expression of both genes is much lower in the thymus than in other tissues. EAF1 and EAF2 are not homologous to EAP20, EAP30, and EAP45, which were purified from rat liver in a multiprotein complex with ELL.20 Although we did not identify these 3 proteins in our yeast 2-hybrid screen, these proteins may not be well represented in bone marrow, which was the source of cDNA for the 2-hybrid library. Using monoclonal antibodies generated to EAF2 followed by detection with an affinity-purified polyclonal antiserum to ELL, we coimmunoprecipitated the endogenous ELL and EAF2 proteins, demonstrating that these proteins have a direct physical interaction in vivo. In transient transfections of epitope-tagged ELL2, we also found that endogenous EAF2 has the potential to bind to ELL2, suggesting the potential for complexes involving multiple members of the ELL and EAF families.
Using the BLASTP and the DeCypher II algorithms to compare the predicted amino acid sequence of EAF2 with that of known proteins, we identified several translocation partner proteins of MLL including AF4, AF5q31, LAF4, ENL, and AF9. AF4, AF5q31, and LAF4 are highly homologous to each other, as are ENL and AF9. The amino acid similarity of these proteins to EAF1 and EAF2 is limited to a region that is rich in serine, aspartic acid, and glutamic acid residues. Moreover, a transcriptional activation domain has been mapped to this region in each of these proteins. However, this region of similarity represents a relatively small domain within each of these proteins.
In view of the amino acid similarity of EAF1 to the transcriptional activation domains of several MLL partner proteins, we examined EAF2 for the potential to function as a transactivator. We found that the full-length EAF2 protein could function as an activator of transcription. This activity mapped to the C-terminal one third of EAF2, which contains the region of similarity to AF4 and ENL family members. Compared with the full-length EAF2 protein, the potency of transcriptional activation was greater in the C-terminal one third of EAF2. A direct comparison of the corresponding regions of EAF2 and EAF1 revealed that the transcriptional activation potential of these proteins is quite similar.
Previously, we observed that EAF1 colocalized with ELL in nuclear speckles. Confocal microscopy revealed that ELL and EAF2 also colocalized in a nuclear speckled distribution. This pattern was observed with each of 3 EAF2 monoclonal antibodies. At this time, it is not clear whether these nuclear speckles represent storage forms of these proteins or a distinct substructure within the nucleus. In dividing cells, ELL, EAF1, and EAF2 were distributed diffusely, consistent with the dissolution of the nuclear membrane during cell division. The disappearance of the speckles in dividing cells may be associated with the lack of transcription by Pol II during mitosis. We did not hybridize cells with the monoclonal antibodies to EAF1 and EAF2 simultaneously, as the 2 antibodies would not be distinguishable using antimouse secondary antibodies. Thus, subtle distinctions in their subcellular distribution may exist. However, we observed a similar pattern of colocalization with ELL for both proteins. In addition, all the speckles that were observed with the ELL antibody were colocal with either EAF1 or EAF2. These data suggest that either ELL binds to EAF1 and EAF2 simultaneously or that distinct ELL/EAF1 or ELL/EAF2 complexes localize in the same speckles within the nucleus.
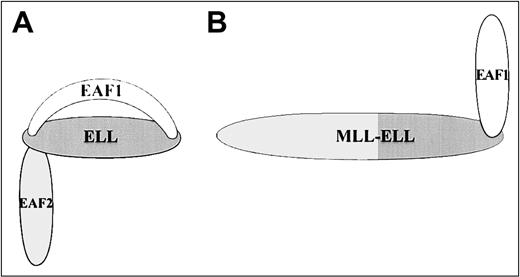
We have observed a high level of homology between EAF1 and EAF2, a similar tissue distribution with the exception of the liver and ovary, and a similar colocalization with ELL in nuclear speckles. Functional assays with EAF1 and EAF2 also have resulted in similar phenotypes with both proteins capable of transcriptional activation and of immortalization of hematopoietic cells when fused to MLL. These data suggest a potential redundancy in these 2 proteins. However, we observed a striking difference in the pattern of binding to ELL. ELL contains 2 separable binding domains for EAF1, with one in the amino-terminus and the other in the carboxy-terminus. In contrast, EAF2 binds only to the amino-terminus of ELL (Figure11). Although the functional significance of this distinction is not yet clear, one consequence is that the MLL-ELL fusion protein would retain an interaction domain for EAF1 but not for EAF2. In addition, the potential for competition exists for binding by EAF1 and EAF2 at the amino-terminal interaction domain of ELL but not at the carboxy-terminal interaction domain. Thus, MLL-ELL disrupts the normal network of protein-protein interactions among ELL, EAF1, and EAF2.
Model of ELL protein-protein interactions with EAF1 and EAF2.
(A) EAF1 and EAF2 bind to the amino-terminus of the wild-type ELL protein, but only EAF1 binds to the carboxy-terminus of ELL. (B) The MLL-ELL fusion protein disrupts the amino-terminal protein-protein interaction domain of ELL but retains the carboxy-terminal EAF1 interaction domain.
Model of ELL protein-protein interactions with EAF1 and EAF2.
(A) EAF1 and EAF2 bind to the amino-terminus of the wild-type ELL protein, but only EAF1 binds to the carboxy-terminus of ELL. (B) The MLL-ELL fusion protein disrupts the amino-terminal protein-protein interaction domain of ELL but retains the carboxy-terminal EAF1 interaction domain.
Using a hematopoietic progenitor cell immortalization assay, we recently performed a structure-function analysis of the ELLgene.16 Whereas the elongation domain of ELL was dispensable for immortalization; fusion of the carboxy-terminus of ELL to MLL was necessary and sufficient to immortalize hematopoietic progenitor cells. In contrast, a construct containing the amino-terminus of MLL alone and fusions of the amino-terminus of MLL to ELL fragments lacking the EAF1 interaction domain were incapable of immortalizing hematopoietic cells. The carboxy-terminus of ELL contains an EAF1 interaction domain, suggesting that retention of this protein-protein interaction domain may be relevant to leukemic transformation by MLL-ELL. The region of ELL that interacts with EAF1 corresponded precisely to the region necessary for immortalization. To examine whether this interaction with EAF1 was critical to immortalization, we prepared a heterologous fusion of MLL to EAF1. We observed that this MLL-EAF1 fusion protein immortalized hematopoietic progenitor cells in vitro and induced AML in mice. Similarly, we have now observed that a heterologous MLL-EAF2 fusion protein is also capable of immortalizing hematopoietic progenitor cells in vitro. At this time, no chromosome translocations affecting either EAF1 or EAF2 have been described. Although the MLL-ELL fusion protein does not retain the capacity to bind to EAF2, the high homology between EAF1 and EAF2 suggests that both proteins contribute a similar immortalizing function to the heterologous fusions with MLL. At this time, the functional contributions of EAF1 or EAF2 to this MLL fusion remain unclear. However, both EAF2 and EAF1 contain transactivation domains that are similar to activation domains in other MLL partner proteins. The contribution of a transactivation domain of this class may be a critical feature of multiple MLL fusion proteins. In addition, the MLL-EAF2 construct retains the capacity to bind to ELL, which may indirectly recruit EAF1 to a multiprotein complex. In addition to the fusion of MLL to a large number of diverse partner proteins, disruption of the MLL protein by internal tandem duplications also results in leukemia. Thus, the various subtypes of MLL leukemia may affect several independent pathways that regulate normal hematopoiesis. Further studies will be necessary to dissect the critical contributions of ELL, EAF1, and EAF2 to leukemogenesis.
We would like to thank Carol McShan and the University of Chicago Frank Fitch Monoclonal Antibody Facility (supported by grant CA14599 from the National Cancer Institute) and the Al Robin Laser Scanning Confocal Microscopy Core of the University of Chicago Digestive Disease Center.
Prepublished online as Blood First Edition Paper, November 21, 2002; DOI 10.1182/blood-2002- 06-1664.
Supported by grant CA78431 from the National Institutes of Health, a Scholar Award from the Leukemia and Lymphoma Society, and the family of Robert A. Chapski.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michael J. Thirman, University of Chicago, Section of Hematology/ Oncology, 5841 S Maryland Ave, MC2115, Chicago, IL 60637; e-mail:mthirman@medicine.bsd.uchicago.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal