Cellular iron uptake in most tissues occurs via endocytosis of diferric transferrin (Tf) bound to the transferrin receptor (TfR). Recently, a second transferrin receptor, transferrin receptor 2 (TfR2), has been identified and shown to play a critical role in iron metabolism. TfR2 is capable of Tf-mediated iron uptake and mutations in this gene result in a rare form of hereditary hemochromatosis unrelated to the hereditary hemochromatosis protein, HFE. Unlike TfR, TfR2 expression is not controlled by cellular iron concentrations and little information is currently available regarding the role of TfR2 in cellular iron homeostasis. To investigate the relationship between TfR and TfR2, we performed a series of in vivo and in vitro experiments using antibodies generated to each receptor. Western blots demonstrate that TfR2 protein is expressed strongest in erythroid/myeloid cell lines. Metabolic labeling studies indicate that TfR2 protein levels are approximately 20-fold lower than TfR in these cells. TfR and TfR2 have similar cellular localizations in K562 cells and coimmunoprecipitate to only a very limited extent. Western analysis of the receptors under nonreducing conditions reveals that they can form heterodimers.
Introduction
Iron is an essential nutrient required for a variety of biochemical processes such as respiration, metabolism, and DNA synthesis. To maintain intracellular iron levels, cells possess tightly regulated mechanisms for iron absorption and metabolism. Transferrin (Tf), the major iron transport protein in the blood, is taken up into cells by binding to the transferrin receptor 2 (TfR2). This homodimeric membrane receptor binds 2 Tf molecules and is internalized into endosomes that are acidified, resulting in the release of iron from Tf. Iron is transported across the vesicle membrane for utilization and/or storage within the cell, and the TfR-Tf complex recycles back to the cell surface where apo-Tf is released at the higher pH of blood (pH 7.4; reviewed in Aisen et al1). The TfR plays a critical role in iron homeostasis. The TfR knock-out mouse results in embryonic lethality.2
The recently identified TfR2, a second distinct Tf receptor, is most likely responsible for the non–TfR-mediated uptake of Tf into cells, and it also plays a critical role in iron homeostasis.3 Mutations in this gene are associated with a rare form of hemochromatosis unrelated to mutations in the hereditary hemochromatosis protein, HFE.3 TfR2 can support growth of a transfected Chinese hamster ovary cell line lacking endogenous transferrin receptors when given Tf as an iron source.4However, TfR2 expression is not sufficient to replace the function of TfR, because mice in which the TfR gene has been deleted die as embryos.2TfR2, like TfR, is a type II membrane glycoprotein with a large C-terminal ectodomain and a small N-terminal cytoplasmic domain.5,6 TfR2 shares 45% amino acid sequence identity with TfR in the extracellular region, contains a cytoplasmic internalization motif similar to TfR, and has 2 cysteines, which form intersubunit disulfide bonds, in the ectodomain proximal to the transmembrane domain.5 6
Clear differences exist between the 2 transferrin receptors despite their similarities. Both receptors bind diferric Tf better than apotransferrin at neutral pH, however the affinity of TfR2 for Tf is approximately 25-fold lower than that of TfR.5 While TfR and HFE are associated in the placenta and transfected cells,7,8 and in vitro binding assays demonstrate that the ectodomain of TfR binds to HFE with nM affinity,9-11 the TfR2 ectodomain does not detectably bind to HFE.12 In humans and mice, TfR2 is expressed predominantly in liver and erythroid cells, while TfR is expressed in a wider range of tissues.5,6,13 Even though the ectodomains of the 2 TfRs are similar, their cytoplasmic domains share no sequence homology. TfR expression is controlled primarily at the posttranscriptional level in response to cellular iron levels, while TfR2 expression is not influenced by changes in cellular iron levels.4-6 TfR2 expression is controlled at the transcriptional level by the erythroid transcription factor GATA-1.14
Although TfR2 can mediate cellular iron uptake in transfected cells, little is known about its physiologic function and potential interaction with TfR in cell lines that express both receptors. In this study, we investigated the interaction of TfR and TfR2 from the K562 chronic myelogenous cell line with erythrocytic features and from human liver tissue, and we compared the quantities and cellular localization of each receptor. TfR is more abundant than TfR2 in K562 cells while the reverse is true in human liver. We found that in K562 cells TfR and TfR2 colocalize, but coprecipitate to only a limited extent. In liver, only limited coprecipitation of the 2 receptors is detected. These data suggest that homotypic more than heterotypic interactions at the dimer interface are favored.
Materials and methods
Generation of monoclonal antibodies to TfR and TfR2
Soluble versions of human TfR and TfR2 were expressed separately in a lytic baculovirus/insect cell expression system as described previously.9,12 Briefly, constructs encoding the ectodomain of TfR or TfR2 were joined to a gene segment encoding the leader peptide from the baculovirus protein GP67, a 6xHis-tag, and a Factor Xa cleavage site in a modified form of the pAcGP67A expression vector (Pharmingen, San Diego, CA). Recombinant virus was generated by cotransfection of the transfer vector with linearized viral DNA (Baculogold; Pharmingen). TfR or TfR2 was purified from supernatants of baculovirus-infected High 5 cells using nickel-nitrilotriacetic acid chromatography (Ni-NTA Superflow; Qiagen, Valencia, CA) followed by gel filtration chromatography using a Superdex 200 fast protein liquid chromatography column (Amersham Pharmacia Biotech, Piscataway, NJ). Monoclonal antibodies 3B8 2A1 and 9F8 1C11 were generated against the purified ectodomains of the human TfR and human TfR2, respectively. Female BALB/c mice (aged 5 weeks) were primed and boosted twice (at 2-week intervals) by intraperitoneal injection of 100 μg of the purified ectodomain of TfR2 or TfR in adjuvant. Serum was screened one week after each injection by enzyme-linked immunoassay (ELISA) as described.15 At 3 days preceding the fusion, one mouse was boosted with 100 μg of purified TfR2 or TfR. Splenocytes from the boosted mouse were fused with HL-1 murine myeloma cells. Media from the hybridoma cultures were tested for antibodies against TfR2 or TfR by ELISA and subsequently by Western blotting. After subcloning positive clones at clonal density, ascites tumors were produced in pristine-primed BALB/c mice. These monoclonals were selected by ELISA assay and then screened for their ability to detect TfR2 or TfR by Western blot analysis. Both antibodies were the immunoglobulin G1 (IgG1), κ subtype. Antibodies to TfR2 did not interact with the ectodomain of TfR by either Western blot or ELISA analysis. The opposite was also true.
Cell lines
HepG2 cells (human hepatocarcinoma) were obtained from the Vollum Institute (Portland, OR), Huh7 cells (human hepatocarcinoma) were kindly provided by Dr Philip Aisen (Einstein University, Bronx, NY). TRVb1 and TRVb cells were gifts of Dr Tim McGraw (Cornell Medical College, New York, NY). The TRVb2 cell line was generated by transfection of TRVb cells with pCDNA 3.1 encoding TfR2 with a FLAG epitope on the N-terminus, selected with G418, and subcloned as described previously.16 The plasmid was the gift of Drs Koeffler and Kawabata (University of California, Los Angeles). All other cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA).
Immunodetection
K562 cells were maintained in RPMI-1640 (Sigma, St Louis, MO) supplemented with 10% fetal bovine serum. Cells were collected and counted on a hemocytometer, washed 3 times with ice-cold phosphate-buffered saline (PBS; pH 7.4) and lysed at a concentration of 1 × 107 cells/mL in NET-Triton buffer (150 mM NaCl, 5 mM EDTA [ethylenediaminetetraacetic acid], 10 mM Tris, pH 7.4, 1% Triton X-100). Cell extracts were incubated with 2 × Laemmli buffer17 and subjected to electrophoresis on 8% polyacrylamide gels under both reducing and nonreducing denaturing conditions. Separated proteins were transferred to nitrocellulose and blocked overnight at 4°C with 5% milk in 0.01 M Tris-HCl, 0.15 M NaCl, pH 7.4, plus 0.05% Tween-20. Immunoblot analysis was performed using either a sheep anti-TfR serum (1:10 000 dilution)18 or the monoclonal anti-TfR2 (9F8 1C11) antibody (1:10 000 dilution) followed by the appropriate secondary antibody conjugated to horseradish peroxidase and chemiluminescence (Supersignal; Pierce, Rockford, IL) per the manufacturer's directions.
Immunoprecipitation
K562 cells were washed 3 times with ice-cold PBS and lysed with NET-Triton buffer (0.05 M Tris-Cl, 0.15 M NaCl, 5 mM EDTA, pH 7.4, 1% Triton X-100) followed by centrifugation at 2000g for 5 minutes to remove nuclei. Cell lysates were incubated for 60 minutes at 4°C with either 25 μL of protein A–Sepharose (Amersham Pharmacia Biotech) or 25 μL of protein A–Sepharose coated with affinity-purified rabbit anti–mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA), and 1.5 μL of either sheep anti-TfR serum or mouse anti-TfR2. The pellet was resuspended into 100 μL NET-Triton buffer, layered on top of 1 mL of the same buffer with 15% sucrose, and pelleted. Samples were eluted in 30 μL of 2 × Laemmli buffer,17subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis on an 8% polyacrylamide gel, transferred to nitrocellulose, and immunodetected for either TfR or TfR2.
Quantitation of TfR and TfR2
Subconfluent K562 cells were incubated overnight at 37°C in RPMI-1640 medium without methionine (Life Technologies, Invitrogen, Carlsbad, CA) with 50 μCi (1.85 MBq) of35S-methionine/cysteine with 10% fetal bovine serum. The cells were then washed 3 times with ice-cold PBS and lysed in NET-Triton, and the nuclei pelleted. Cell extracts were subjected to immunoprecipitation with either sheep anti-TfR or mouse anti-TfR2 antibody as described in “Immunoprecipitation” and analyzed by SDS-PAGE on an 8% acrylamide gel under reducing and denaturing conditions. Gels were fixed, treated with Amplify (Amersham Pharmacia Biotech) for 30 minutes, dried, and exposed to a PhosphorImager screen (Amersham Pharmacia Biotech). Quantitation of the amount of TfR and TfR2 took into account the differences in methionine and cysteine composition of the 2 receptors (22 Met and Cys for TfR and 15, for TfR2).
Interaction of soluble HFE with cell extracts
Cell extracts from K562 cells were incubated with the ectodomain of purified recombinant HFE/β2 microglobulin (final concentration 1 μM) for 60 minutes at 4°C (K562 + HFE) and immunoprecipitated using sheep anti-TfR serum and Staphylococcus aureus (Pansorbin; Calbiochem, San Diego, CA). The ectodomain of purified recombinant HFE/β2 microglobulin was generated as previously described.9 Immunoprecipitated proteins were subjected to SDS-PAGE on an 8% acrylamide gel, transferred to nitrocellulose, and immunodetected for TfR2 or TfR.
Immunohistochemistry
K562 cells were fixed in 4% paraformaldehyde, blocked with 2.5 mg/mL bovine serum albumin (BSA), then incubated with sheep anti-TfR (1:50) and mouse anti-TfR2 antibody (1:300) for 1 hour at room temperature. Cells were layered onto 500 μL of fetal bovine serum, centrifuged at 1000gfor 2 minutes, and resuspended, followed by incubation for 60 minutes with both Alexa 488 conjugated donkey antisheep antibody (1:500) (Molecular Probes, Eugene, OR) and donkey anti–mouse IgG (Jackson ImmnunoResearch Laboratories) conjugated to Alexa 594 using an Alexa Fluor 594 Protein Labeling Kit (1:100) (Molecular Probes). Cells were again layered on top of 500 μL fetal bovine serum, pelleted, washed with PBS, and mounted on slides using Prolong Antifade (Molecular Probes). Images were obtained using a BioRad 1024 ES laser scanning confocal system (Hercules, CA) on a Nikon Eclipse TE300 microscope (Melville, NY) with a × 60 oil immersion Planapo objective. Permeabilized cells were treated with NET-Triton after fixation and prior to incubation with antibodies.
Results
Characterization of TfR2 expression
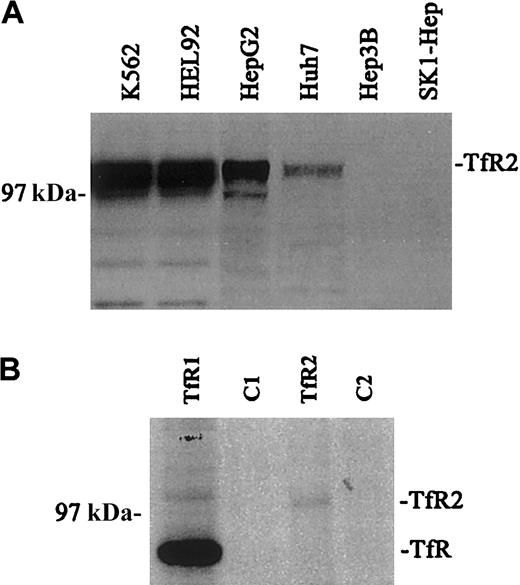
The relative amounts of TfR2 in a variety of human cell lines were visualized by Western blot analysis using the mouse monoclonal antibody (9F8 1C11) against the ectodomain of human TfR2 (Figure 1A). Closely migrating bands (2-3) of between approximately 97 and 105 kDa were observed for TfR2. Multiple bands have been observed previously and have been attributed to heterogeneity in glycosylation.4 The TfR2 signal was strongest in K562 and HEL 92 cells (erythroleukemia cell lines), moderate in HepG2 and Huh7 cells (hepatoblastoma cell lines), and undetectable in the Hep3B and SK1 Hep cell lines (hepatoblastoma cell lines). These results correlate with previous work, which demonstrated that TfR2 mRNA was expressed strongly in both K562 and HepG2 cells by Northern blot and reverse transcriptase–polymerase chain reaction (RT-PCR) analysis.5 In contrast, TfR protein levels were strong and uniform in these and other cell lines tested (data not shown).
Expression of TfR and TfR2.
(A) The relative amounts of TfR2 were visualized in a variety of human cell lines by Western blots. Equal amounts of cell extracts (25 μg) were loaded onto gels and subjected to SDS-PAGE. TfR2 was detected with the mouse monoclonal antibody to human TfR2 (9F8 1C11) followed by a goat antimouse/horseradish peroxidase (HRP) secondary antibody. All blots were developed by chemiluminescence (Pierce): K562, HEL 92 (erythroid leukemia), HepG2, Huh7, Hep 3B, and SK-1 Hep (hepatoma). The TfR2 signal produces multiple bands between 97 and 105 kDa. (B) Quantitative immunoprecipitation of TfR and TfR2. K562 cells were labeled overnight with 50 μCi (1.85 MBq) 35S-methionine, lysed, and quantitatively immunoprecipitated with either a sheep antihuman TfR serum and protein A–Sepharose (Pharmacia) (TfR); cell extract and protein A–Sepharose as a control (C1); the TfR2 monoclonal antibody (9F8 1C11) and rabbit anti–mouse IgG–coated protein A–Sepharose (TfR2); or cell extract and rabbit antimouse coated protein A–Sepharose as a control (C2). Immunoprecipitated proteins were eluted with 2 × Laemmli buffer, run on an SDS-8% polyacrylamide gel, dried, and exposed to film. The relative amount of radioactivity in each band was determined by PhosphorImager analysis correcting for the Met/Cys content of each TfR. Reimmunoprecipitation of the supernatants of the immunoprecipitates showed that all of TfR and TfR2 were bound in the first immunoprecipitation (results not shown).
Expression of TfR and TfR2.
(A) The relative amounts of TfR2 were visualized in a variety of human cell lines by Western blots. Equal amounts of cell extracts (25 μg) were loaded onto gels and subjected to SDS-PAGE. TfR2 was detected with the mouse monoclonal antibody to human TfR2 (9F8 1C11) followed by a goat antimouse/horseradish peroxidase (HRP) secondary antibody. All blots were developed by chemiluminescence (Pierce): K562, HEL 92 (erythroid leukemia), HepG2, Huh7, Hep 3B, and SK-1 Hep (hepatoma). The TfR2 signal produces multiple bands between 97 and 105 kDa. (B) Quantitative immunoprecipitation of TfR and TfR2. K562 cells were labeled overnight with 50 μCi (1.85 MBq) 35S-methionine, lysed, and quantitatively immunoprecipitated with either a sheep antihuman TfR serum and protein A–Sepharose (Pharmacia) (TfR); cell extract and protein A–Sepharose as a control (C1); the TfR2 monoclonal antibody (9F8 1C11) and rabbit anti–mouse IgG–coated protein A–Sepharose (TfR2); or cell extract and rabbit antimouse coated protein A–Sepharose as a control (C2). Immunoprecipitated proteins were eluted with 2 × Laemmli buffer, run on an SDS-8% polyacrylamide gel, dried, and exposed to film. The relative amount of radioactivity in each band was determined by PhosphorImager analysis correcting for the Met/Cys content of each TfR. Reimmunoprecipitation of the supernatants of the immunoprecipitates showed that all of TfR and TfR2 were bound in the first immunoprecipitation (results not shown).
Quantitation of TfR and TfR2 in K562 cells
To measure the relative amounts of TfR and TfR2 protein, cell lysates from35S-methionine/cysteine-labeled K562 cells were immunoprecipitated with either a sheep antihuman TfR antiserum or the mouse antihuman TfR2 monoclonal antibody (9F8 1C11), separated on a 8% acrylamide gel under reducing and denaturing conditions, dried, and exposed to a PhosphorImager (Figure 1B). The amount of TfR2 protein is approximately 20 times less than TfR protein. Background of the35S-labeled immunoprecipitates prevented the quantitation of the amount of each receptor that formed heterodimers. We estimate that if 5% of TfR formed heterodimers with TfR2 then TfR should have been detectable in the TfR2 immunoprecipitates. If less than 25% of the TfR2 coprecipitated with TfR1 then the heterodimers would not be detectable by this method. Thus although the 2 TfR receptors coprecipitate in K562 cells, the coprecipitated material accounts for only less than 5% of the total amount of receptors.
TfR and TfR2 association
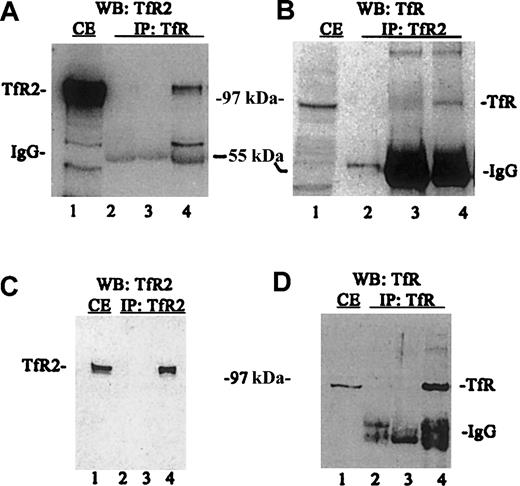
A series of immunoprecipitations and Western blots of the immunoprecipitated proteins were performed using TfR and TfR2 antibodies to determine the relative amounts of each protein in this cell line (Figure2). When TfR was immunoprecipitated from K562 cell lysates followed by Western blotting for TfR2, TfR2 clearly coimmunoprecipitates with TfR (Figure 2A, lane 4). TfR2 also coprecipitates with TfR (Figure2B, lane 4), as demonstrated by conducting the experiment in the reverse order (immunoprecipitate TfR2 and immunoblot for TfR). The coprecipitation does not result from an artifactual association of TfR or TfR2 with reagents used in the immunoprecipitation, since TfR does not bind protein G–Sepharose in the absence of antibody (Figure2B, lane 3), TfR2 does not bind S aureus in the absence of antibody (Figure 2A, lane 3) and both S aureus and protein G–Sepharose contain no immunoreactive bands (Figure 2A, lane 2 and 2B, lane 2, respectively). To measure the total amount of TfR2 and TfR that immunoprecipitated, a series of control blots were performed (Figure 2C-D). Immunoprecipitation with TfR2 antibody andS aureus, followed by immunodetection for TfR2, shows that TfR2 can be quantitatively precipitated with this antibody (Figure 2C, lanes 1,4). Similarly, TfR can be quantitatively precipitated from lysates (Figure 2D, lanes 1,4). These results indicate that that homodimer formation is strongly preferred.
TfR and TfR2 are associated in vitro.
Cell extracts from K562 cells were solubilized (1 × 107cells/mL) as described in “Materials and methods,” immunoprecipitated, subjected to SDS-PAGE, transferred to nitrocellulose, and immunodetected using TfR (WB:TfR) and TfR2 antibodies (WB:TfR2). (A) K562 cell extracts (50 mL) were immunoprecipitated with a sheep antihuman TfR antiserum and S aureus (IP:TfR), and detected by Western blotting using the mouse monoclonal antibody to human TfR2 (9F8 1C11) and an antimouse/HRP-conjugated secondary antibody. All blots were visualized by chemiluminescence (Pierce). Lanes are as follows: (1) K562 extract (25 mL); (2) S aureus and TfR antibody alone; (3) S aureus and K562 extract alone; and (4) S aureus,anti-TfR serum, and K562 extract. (B) K562 extract from 5 × 105 cells was immunoprecipitated with the monoclonal TfR2 antibody (9F8 1C11) and protein G–Sepharose (IP:TfR2) and detected with sheep anti-TfR serum and swine antisheep/HRP-conjugated secondary antibody on Western blots. Lanes are as follows: (1) K562 extract (25 mL); (2) protein G–Sepharose and TfR2 monoclonal antibody (9F8 1C11) only; (3) protein G–Sepharose and K562 extract (50 mL) only; and (4) protein G–Sepharose, TfR2 monoclonal antibody (9F8 1C11), and K562 extract (50 mL). (C) K562 extract (50 mL) immunoprecipitated with anti-TfR2 monoclonal antibody (9F8 1C11) (IP:TfR2) and immunodetected for TfR2 by Western blots. Lanes are as follows: (1) K562 extract (25 mL); (2) TfR2 monoclonal antibody (9F8 1C11) isolated with protein G–Sepharose; (3) K562 extract (50 mL) isolated with S aureus alone; and (4) TfR2 isolated from K562 extracts (50 mL) with anti-TfR2 monoclonal antibody (9F8 1C11) and protein G–Sepharose. (D) K562 extract (50 mL) immunoprecipitated with anti-TfR (IP:TfR) and Western blotted for TfR (WB:TfR). Lanes are as follows: (1) K562 extract (25 mL); (2) S aureus and anti-TfR serum only; (3) K562 extract (50 mL) incubated with S aureus only; and (4) K562 extract (50 mL) immunoprecipitated with anti-TfR serum and S aureus. IP indicates immunoprecipitated; WB, Western blot; and CE, cell extract.
TfR and TfR2 are associated in vitro.
Cell extracts from K562 cells were solubilized (1 × 107cells/mL) as described in “Materials and methods,” immunoprecipitated, subjected to SDS-PAGE, transferred to nitrocellulose, and immunodetected using TfR (WB:TfR) and TfR2 antibodies (WB:TfR2). (A) K562 cell extracts (50 mL) were immunoprecipitated with a sheep antihuman TfR antiserum and S aureus (IP:TfR), and detected by Western blotting using the mouse monoclonal antibody to human TfR2 (9F8 1C11) and an antimouse/HRP-conjugated secondary antibody. All blots were visualized by chemiluminescence (Pierce). Lanes are as follows: (1) K562 extract (25 mL); (2) S aureus and TfR antibody alone; (3) S aureus and K562 extract alone; and (4) S aureus,anti-TfR serum, and K562 extract. (B) K562 extract from 5 × 105 cells was immunoprecipitated with the monoclonal TfR2 antibody (9F8 1C11) and protein G–Sepharose (IP:TfR2) and detected with sheep anti-TfR serum and swine antisheep/HRP-conjugated secondary antibody on Western blots. Lanes are as follows: (1) K562 extract (25 mL); (2) protein G–Sepharose and TfR2 monoclonal antibody (9F8 1C11) only; (3) protein G–Sepharose and K562 extract (50 mL) only; and (4) protein G–Sepharose, TfR2 monoclonal antibody (9F8 1C11), and K562 extract (50 mL). (C) K562 extract (50 mL) immunoprecipitated with anti-TfR2 monoclonal antibody (9F8 1C11) (IP:TfR2) and immunodetected for TfR2 by Western blots. Lanes are as follows: (1) K562 extract (25 mL); (2) TfR2 monoclonal antibody (9F8 1C11) isolated with protein G–Sepharose; (3) K562 extract (50 mL) isolated with S aureus alone; and (4) TfR2 isolated from K562 extracts (50 mL) with anti-TfR2 monoclonal antibody (9F8 1C11) and protein G–Sepharose. (D) K562 extract (50 mL) immunoprecipitated with anti-TfR (IP:TfR) and Western blotted for TfR (WB:TfR). Lanes are as follows: (1) K562 extract (25 mL); (2) S aureus and anti-TfR serum only; (3) K562 extract (50 mL) incubated with S aureus only; and (4) K562 extract (50 mL) immunoprecipitated with anti-TfR serum and S aureus. IP indicates immunoprecipitated; WB, Western blot; and CE, cell extract.
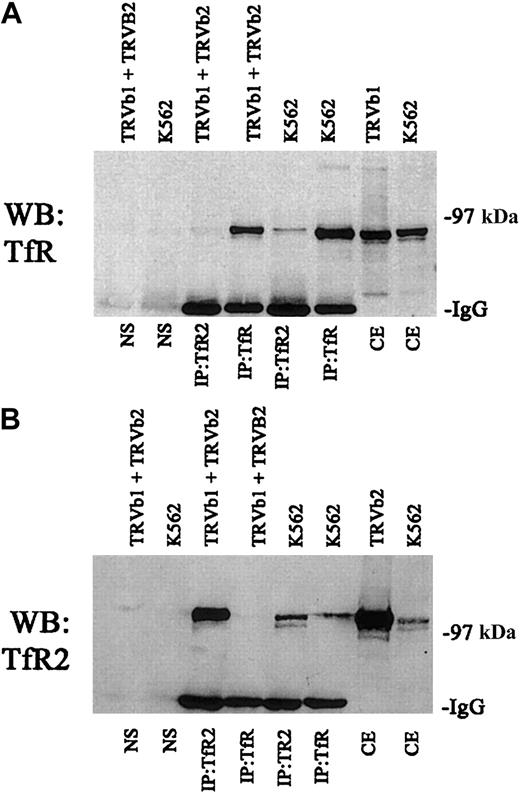
We also tested to determine if the 2 receptors had to be expressed in the same cell in order to associate or if higher order complexes formed only after the cells were lysed. For these experiments, we transfected TRVb cells, a Chinese hamster ovary cell line lacking endogenous transferrin receptor.19 A series of immunoprecipitations were carried out with TRVb cells stably transfected with TfR (TRVb1) or TfR2 (TRVb2) (Figure3). When cell lysates from these 2 cell lines were mixed and immunoprecipitated with anti-TfR2, no TfR could be detected (Figure 3A). Similarly, immunoprecipitation of a mixture of cell lysates from the 2 cell lines with anti-TfR shows no detectable TfR2 in the immunoprecipitates (Figure 3B). These results indicate that the 2 homodimers generated in separate cell lines do not form higher order complexes after solubilization.
Specificity of interaction between TfR and TfR2.
A series of control immunoprecipitations for TfR and TfR2 was performed. Fifty micrograms of extracts from K562 cells, TRVb cells transfected with a wild-type TfR plasmid (TRVb1), TRVb cells transfected with a wild-type TfR2 plasmid (TRVb2), and a mixture of TRVb1 and TRVb2 cells were immunoprecipitated using either the TfR mouse monoclonal antibody or the TfR2 mouse monoclonal antibody and S aureus precoated with an affinity-purified rabbit antimouse antibody (Jackson Immunolabs). Immunoprecipitates and 16.7 μg of cell extracts (equivalent to one third of the amount immunoprecipitated) were run on an 8% gel under denaturing and reducing conditions, transferred to nitrocellulose, and immunodetected for TfR or TfR2 (A and B, respectively). The legend above each blot indicates the cell lines in each lane and the legend below each blot indicates the treatment of each cell line and the antibodies used for the immunoprecipitations. NS indicates cell extracts combined only with precoated S aureus; CE, cell extracts only. These results indicate that anti-TfR cannot immunoprecipitate TfR2 by itself and vice versa.
Specificity of interaction between TfR and TfR2.
A series of control immunoprecipitations for TfR and TfR2 was performed. Fifty micrograms of extracts from K562 cells, TRVb cells transfected with a wild-type TfR plasmid (TRVb1), TRVb cells transfected with a wild-type TfR2 plasmid (TRVb2), and a mixture of TRVb1 and TRVb2 cells were immunoprecipitated using either the TfR mouse monoclonal antibody or the TfR2 mouse monoclonal antibody and S aureus precoated with an affinity-purified rabbit antimouse antibody (Jackson Immunolabs). Immunoprecipitates and 16.7 μg of cell extracts (equivalent to one third of the amount immunoprecipitated) were run on an 8% gel under denaturing and reducing conditions, transferred to nitrocellulose, and immunodetected for TfR or TfR2 (A and B, respectively). The legend above each blot indicates the cell lines in each lane and the legend below each blot indicates the treatment of each cell line and the antibodies used for the immunoprecipitations. NS indicates cell extracts combined only with precoated S aureus; CE, cell extracts only. These results indicate that anti-TfR cannot immunoprecipitate TfR2 by itself and vice versa.
Colocalization of TfR and TfR2 in K562 cells
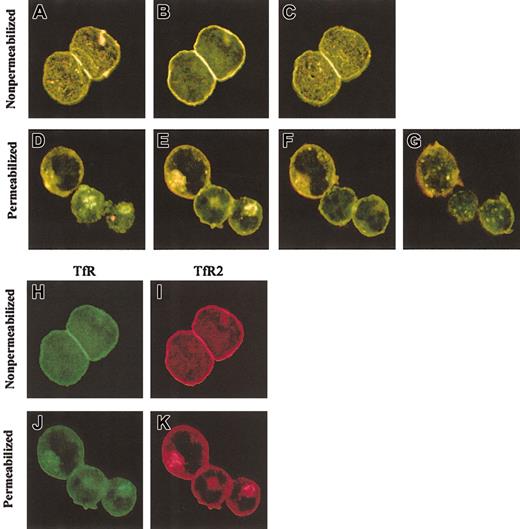
Confocal microscopy was used to examine the intracellular locations of TfR and TfR2. K562 cells were fixed and incubated with either a sheep anti-TfR serum or the mouse anti-TfR2 antibody, followed by appropriate fluorescent secondary antibodies (Figure4). Prior experiments (Figures 1-2) demonstrated that TfR and TfR2 are both expressed in erythroid and liver cell lines and that they can associate to form heterodimers; yet the extent to which these proteins share similar subcellular locations was unknown. Confocal images of stained cells demonstrate that TfR and TfR2 localize in overlapping subcellular compartments. In both nonpermeabilized cells (Figure 4A-C) and cells permeabilized with Triton X-100 (Figure 4D-E,K), TfR and TfR2 have similar localizations. Both receptors are expressed on the cell surface (Figure 4 H-I) and appear to colocalize (Figure 4A-C,H-I). Inside the cells, both TfR and TfR2 localize to punctate perinuclear compartments (Figure 4E-F, J-K). Previous results from our laboratory and others demonstrated that TfR localizes to perinuclear recycling endosomes,20 21 which in combination with the present results suggests that TfR and TfR2 utilize the same endosomes and traffic together in K562 cells.
Colocalization of TfR and TfR2 in K562 cells.
K562 cells were fixed and labeled with mouse anti-TfR2 monoclonal antibody (9F8 1C11) and sheep anti-TfR serum, followed by Alexa 594 antimouse (red) and Alexa 488 antisheep (green). Merged sections of nonpermeabilized cells are shown in panels A to C. Yellow indicates overlapping fluorescence. Sections are 2.0 μm apart. Panels H and I, respectively, show the separate TfR (green) and TfR2 (red) channels of panel B. Merged sections of permeabilized cells (D-G) also indicate colocalization in perinuclear compartments. Sections in panels D to G are 1.5 μm apart; panels J and K are separate TfR (green) and TfR2 (red) channels of image in panel E. Images were captured with a × 60 oil immersion lens as described in “Materials and methods.”
Colocalization of TfR and TfR2 in K562 cells.
K562 cells were fixed and labeled with mouse anti-TfR2 monoclonal antibody (9F8 1C11) and sheep anti-TfR serum, followed by Alexa 594 antimouse (red) and Alexa 488 antisheep (green). Merged sections of nonpermeabilized cells are shown in panels A to C. Yellow indicates overlapping fluorescence. Sections are 2.0 μm apart. Panels H and I, respectively, show the separate TfR (green) and TfR2 (red) channels of panel B. Merged sections of permeabilized cells (D-G) also indicate colocalization in perinuclear compartments. Sections in panels D to G are 1.5 μm apart; panels J and K are separate TfR (green) and TfR2 (red) channels of image in panel E. Images were captured with a × 60 oil immersion lens as described in “Materials and methods.”
HFE and Tf do not prevent association between TfR and TfR2
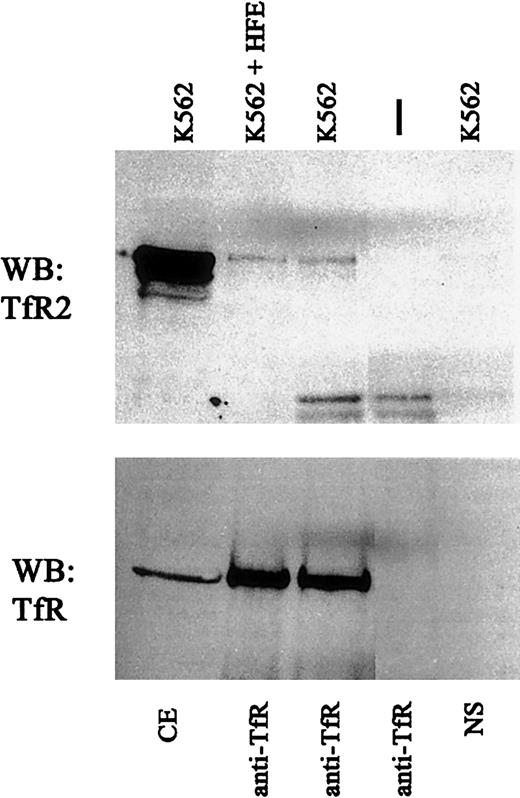
Crystallographic studies of the HFE-TfR complex reveal that the HFE binding site on TfR comprises 2 helices on the outer edge of the helical domain.22 In vitro studies using the ectodomains of TfR, HFE, and Tf suggest that HFE and Tf compete with each other for binding to TfR23 and are consistent with the demonstration that site-directed mutagenesis of the HFE binding site on TfR affects Tf binding.11 We wanted to test whether the TfR2 interaction site on TfR overlapped with the region on the TfR helical domain identified as the HFE and Tf binding site. Extracts of K562 cells were incubated in the presence of 100 μM soluble recombinant HFE,10 and the amount of TfR2 associated with TfR was evaluated. At this concentration, HFE binds to TfR and competes with Tf binding. The extracts were immunoprecipitated with either anti-TfR2 and probed with anti-TfR or vice versa. No decrease in the TfR-TfR2 interaction was detected (Figure5). Similar results were obtained upon addition of Tf (results not shown), suggesting that TfR and TfR2 do not interact with each other at the HFE-TfR interface.
HFE does not alter TfR-TfR2 interaction.
Cell extracts from K562 cells were incubated with 1 μM purified recombinant HFE for 60 minutes at 4°C (K562 + HFE) and immunoprecipitated using sheep anti-TfR serum andS aureus (IP:TfR). Immunoprecipitated proteins were subjected to SDS-PAGE on an 8% gel, transferred to nitrocellulose, and immunodetected for TfR2 (upper panel) (WB:TfR2) and TfR (lower panel) (WB:TfR). NS indicates background control using K562 cell extracts and S aureus without antibody; vertical dash, no cell extract with anti-TfR and S aureusonly; and CE, cell extract.
HFE does not alter TfR-TfR2 interaction.
Cell extracts from K562 cells were incubated with 1 μM purified recombinant HFE for 60 minutes at 4°C (K562 + HFE) and immunoprecipitated using sheep anti-TfR serum andS aureus (IP:TfR). Immunoprecipitated proteins were subjected to SDS-PAGE on an 8% gel, transferred to nitrocellulose, and immunodetected for TfR2 (upper panel) (WB:TfR2) and TfR (lower panel) (WB:TfR). NS indicates background control using K562 cell extracts and S aureus without antibody; vertical dash, no cell extract with anti-TfR and S aureusonly; and CE, cell extract.
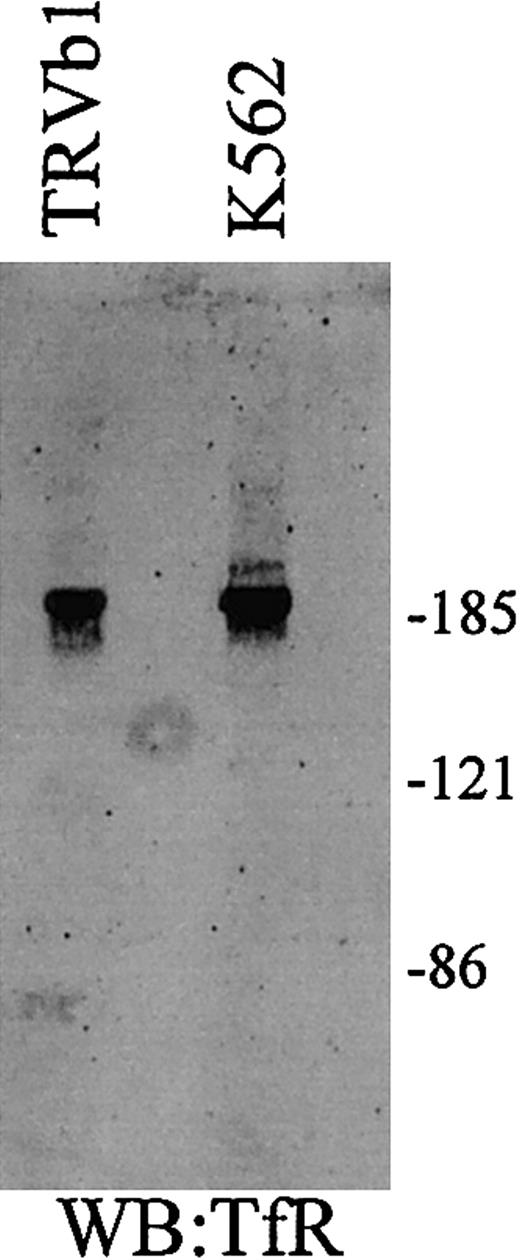
TfR and TfR2 interaction under nonreducing conditions
Because TfR and TfR2 each contain cysteine residues that are used for intersubunit disulfide bonds, a possible mode of association between TfR and TfR2 is the formation of covalent, disulfide-linked heterodimers (Figure 6). To determine whether covalent TfR-TfR2 heterodimers can form, cell lysates from K562 and TRVb1 cells were examined under nonreducing conditions and TfR and TfR2 were immunodetected on Western blots. A single homodimer band will be seen at approximately 186 kDa and a monomeric band will be detected at approximately 93 kDa for TfR if the receptors interact yet do not form intersubunit disulfide bonds. The migration pattern of TfR2 under nonreducing conditions is more complicated because it migrates as a doublet under reducing conditions. If TfR and TfR2 form intersubunit disulfide bonds, then TfR should be detected as a doublet with the lower molecular mass at approximately 186 kDa and little or no monomer. The evidence supports this possibility. Under nonreducing conditions a doublet of the TfR is detected in K562 cell extracts. No such doublet is seen in TRVb1 cells expressing TfR only (Figure6).
TfR and TfR2 under nonreducing conditions.
Cell extracts (25 μg) from TfR-transfected TRVb cells (TRVb1) and K562 cells were separated on an 8% acrylamide gel under denaturing but nonreducing conditions, transferred to nitrocellulose, and Western blotted for TfR and TfR2. TfR homodimer is approximately 190 kDa, while the TfR2 homodimer is a doublet of approximately 200 and 210 kDa.
TfR and TfR2 under nonreducing conditions.
Cell extracts (25 μg) from TfR-transfected TRVb cells (TRVb1) and K562 cells were separated on an 8% acrylamide gel under denaturing but nonreducing conditions, transferred to nitrocellulose, and Western blotted for TfR and TfR2. TfR homodimer is approximately 190 kDa, while the TfR2 homodimer is a doublet of approximately 200 and 210 kDa.
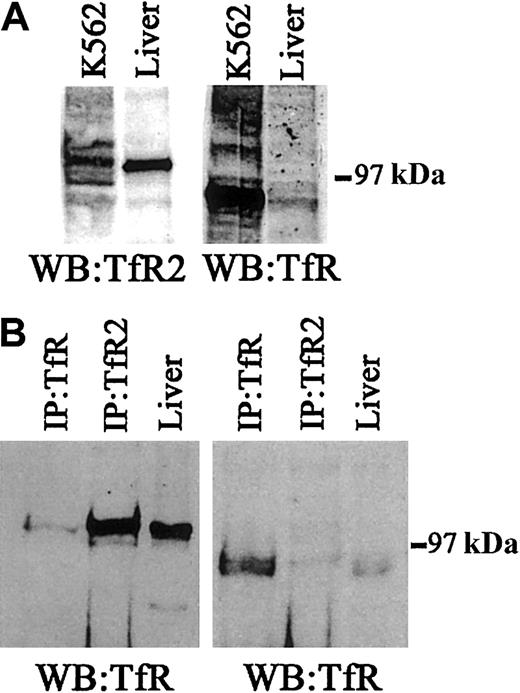
Interaction of TfR and TfR2 in human liver tissue
Because TfR and TfR2 interact with each other in K562 cells we wanted to determine whether this interaction could be detected in the liver, the tissue that has the highest concentration of TfR2 mRNA. Western blots were used to evaluate the relative levels of TfR2 in K562 cells and liver tissue. TfR2 is much more abundant in liver compared with K562 cells. The opposite is true for TfR. TfR is barely detectable in liver and easily detected in K562 cells (Figure7A). Similar to K562 cells, small but detectable amounts of TfR2 and TfR coprecipitate, indicating that the 2 receptors interact in liver as well as K562 cells but, again, to only a limited extent.
TfR and TfR2 protein expression in human liver.
(A) To determine the relative levels of TfR and TfR2 protein in human liver tissue and K562 cells, 50 μg of cell extracts were Western blotted for either TfR (WB:TfR) or TfR2 (WB:TfR2). The panel on the left was developed using a mouse monoclonal antibody to human TfR2 (1:10 000), while the panel on the right was developed using a mouse monoclonal antibody to TfR (1:10 000). (B) To determine whether TfR and TfR2 interact in human liver, a series of immunoprecipitations for either TfR or TfR2, followed by immunoblotting for TfR or TfR2, was performed. Human liver extract (60 μg) was immunoprecipitated with either a sheep polyclonal antibody to TfR (1:100) and 30 μL of S aureus(IP:TfR) or with a mouse monoclonal antibody to TfR2 (1:120) and 30 μL of protein G–Sepharose (IP:TfR2), or the extract was loaded onto the gel (liver). Proteins were separated on an 8% acrylamide gel under denaturing and reducing conditions, transferred to nitrocellulose, and immunoblotted for either TfR (right panel) or TfR2 (left panel).
TfR and TfR2 protein expression in human liver.
(A) To determine the relative levels of TfR and TfR2 protein in human liver tissue and K562 cells, 50 μg of cell extracts were Western blotted for either TfR (WB:TfR) or TfR2 (WB:TfR2). The panel on the left was developed using a mouse monoclonal antibody to human TfR2 (1:10 000), while the panel on the right was developed using a mouse monoclonal antibody to TfR (1:10 000). (B) To determine whether TfR and TfR2 interact in human liver, a series of immunoprecipitations for either TfR or TfR2, followed by immunoblotting for TfR or TfR2, was performed. Human liver extract (60 μg) was immunoprecipitated with either a sheep polyclonal antibody to TfR (1:100) and 30 μL of S aureus(IP:TfR) or with a mouse monoclonal antibody to TfR2 (1:120) and 30 μL of protein G–Sepharose (IP:TfR2), or the extract was loaded onto the gel (liver). Proteins were separated on an 8% acrylamide gel under denaturing and reducing conditions, transferred to nitrocellulose, and immunoblotted for either TfR (right panel) or TfR2 (left panel).
Discussion
Tf is the major iron transport protein in the blood and was originally thought to bind to and be taken up into cells via only a single receptor, TfR. Recently however, 2 more Tf binding receptors have been identified, TfR25,6 and the cubulin-megalin complex.24 These 2 receptors for Tf are tissue specific, whereas TfR appears to be expressed at least at low levels ubiquitously. TfR2 is expressed mainly in the liver and erythropoietic cells, and the cubulin-megalin complex is localized in the kidneys and rat yolk sac.24
Each receptor presumably has a different function in the regulation of iron homeostasis in the body. TfR regulates the uptake of iron into cells and itself is regulated by intracellular iron concentrations and by the proliferation status of the cells. The cubulin-megalin complex appears to be essential in the scavenging of iron, which would otherwise be excreted by the kidneys. The function of TfR2 has not been elucidated to date. It likely plays a major role in iron homeostasis in the body because mutations in this gene result in a rare form of hereditary hemochromatosis with iron accumulation in the liver, heart, and pancreas and high levels of Tf iron saturation.3
The interactions between these 3 transferrin receptors have not been examined. There is probably no interaction between the cubulin-megalin complex and TfR. They are located in different regions of the cell: the cubulin-megalin complex apically oriented, and the TfR basolaterally oriented. Here we used a cell line that endogenously expresses both TfR and TfR2 in order to study their interaction. We found the highest levels of TfR2 protein present in K562 and HEL92 erythroleukemia cell lines, with lesser amounts in HepG2 and Huh7 liver cell lines, correlating with previous work by Kawabata et al, which demonstrated high levels of TfR2 mRNA expression in erythroid precursors and liver.5,14 In K562 cells, TfR2 levels are approximately 10 times lower than TfR levels; however, the 2 receptors have similar cellular distributions, with most of both receptors in the cell interior. Being 10 times more abundant and possessing a higher affinity for Tf, TfR is responsible for the majority of iron uptake in this cell line. However, TfR2 mRNA and protein levels are more abundant than TfR mRNA and protein levels in liver and immature erythroid precursors, suggesting that TfR2 protein predominates in these cells.4 6
We have demonstrated an interaction between TfR and TfR2. Studies using coprecipitation and nonreducing SDS-PAGE show that the transferrin receptors interact and form heterodimers, and this in vitro interaction is supported by the similar colocalization of TfR and TfR2 in vivo. On the cell surface both receptors appear diffuse and in similar locations, while on the cell interior both receptors colocalize to punctate vesicles, likely the recycling endosome. These results suggest that similar mechanisms are used in the internalization and trafficking of each receptor within the cell, even though the amino acid sequences of the cytoplasmic domains of each receptor share no similarity other than both containing tyrosine-based internalization motifs.
HFE and Tf do not prevent TfR-TfR2 association, suggesting TfR2 binds to TfR somewhere other than the common HFE and Tf binding site on TfR. Another possibility is that TfR and TfR2 formed mixed heterodimers. This suggestion is supported by the observation of the mixed covalent heterodimers and by the fact that many of the residues at the crystallographically determined TfR dimerization interface are conserved in TfR2 (Table 1).
TfR dimer interface residues and their TfR2 counterparts
| TfR position . | TfR residue . | TfR2 residue . | TfR2 position . |
|---|---|---|---|
| 182 | Trp | Trp | 192 |
| 183 | Arg | Thr | 193 |
| 185 | Gln | Thr | 196 |
| 312 | Gly | Gly | 334 |
| 313 | Phe | Phe | 335 |
| 314 | Pro | Pro | 336 |
| 315 | Ser | Ser | 337 |
| 316 | Phe | Phe | 338 |
| 317 | Asn | Asn | 339 |
| 320 | Gln | Gln | 342 |
| 321 | Phe | Phe | 343 |
| 322 | Pro | Pro | 344 |
| 400 | Asp | Asp | 424 |
| 402 | Tyr | Tyr | 426 |
| 449 | Ile | Leu | 472 |
| 466 | Trp | Trp | 489 |
| 469 | Gly | Gly | 492 |
| 470 | Tyr | Tyr | 493 |
| 471 | Leu | Leu | 494 |
| 472 | Ser | Ser | 495 |
| 473 | Ser | Val* | 496 |
| 474 | Leu | Leu | 497 |
| 476 | Leu | Leu | 499 |
| 477 | Lys | Lys | 500 |
| 637 | Leu | Leu | 669 |
| 638 | Ser | Thr | 670 |
| 639 | Leu | Leu | 671 |
| 641 | Trp | Trp | 673 |
| 667 | Asp | Asp | 699 |
| 668 | Arg | Glu* | 700 |
| 669 | Phe | Arg* | 701 |
| 672 | Lys | Arg | 704 |
| 673 | Lys | Met* | 705 |
| 676 | Asp | Val | 708 |
| 680 | Arg | Arg | 712 |
| 683 | Tyr | Phe | 715 |
| 684 | His | Tyr | 716 |
| 688 | Pro | Gln* | 720 |
| 689 | Tyr | Tyr | 721 |
| 691 | Ser | Ser | 723 |
| 692 | Pro | Pro | 724 |
| 693 | Lys | Ala | 725 |
| 733 | Asn | Arg* | 774 |
| 735 | Leu | Leu | 776 |
| 736 | Ala | Ala | 777 |
| 737 | Leu | Leu | 778 |
| 740 | Trp | Trp | 781 |
| 744 | Gly | Gly | 785 |
| 747 | Asn | Asn | 788 |
| 748 | Ala | Ala | 789 |
| 753 | Val | Val | 794 |
| 754 | Trp | Trp | 795 |
| 755 | Asp | Asn | 796 |
| 756 | Ile | Ile | 797 |
| 758 | Asn | Asn | 799 |
| 759 | Glu | Asn | 800 |
| 760 | Phe | Phe | 801 |
| TfR position . | TfR residue . | TfR2 residue . | TfR2 position . |
|---|---|---|---|
| 182 | Trp | Trp | 192 |
| 183 | Arg | Thr | 193 |
| 185 | Gln | Thr | 196 |
| 312 | Gly | Gly | 334 |
| 313 | Phe | Phe | 335 |
| 314 | Pro | Pro | 336 |
| 315 | Ser | Ser | 337 |
| 316 | Phe | Phe | 338 |
| 317 | Asn | Asn | 339 |
| 320 | Gln | Gln | 342 |
| 321 | Phe | Phe | 343 |
| 322 | Pro | Pro | 344 |
| 400 | Asp | Asp | 424 |
| 402 | Tyr | Tyr | 426 |
| 449 | Ile | Leu | 472 |
| 466 | Trp | Trp | 489 |
| 469 | Gly | Gly | 492 |
| 470 | Tyr | Tyr | 493 |
| 471 | Leu | Leu | 494 |
| 472 | Ser | Ser | 495 |
| 473 | Ser | Val* | 496 |
| 474 | Leu | Leu | 497 |
| 476 | Leu | Leu | 499 |
| 477 | Lys | Lys | 500 |
| 637 | Leu | Leu | 669 |
| 638 | Ser | Thr | 670 |
| 639 | Leu | Leu | 671 |
| 641 | Trp | Trp | 673 |
| 667 | Asp | Asp | 699 |
| 668 | Arg | Glu* | 700 |
| 669 | Phe | Arg* | 701 |
| 672 | Lys | Arg | 704 |
| 673 | Lys | Met* | 705 |
| 676 | Asp | Val | 708 |
| 680 | Arg | Arg | 712 |
| 683 | Tyr | Phe | 715 |
| 684 | His | Tyr | 716 |
| 688 | Pro | Gln* | 720 |
| 689 | Tyr | Tyr | 721 |
| 691 | Ser | Ser | 723 |
| 692 | Pro | Pro | 724 |
| 693 | Lys | Ala | 725 |
| 733 | Asn | Arg* | 774 |
| 735 | Leu | Leu | 776 |
| 736 | Ala | Ala | 777 |
| 737 | Leu | Leu | 778 |
| 740 | Trp | Trp | 781 |
| 744 | Gly | Gly | 785 |
| 747 | Asn | Asn | 788 |
| 748 | Ala | Ala | 789 |
| 753 | Val | Val | 794 |
| 754 | Trp | Trp | 795 |
| 755 | Asp | Asn | 796 |
| 756 | Ile | Ile | 797 |
| 758 | Asn | Asn | 799 |
| 759 | Glu | Asn | 800 |
| 760 | Phe | Phe | 801 |
Residues in the TfR dimer interface were identified by contact analysis in CNS25 of the unliganded TfR structure (1CX8.pdb)26 using a probe radius of 1.4 Å and a distance cutoff of 4 Å. The interface is 70% identical between TfR and TfR2, consistent with the observation of TfR-TfR2 heterodimers. Nonconservative substitutions are underlined and semiconservative substitutions are italicized. Nonconservative substitutions with an asterisk could probably be accommodated in the dimer interface, as determined by inspection of the TfR structure. For example, the substitutions at TfR position 668 and 669 would disrupt a cation-π interaction between an arginine and a phenylalanine and replace it with a salt bridge between a glutamate and an arginine.
How the interaction between these 2 transferrin receptors affects their function remains to be studied. The subunits of TfR and TfR2 do not interact equally well with each other. TfR and TfR2 preferentially form homodimers, and only a small percentage form heterodimers. From crystallographic studies, HFE interacts with TfR via a hydrophobic region on the ectodomain of the receptor, competing with Tf for binding to TfR.9 23 Because TfR2 is not regulated in response to changes in cellular iron levels and does not interact with HFE, the TfR-TfR2 interaction could function as a potential mechanism to regulate iron uptake in unique ways, perhaps by utilizing signaling pathways. If this were the case, then the heterodimers could be potentially important in regulating signaling. Additional studies are required to fully understand the role of TfR2 and the relationship of TfR2 with TfR.
We would like to thank Dr Tim McGraw, Cornell Medical College for the TRVb and the TRVb1 cell lines; Drs Kawabata and Koeffler for the human TfR2 plasmid; Anthony P. West for the TfR2 used in the immunizations; Thomas O'Hare and Greg Wiens for critical comments on the manuscript; and Aeisha D. Robb and Marianne Wessling-Resnick for insightful discussions.
Prepublished online as Blood First Edition Paper, October 24, 2002; DOI 10.1182/blood-2002-09-2742.
Supported by National Institutes of Health grant DK 40608 (C.A.E.) and Howard Hughes Medical Center (P.J.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Caroline A. Enns, Department of Cell and Developmental Biology L215, Oregon Health and Science University, 3181 SW Sam Jackson Park Rd, Portland, OR 97201-3098; e-mail:ennsca@ohsu.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal