Pharmacologic stimulation of fetal hemoglobin (HbF) expression may be a promising approach for the treatment of β-thalassemia. In this study, we have investigated the HbF-inducing activity and molecular mechanisms of specific histone deacetylase (HDAC) inhibitors in human K562 erythroleukemia cells. Apicidin was the most potent agent compared with other HDAC inhibitors (trichostatin A, MS-275, HC-toxin, suberoylanilide hydroxamic acid [SAHA]) and previously tested compounds (butyrate, phenylbutyrate, isobutyramide, hydroxyurea, 5-aza-cytidine), leading to a 10-fold stimulation of HbF expression at nanomolar to micromolar concentrations. Hyperacetylation of histones correlated with the ability of HDAC inhibitors to stimulate HbF synthesis. Furthermore, analysis of different mitogen-activated protein (MAP) kinase signaling pathways revealed that p38 signaling was activated following apicidin treatment of cells and that inhibition of this pathway abolished the HbF-inducing effect of apicidin. Additionally, activation of the Aγ-globin promoter by apicidin could be inhibited by p38 inhibitor SB203580. In summary, the novel HDAC inhibitor apicidin was found to be a potent inducer of HbF synthesis in K562 cells. The present data outline the role of histone hyperacetylation and p38 MAP kinase signaling as molecular targets for pharmacologic stimulation of HbF production in erythroid cells.
Introduction
Severe β-thalassemia (thalassemia major, Cooley anemia) is characterized by insufficient production of adult β-globin chains with subsequent excess of α-globin chains leading to ineffective erythropoiesis, intramedullar degradation of erythroid cells, and lifelong transfusion requirement of affected patients.1 One molecular treatment strategy of this disease comprises the reactivation of fetal γ-globin production to substitute for the lack of β-globin chains and to correct the imbalance of α/non-α chains.2 Several pharmacologic agents such as 5-azacytidine, hydroxyurea, erythropoietin, butyrate derivatives and combinations of these drugs have been shown to possess γ-globin chain–inducing activity.3
Among these compounds, butyrate analogues have been studied over many years now, and clinical benefit in some patients with β-thalassemia has been reported.4-8 However, several pharmacologic problems are associated with these substances. First, many analogues have a very short half-life, requiring continuous intravenous application. Second, butyrate derivatives need a plasma concentration in the millimolar range to be effective, requiring large amounts of drugs to be applied. Third, induction of γ-globin chain is relatively weak, which might account for the inconsistent clinical effects observed. Fourth, many derivatives have an offensive odor, making these compounds less suitable for a long-term patient compliance. Therefore, identification of new agents that might be able to stimulate γ-globin chain via similar molecular mechanisms as butyrates but are more efficient at lower concentrations is warranted. A reasonable approach would be to investigate compounds that mimic the molecular effects of butyrate on erythroid cells.
Butyrate has been found to possess inhibitory activity on histone deacetylases, leading to hyperacetylation of ε-amino groups of lysine residues in histones.9-11 This in turn causes a decreased association of basic core histone proteins with the DNA, rendering certain genes more accessible to the transcriptional machinery. In fact, trichostatin A, a specific histone deacetylase (HDAC) inhibitor was found to possess some fetal hemoglobulin (HbF)–inducing activity in human and mouse erythroleukemia cells,12 13 suggesting that the histone deacetylase–inhibiting properties of butyrate contribute to its mode of action.
In this report, we have investigated several specific HDAC inhibitors with respect to their HbF-inducing activity in K562 cells. Apicidin was by far the most efficient HbF-inducing agent at nanomolar to micromolar concentrations. Our data further demonstrate that, in addition to HDAC inhibition, p38 mitogen-activated protein (MAP) kinase signaling appears to be involved in apicidin-mediated stimulation of HbF synthesis.
Materials and methods
Cell culture
The human leukemia cell line K562 was obtained from American Type Culture Collection (ATCC), Philadelphia, PA (CCL-243). Cells were cultured in RPMI containing 10% fetal calf serum with addition of penicillin/streptomycin. For experiments, cells were seeded at a density of 4 × 105 cells/4 mL in 35-mm dishes and cultured for 4 days in the presence or absence of the inducing/inhibiting agents as indicated. Viable cell counts were determined by using trypan-blue dye exclusion test. For measurement of fetal hemoglobin and total hemoglobin, cells were centrifuged and washed with phosphate-buffered saline (PBS), and the cell pellet was resuspended in lysis buffer (100 mM potassium phosphate, pH 7.8, 0.2% Triton X-100) and incubated 10 minutes at room temperature. After pelleting cellular debris, the supernatant was collected and stored at −20°C until further use. For MAP kinase immunoblot analysis, cell pellets were directly lysed in sodium dodecyl sulfate (SDS) sample buffer (62.5 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 6.8, 2% SDS, 10% glycerol, 50 mM DTT [dithiothreitol], 0.1% bromophenol blue), incubated for 5 minutes at 95°C, cooled on ice for 5 minutes, and stored at −20°C until further use.
Reporter gene experiments
A 1436-bp fragment of the human Aγ-globin promoter (+64 to −1372 relative to the start site of transcription) was amplified from human genomic DNA using standard polymerase chain reaction (PCR) methods. The promoter fragment was cloned into theNheI/XhoI site of the reporter gene plasmid pGL3 basic (Promega, Madison, WI). The correct sequence of the construct was verified by automated DNA sequencing. Transient transfection of K562 cells was done by lipofection using 3 μL lipofectamine 2000 (Gibco-Life Technologies, Karlsruhe, Germany) per microgram DNA. A stock suspension of 6 × 106 K562 cells/6 mL was transfected with 12 μg DNA according to the manufacturer's protocol. After 24 hours of transfection, the transfected stock suspension was divided into 500-μL aliquots, diluted 1:4 with RPMI medium containing 10% fetal calf serum (FCS), and cultured in the presence or absence of 0.5 μM apicidin for the various times indicated. Apicidin-treated and untreated control cells were harvested in parallel at the different time points indicated; luciferase activity was determined by the luciferase assay kit (Promega) as described previously14 and normalized by protein concentration of lysates. Normalized luciferase activity from apicidin-treated cells was corrected by the activity of untreated control cells harvested at the same time points to take into account basal, unstimulated reporter gene transcription. Because transfection was carried out in a stock suspension of cells before splitting into aliquots, there was no need to correct for different transfection efficacies by cotransfection of a constitutively expressed plasmid. To rule out direct activation of the luciferase gene or enzyme by apicidin, we performed control experiments by transfecting cells with the promoterless pGL3basic luciferase plasmid. Apicidin-treated cells did not show significant increase of luciferase activity compared with untreated control cells exhibiting a basal luciferase activity. Furthermore, direct incubation of luciferase activity containing cell lysates with apicidin did not stimulate enzyme activity.
Determination of total hemoglobin and HbF
Hemoglobin concentration was determined by using the plasma hemoglobin kit from Sigma (St Louis, MO) according to the manufacturer's instructions. This assay is based on the catalytic action of any hemoglobin on the oxidation of benzidine by hydrogen peroxide. After measurement of protein concentration of the lysate by the Coomassie method, nanogram hemoglobin per microgram total cellular protein was calculated.
Concentration of fetal hemoglobin was measured by enzyme-linked immunosorbent assay (ELISA) based on a 2-antibody sandwich principle as follows: microtiter plates were coated at 37°C for 1 hour with 100 μL sheep antihemoglobin F antibody (1 mg/mL; Bethyl Laboratories, Montgomery, TX) diluted 1:1000 in 100 mM Na2CO3/NaHCO3, pH 9.6. After washing 4 times with Tris-buffered saline containing 0.02% (vol/vol) Tween-20 (TBS-T), unspecific binding sites were blocked with 200 μL 40 mM Tris/HCl, pH 7.6, 137 mM NaCl, 0.02% (vol/vol) Tween-20, 3% (wt/vol) bovine serum albumin at 4°C for 12 hours. After washing 4 times with TBS-T, 100 μL K562 cell lysate, diluted 1:10.000 with lysate buffer (40 mM Tris/HCl, pH 7.6, 137 mM NaCl, 0.02% [vol/vol] Tween-20, 0.5% [wt/vol] bovine serum albumin) was applied to each well and incubated at room temperature for 1 hour. After washing 4 times with TBS-T, 100 μL mouse-antihuman hemoglobin γ-chain antibody (Accurate Chemical, New York) diluted 1:10 000 with lysate buffer was added at room temperature for 1 hour. After washing 4 times with TBS-T, wells were incubated with 100 μL peroxidase-linked sheep-antimouse immunoglobulin (Amersham-Pharmacia, Piscataway, NJ) and diluted 1:1000 with lysate buffer at room temperature for 1 hour. After washing 4 times with TBS-T, bound antibody was detected by addition of 100 μL substrate solution (BM Blue POD [peroxidase] substrate; Boehringer Mannheim, Mannheim, Germany). Substrate reaction was stopped by addition of 25 μL 1 M H2SO4, and color reaction was determined at 450 nm (690 nm as reference) in an ELISA reader. As HbF standards, we used erythrocyte lysates obtained from a premature newborn shown to consist of pure HbF by cellulose acetate electrophoresis. The ELISA was linear at HbF concentrations ranging from 125 pg/mL to 2.5 ng/mL and was about 1000-fold more sensitive for detection of HbF than of adult hemoglobin (HbA).
Quantitative RT-PCR
Quantification of mRNA expression was done by real-time reverse transcription (RT)–PCR with Sybr Green using an ABI Prism 7700 thermal cycler (Perkin-Elmer Applied Biosystems, Foster City, CA). Total RNA was prepared from 106 cells with the RNeasy-kit from Quiagen (Chatsworth, CA) according to the manufacturer's instructions. For reverse transcription (RT), 1 μg total RNA was randomly primed for 10 minutes at 60°C and then subjected to reverse transcription for 1 hour at 37°C by using Moloney murine leukemia virus (MMLV)–RT from Invitrogen (Karlsruhe, Germany). For PCR: cDNA aliquots were quantified for globin gene expression by using the threshold cycle (Ct) method normalized for the house keeping gene β-actin. The following exon-spanning primer sequences were used (5′-3′ orientation): β-actin (cDNA amplicon length 151 bp), GCATCCCCCAAAGTTCACAA (forward) and AGGACTGGGCCATTCTCCTT (reverse); α-globin (cDNA amplicon length 372 bp), GACAAGACCAACGTCAAGGCCGCC (forward) and CAGGAACTTGTCCAGGGAGGC (reverse); γ-globin (cDNA amplicon length 489 bp), ACTCGCTTCTGGAACGTCTGA (forward) and GTATCTGGAGGACAGGGCACT (reverse). PCR was performed in triplicates using the qPCR Mastermix for Sybr Green 1 kit from Eurogentic (Seraing, Belgium) with the following protocol: after initial denaturing of the cDNA (10 minutes at 95°C), a 2-step PCR was performed (15 seconds at 95°C, 1 minute at 60°C, 40 cycles). Dilution experiments were performed to ensure similar efficiency of the PCRs, and standard curves were calculated referring the Ct (the PCR cycle at which a specific fluorescence becomes detectable) to the log of each cDNA dilution step. Specific amplification was verified by generation of a melting curve as well as agarose gel electrophoresis. Threshold cycle (Ct) values obtained for γ- and α-globin were normalized by corresponding Ct values of β-actin. Results from apicidin-treated cells were expressed relative to untreated control cells.
Materials
The following HDAC inhibitors were used: TSA (trichostatin A; Calbiochem, San Diego, CA), SAHA (suberoylanilide hydroxamic acid; Calbiochem), MS-275 (N-(2-aminophenyl)-4-[pyridine-3-ylmethoxycarbonyl)aminomethyl]benzamide; Calbiochem), HC-toxin (cyclo-D-Pro-L-Ala-D-L-2-amino-8-oxo-9, 10-epoxydecanoic acid; Sigma), apicidin (cyclo-[L- (2-amino-8-oxodecanoyl)-L-(N-methoxytryptophan)-L-isoleucyl-D-pipecolinyl); Calbiochem). The substances were dissolved in dimethyl sulfoxide (DMSO) or ethanol as recommended by the supplier and added to the culture medium to give the final concentrations as indicated. The final concentration of DMSO and ethanol in the culture medium was kept below 0.1% (vol/vol). At this concentration, the solvents did not influence hemoglobin synthesis of K562 cells. Furthermore, the respective volume of solvent was used in all control experiments. Sodium butyrate, isobutyramide, sodium valproate, hemin, hydroxyurea, 5-aza-cytidine, and sodium phenylacetate were purchased from Sigma. Sheep antihemoglobin F antibody was purchased from Bethyl Laboratories, and mouse antihuman-hemoglobin-γ chain antibody was from Accurate Chemical. The following antibodies were obtained from Calbiochem: anti-ERK1/2 (extracellular signal-related kinase 1/2) phosphorylated, anti-Jun N-terminal kinase (JNK)1/2 phosphorylated, and anti-p38 total. Antibodies obtained from Sigma were anti-p38 phosphorylated, anti-ERK1/2 total, and JNK1/2 total. The p38-specific inhibitor SB203580 was from Calbiochem, and the ERK pathway inhibitor UO126 was from Promega. Antiacetyl H4 histone antibodies were from Upstate Biotechnology (Lake Placid, NY).
Immunoblot analysis
Detection of phosphorylated MAP kinase proteins.
Cell lysates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) using 10% polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA) by using a semidry electroblot chamber. Transfer of proteins was assessed by ponceau-red staining. Membranes were blocked in tris-buffered saline, pH 7.4, containing 0.1% Tween-20 and 5% bovine serum albumin for 1 hour at room temperature. Incubations with primary antibodies were carried out at 4°C overnight by using antibody dilutions as recommended by the manufacturer in tris-buffered saline, pH 7.4, 0.1% Tween-20. Following 1 hour of incubation with goat-antirabbit peroxidase-conjugated antibody (Promega) at room temperature, proteins were detected by the electrogenerated chemiluminescence (ECL) method (Amersham-Pharmacia) according to the manufacturer's instructions. Blots were stripped at 50°C for 30 minutes in 100 mM 2-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl, pH 6.7, and reprobed as indicated.
Detection of acetylated H4 histone proteins.
Histones were purified from nuclear proteins by acid extraction as described.15 Briefly, 2 × 106 K562 cells were collected by centrifugation, washed with phosphate-buffered saline, and resuspended in 1 mL ice-cold lysis buffer (10 mM Tris/HCl, pH 6.5, 50 mM sodium bisulfite, 1% [vol/vol] Triton X-100, 10 mM MgCl2, 8.6% sucrose). Cells were disrupted by using a dounce homogenizer, and nuclei were pelleted by centrifugation for 10 minutes at 1000g. Pellets were washed 3 times with lysis buffer and once with 10 mM Tris/HCl, pH 7.4, 13 mM EDTA (ethylenediaminetetraacetic acid). Pellets were then dissolved in 100 μL ice-cold water by vortexing. Acid extraction of nuclear histone proteins was carried out by adding 7 μL 6 N H2SO4 to give a final concentration of 0.4 N H2SO4 and incubated at 4°C for at least 1 hour. After pelleting acid-insoluble proteins (5 minutes full speed, microfuge), supernatants were collected and 1 mL ice-cold acetone was added, and acid-soluble proteins were precipitated at −20°C overnight. After pelleting for 5 minutes at full speed in a microfuge, proteins were air dried for 5 to 10 minutes and dissolved in 50 μL water. For detection of acetylated histones, acid-extracted nuclear proteins were separated by 15% SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane by electroblotting. The blotted membrane was blocked with freshly prepared TBS containing 3% nonfat dry milk (TBS-milk) for 1 hour at room temperature. The nitrocellulose membrane was incubated with 1:2000 dilution of antiacetyl histone H4, chromatin immunoprecipitation (ChIP) grade (Upstate Biotechnology) in TBS-milk, overnight at 4°C. The membrane was washed 3 times with water and incubated with donkey antirabbit immunoglobulin G (IgG) 1:10 000 in TBS-milk for 1.5 hours at room temperature. Blots were washed 3 times with water and once with TBS-0.05% Tween 20 for 5 minutes. After rinsing the membrane with 4 changes of water, detection of bound antibodies was carried out by using the ECL detection Kit (Amersham-Pharmacia) according to the manufacturer's instructions.
Results
Stimulation of fetal hemoglobin production by HDAC inhibitors
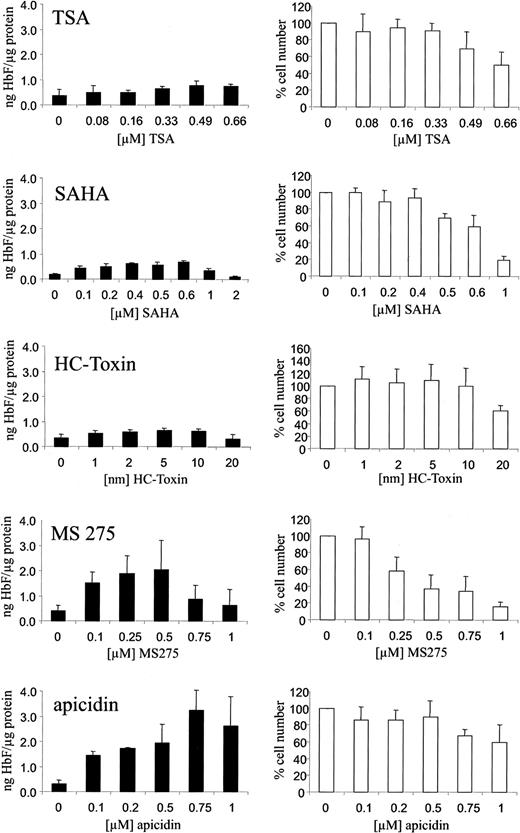
We have investigated the HbF-stimulating potential of the HDAC inhibitors trichostatin A, SAHA, HC-toxin, MS-275, and apicidin in human K562 erythroleukemia cells. These cells are widely used as an in vitro model system for the investigation of compounds with HbF-inducing activity. Cells were cultured with increasing concentrations of the respective HDAC inhibitor for 4 days. HbF concentrations in total cellular extracts were determined by ELISA as described. Figure 1 shows that the HbF-inducing potential of the HDAC inhibitors varied significantly. Whereas trichostatin A, SAHA, and HC-toxin showed relatively weak stimulation, apicidin increased HbF synthesis up to 10-fold compared with untreated control cells at a concentration of 0.1 to 1 μM (Figure 1, black bars). At the concentrations effective in stimulating HbF synthesis, inhibition of cell proliferation also varied significantly (Figure 1, white bars). Apicidin showed relative low cytotoxicity in this regard.
Induction of HbF synthesis in K562 cells by HDAC inhibitors.
Cells were treated with increasing concentrations of the HDAC inhibitors TSA, SAHA, HC-toxin, MS-275, apicidin, or solvent only (0 value) for 4 days. HbF was determined from total cellular extracts by ELISA, normalized by total protein concentrations of extracts, and expressed as nanogram HbF per microgram protein (black bars). Cytotoxicity was assessed by counting cell numbers (white bars). Cell numbers were expressed relative to untreated cells (100%). Each experiment was performed at least 4 times, and standard errors were calculated as indicated.
Induction of HbF synthesis in K562 cells by HDAC inhibitors.
Cells were treated with increasing concentrations of the HDAC inhibitors TSA, SAHA, HC-toxin, MS-275, apicidin, or solvent only (0 value) for 4 days. HbF was determined from total cellular extracts by ELISA, normalized by total protein concentrations of extracts, and expressed as nanogram HbF per microgram protein (black bars). Cytotoxicity was assessed by counting cell numbers (white bars). Cell numbers were expressed relative to untreated cells (100%). Each experiment was performed at least 4 times, and standard errors were calculated as indicated.
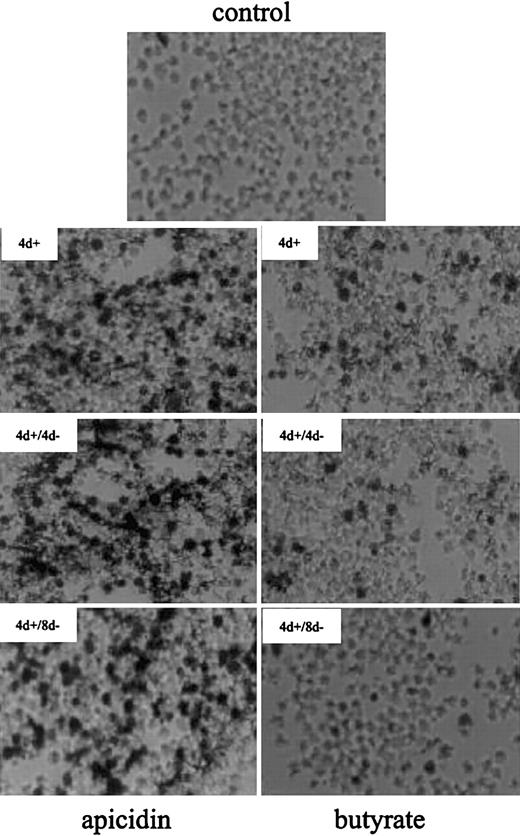
At the cellular level, apicidin increased the number of benzidine-positive (ie, hemoglobin-containing) cells from 3% up to 70% after 4 days of treatment (Figure2). Butyrate, a compound with well-documented hemoglobin-inducing activity in K562 cells, leads to detectable hemoglobin in 20% of cells in the same time. After removal of the compounds, apicidin-treated K562 cells remained benzidine positive for at least another 8 days, whereas benzidine-positivity reverted back to untreated control levels in butyrate-induced cells. This finding suggests that apicidin, in contrast to butyrate, is an irreversible inhibitor of HDAC in vivo, as has been found in HeLa cells.16 Alternatively, the compound may persist in cells much longer than butyrate, resulting in continuous blockage of HDAC activity.
Benzidine staining of K562 cells treated with butyrate and apicidin.
Cells were cultured in the absence (control) or presence of 0.5 μM apicidin (left panels) or 0.6 mM butyrate (right panels) for 4 days (4 d+). Thereafter, the compounds were removed from medium and cultured for another 4 days (4 d+/4 d−) or 8 days (4 d+/8 d−), respectively. After harvesting, intracellular hemoglobin was detected by benzidine staining, and cell smears were subjected to microscopy using × 400 magnification.
Benzidine staining of K562 cells treated with butyrate and apicidin.
Cells were cultured in the absence (control) or presence of 0.5 μM apicidin (left panels) or 0.6 mM butyrate (right panels) for 4 days (4 d+). Thereafter, the compounds were removed from medium and cultured for another 4 days (4 d+/4 d−) or 8 days (4 d+/8 d−), respectively. After harvesting, intracellular hemoglobin was detected by benzidine staining, and cell smears were subjected to microscopy using × 400 magnification.
Comparison of the HDAC inhibitors (Figure3A, black bars) with previously tested agents (Figure 3A, gray bars) at concentrations with maximum HbF-inducing activity revealed that apicidin was much more effective in stimulating HbF-production than butyrate, hydroxyurea, 5-azacytidine, phenylacetate, isobutyramide, and valproic acid. At the concentrations used, the inhibitory action on cell proliferation varied among the different compounds tested (Figure 3B). Again, apicidin revealed relatively low cytotoxicity at a concentration with maximum HbF-inducing activity. To investigate the specificity of HDAC inhibitors on fetal hemoglobin stimulation, we determined the HbF/total hemoglobin ratio (Figure 3C). All HDAC inhibitors investigated increased the proportion of HbF relative to total hemoglobin in K562 cells. Apicidin caused a 3-fold increase of the HbF/Hb ratio compared with untreated control cells.
Comparison of various HbF-inducing compounds on HbF synthesis and globin mRNA expression in K562 cells.
Cells were treated with HDAC inhibitors (black bars) and different HbF-inducing compounds (gray bars) by using concentrations with maximum HbF-inducing activity or solvent only (white bars) for 4 days. HbF per total protein (A) and cell numbers (B) were expressed relative to untreated control cells. (C) HbF per total Hb. Each experiment was performed 4 times, and standard errors were calculated as indicated. Co indicates untreated control cells; But, 0.6 mM butyrate; PA, 2 mM phenylacetate; Iso, 2 mM isobutyramide; Hu, 100 μM hydroxyurea; 5-aC, 5 μM 5-aza-cytidine; and Val, 2 mM valproate. (D) Analysis of globin mRNA expression in untreated (−) and apicidin-treated (+) K562 cells by quantitative real-time RT-PCR. mRNA expression of untreated and apicidin-treated cells was quantified by real-time RT-PCR, values were normalized by β-actin mRNA, and data expressed relative to untreated control cells. Experiments were repeated 3 times and standard errors were calculated.
Comparison of various HbF-inducing compounds on HbF synthesis and globin mRNA expression in K562 cells.
Cells were treated with HDAC inhibitors (black bars) and different HbF-inducing compounds (gray bars) by using concentrations with maximum HbF-inducing activity or solvent only (white bars) for 4 days. HbF per total protein (A) and cell numbers (B) were expressed relative to untreated control cells. (C) HbF per total Hb. Each experiment was performed 4 times, and standard errors were calculated as indicated. Co indicates untreated control cells; But, 0.6 mM butyrate; PA, 2 mM phenylacetate; Iso, 2 mM isobutyramide; Hu, 100 μM hydroxyurea; 5-aC, 5 μM 5-aza-cytidine; and Val, 2 mM valproate. (D) Analysis of globin mRNA expression in untreated (−) and apicidin-treated (+) K562 cells by quantitative real-time RT-PCR. mRNA expression of untreated and apicidin-treated cells was quantified by real-time RT-PCR, values were normalized by β-actin mRNA, and data expressed relative to untreated control cells. Experiments were repeated 3 times and standard errors were calculated.
Because the HbF-inducing activity of apicidin is expected to be related to increased transcription of the γ-globin genes, we measured γ-globin mRNA levels by quantitative real-time RT-PCR. As shown in Figure 3D, apicidin induced a 16-fold increase of γ-globin mRNA expression, but only a 2- to 3-fold increase of α-globin mRNA expression compared with untreated control cells. Similar results were obtained for butyrate (not shown). Thus, apicidin appears to exhibit relative specificity for γ-globin mRNA expression as the protein data suggested (Figure 3C).
Induction of histone hyperacetylation correlates with stimulation of HbF synthesis
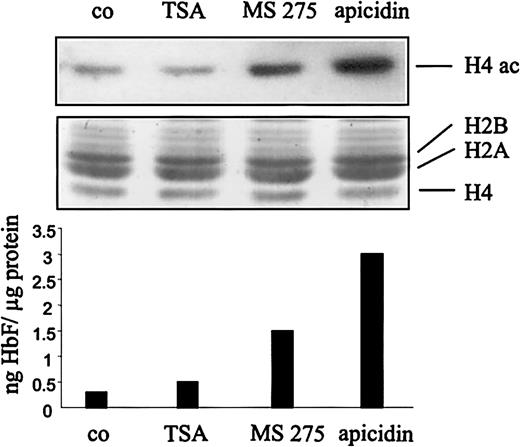
The difference of the HbF-stimulating potential of the HDAC inhibitors tested might be due to their different ability to induce hyperacetylation of histones in K562 cells. To address this question, we have treated cells with a weak (TSA), a medium (MS-275), and a strong HbF inducer (apicidin), respectively, and compared the degree of histone H4 hyperacetylation by Western blot analysis using antiacetyl histone H4 antibodies (Figure 4). The immunoblot indicates that the degree of H4 acetylation correlated well with the potential of the compounds to stimulate HbF synthesis, ie, TSA less than MS-275 less than apicidin.
Induction of histone hyperacetylation correlates with stimulation of HbF synthesis.
K562 cells were treated with the HDAC inhibitors TSA, MS-275, apicidin, or DMSO (co) for 1 hour, respectively. Histone proteins were prepared by acid extraction, and acetylation of histone H4 was determined by Western blot analysis by using a specific antiacetyl H4 histone antibody (top panel). Coomassie staining of the corresponding gel shows equal loading (20 μg) of histone proteins (H2A, H2B, H4) on each lane (middle panel). Bottom panel shows stimulation of HbF synthesis by the respective HDAC inhibitor after 4 days of treatment. H4 ac indicates acetylated histone H4; H2A, H2B, H4; histone proteins H2A, H2B, H4.
Induction of histone hyperacetylation correlates with stimulation of HbF synthesis.
K562 cells were treated with the HDAC inhibitors TSA, MS-275, apicidin, or DMSO (co) for 1 hour, respectively. Histone proteins were prepared by acid extraction, and acetylation of histone H4 was determined by Western blot analysis by using a specific antiacetyl H4 histone antibody (top panel). Coomassie staining of the corresponding gel shows equal loading (20 μg) of histone proteins (H2A, H2B, H4) on each lane (middle panel). Bottom panel shows stimulation of HbF synthesis by the respective HDAC inhibitor after 4 days of treatment. H4 ac indicates acetylated histone H4; H2A, H2B, H4; histone proteins H2A, H2B, H4.
Apicidin modulates MAP kinase signal transduction pathways
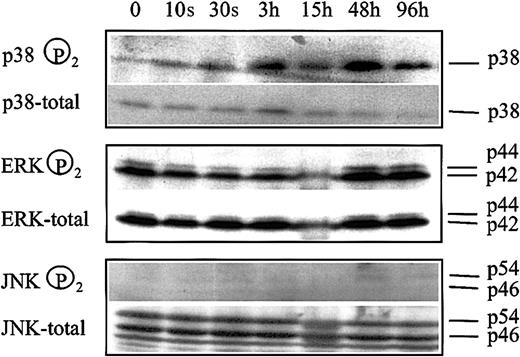
We have previously reported that inhibition of ERK and activation of p38 kinase of the MAP kinase signal transduction system are involved in butyrate-mediated erythroid differentiation of K562 cells.17 If these modulations are associated with the HDAC inhibitory activity of butyrate, apicidin treatment of cells should lead to a similar change in the phosphorylation pattern of MAP kinases. Figure 5 shows that phosphorylation of p38 kinase started to increase 3 hours after addition of apicidin to cells and remained activated for the entire experimental period of 4 days. In contrast, phosphorylation of ERK did not change significantly during the experimental period. Phosphorylation of JNK was not detected, and changes in phosphorylation patterns were not observed. Thus, apicidin activates p38 signaling but has no effect on ERK- or JNK-MAP kinase pathways.
Changes in MAP kinase phosphorylation patterns following apicidin treatment of K562 cells.
Cells were treated with 0.5 μM apicidin for the various times indicated and harvested, and 20 μg cell lysate was subjected to Western blot analysis by using specific antibodies against phosphorylated p38, ERK1/2, and JNK1/2, respectively. Blots were stripped and reprobed with pan antibodies against p38, ERK1/2, and JNK1/2, referred to as total in the figure. Lower panel shows HbF synthesis during the course of the experiment. Similar results were obtained in a second series of experiments.
Changes in MAP kinase phosphorylation patterns following apicidin treatment of K562 cells.
Cells were treated with 0.5 μM apicidin for the various times indicated and harvested, and 20 μg cell lysate was subjected to Western blot analysis by using specific antibodies against phosphorylated p38, ERK1/2, and JNK1/2, respectively. Blots were stripped and reprobed with pan antibodies against p38, ERK1/2, and JNK1/2, referred to as total in the figure. Lower panel shows HbF synthesis during the course of the experiment. Similar results were obtained in a second series of experiments.
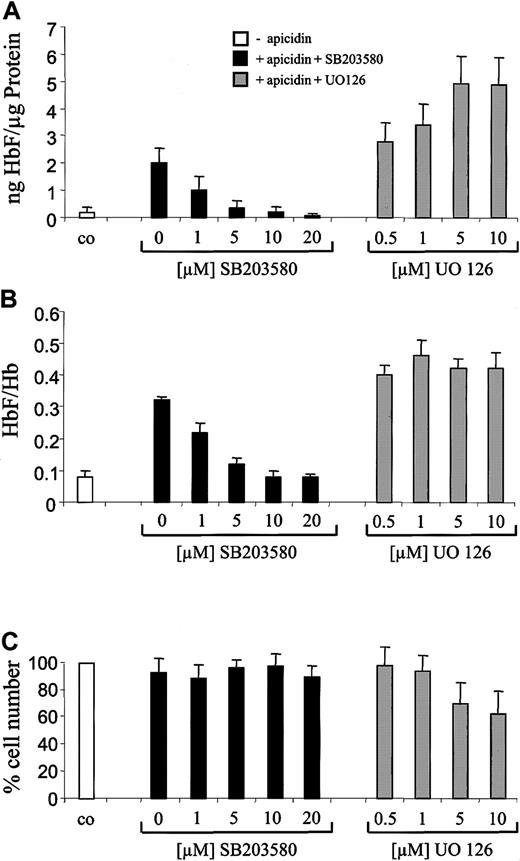
To further investigate the role of p38 signaling, we next examined the influence of the specific p38 inhibitor SB20358018-20 on HbF stimulation by apicidin. Previously, we found that SB203580 inhibited butyrate- but not hemin-induced stimulation of hemoglobin synthesis in K562 cells.17 This finding indicated that this p38 MAP kinase inhibitor is not a general inhibitor of erythroid differentiation in K562 cells. SB203580 inhibited the HbF-inducing effect of apicidin in a concentration-dependent manner (Figure6A, black bars). At a concentration of 5 to 10 μM, SB203580 completely abolished the HbF-inducing effect of apicidin. In contrast, ERK pathway inhibitor UO12621rather increased apicidin-induced HbF synthesis in K562 cells (Figure6A, gray bars). Interestingly, p38 inhibitor SB203580 also reverted the apicidin-induced increment of the HbF/total Hb ratio back to untreated control level (Figure 6B, black bars), whereas ERK inhibitor UO126 did not significantly influence the HbF/Hb ratio (Figure 6B, gray bars), indicating that activation of p38 signaling might be specific for induction of γ-globin gene expression. At the concentrations used, SB203580 did not influence cell proliferation (Figure 6C). UO126 decreased cell numbers by 40% to 50% but was not toxic for K562 cells as evidenced by trypan blue staining (not shown).
Effect of p38 inhibitor SB203580 and ERK inhibitor UO126 on apicidin-mediated induction of HbF synthesis in K562 cells.
Cells were cultured for 4 days in the presence of 0.5 μM apicidin (gray and black bars) or 0.1% (vol/vol) DMSO (white bars). p38-specific inhibitor SB203580 (black bars) or ERK pathway inhibitor UO126 (gray bars) were added in increasing concentrations 1 hour prior to apicidin. After harvesting, cells were lysed, and HbF, total Hb, and total protein concentrations were determined. HbF per total protein (A) and HbF per total hemoglobin (B) were calculated as indicated. Cell numbers relative to untreated control cells (100%) are depicted in (C). Each experiment was performed 4 times, and standard errors were calculated as shown in the figure.
Effect of p38 inhibitor SB203580 and ERK inhibitor UO126 on apicidin-mediated induction of HbF synthesis in K562 cells.
Cells were cultured for 4 days in the presence of 0.5 μM apicidin (gray and black bars) or 0.1% (vol/vol) DMSO (white bars). p38-specific inhibitor SB203580 (black bars) or ERK pathway inhibitor UO126 (gray bars) were added in increasing concentrations 1 hour prior to apicidin. After harvesting, cells were lysed, and HbF, total Hb, and total protein concentrations were determined. HbF per total protein (A) and HbF per total hemoglobin (B) were calculated as indicated. Cell numbers relative to untreated control cells (100%) are depicted in (C). Each experiment was performed 4 times, and standard errors were calculated as shown in the figure.
Apicidin activates Aγ-globin gene promoter
To investigate the influence of apicidin on Aγ-globin promoter activity, we conducted reporter gene experiments. The −1436-bp Aγ-globin promoter fragment was cloned into a luciferase reporter gene plasmid, and the construct was transiently transfected into K562 cells by lipofection. The time course of Aγ-globin promoter activity following apicidin treatment of cells is depicted in Figure7. Apicidin stimulated promoter activity as early as 3 hours after addition to culture medium, and promoter activity peaked after 24 hours of treatment (Figure 7, white bars). Again, inhibition of p38 signaling by SB203580 resulted in inhibition of Aγ-globin promoter activation by apicidin (Figure 7, hatched bars), suggesting that p38 signaling is involved in apicidin-induced activation of the Aγ-globin promoter.
Activation of Aγ-globin gene promoter by apicidin in K562 cells.
Cells were transiently transfected with a 1435-bp Aγ-promoter/luciferase reporter gene construct and subsequently treated with 0.5 μM apicidin for the various times indicated. After determination of luciferase activity from cell lysates as described in “Materials and methods,” values were expressed relative to the activity of the 0 [h] value. Hatched bars show results of transfected cells that have been treated with p38 inhibitor SB203580 prior to addition of apicidin. Each experiment was performed 3 times, and standard errors were calculated as indicated.
Activation of Aγ-globin gene promoter by apicidin in K562 cells.
Cells were transiently transfected with a 1435-bp Aγ-promoter/luciferase reporter gene construct and subsequently treated with 0.5 μM apicidin for the various times indicated. After determination of luciferase activity from cell lysates as described in “Materials and methods,” values were expressed relative to the activity of the 0 [h] value. Hatched bars show results of transfected cells that have been treated with p38 inhibitor SB203580 prior to addition of apicidin. Each experiment was performed 3 times, and standard errors were calculated as indicated.
To investigate the influence of p38 signaling on histone hyperacetylation, we have pretreated K562 cells with SB203580 and then looked for induction of H4 hyperacetylation by apicidin. We observed no influence of inhibition of p38 signaling on histone acetylation (data not shown).
Discussion
Butyrate analogues have long been recognized as inducers of fetal hemoglobin expression in erythroid cells; therefore, these compounds have been used in small clinical trials for the treatment of β-thalassemia.4-8 However, because of rapid metabolism, inconvenient mode of application, and relatively weak HbF-inducing activity of butyrate analogues, alternative substances with HbF-inducing activity are warranted. One approach would be to search for compounds that mimic the molecular action of butyrate. In this regard, butyrate has been shown to inhibit histone deacetylases (HDACs), leading to hyperacetylation of nuclear histones.9-11 Acetylation neutralizes the positively charged histones and subsequently weakens interactions with DNA, resulting in an open nucleosomal configuration.22,23 Such a conformation facilitates access for transcriptional regulators,24,25 and histone acetylation patterns have recently been shown to play a role in the developmental control of murine β-globin gene expression.26 Furthermore, the specific HDAC inhibitors trichostatin A, trapoxin, and HC-toxin have been found to induce γ-globin gene expression in erythroid cells.12,27 However, the HbF-inducing potential of these compounds is relatively weak, being in the order of 1.5- to 2-fold over HbF production in untreated control cells. The hemoglobin-inducing potential of these specific HDAC inhibitors is even lower compared with arginine butyrate in K562 cells.12
In the present paper, we show that a recently identified HDAC inhibitor, apicidin, is a very potent HbF-inducing compound. Apicidin was originally isolated as a fungal metabolite from Fusariumspecies that exhibits broad spectrum antiprotozoal activity by inhibiting parasite histone deacetylases.28 It has been shown to induce morphologic changes in tumor cells and to induce expression of the cell cycle–regulating proteins p21WAF1and gelsolin.16 Compared with other HDAC inhibitors, apicidin has a relative low IC50 (concentration that inhibits 50%) of 0.7 nM28 and 5 nM,16indicative of a high HDAC affinity. We found that apicidin strongly induced hyperacetylation of H4 histones in K562 erythroid cells and was the most potent inducer of HbF synthesis compared with the HDAC inhibitors TSA, HC-toxin, SAHA, MS-275, and butyrate. Furthermore, apicidin caused a 3-fold increase in HbF/total Hb ratio at the protein level and induced γ-globin mRNA expression 16-fold, whereas α-globin mRNA was stimulated only 2- to 3-fold. This finding was indicative of specific action on γ-globin gene expression. At the concentration with maximum HbF-inducing activity, apicidin showed relatively low cytotoxicity. Taken together, these data suggest that apicidin could be an effective HbF inducer in vivo, and further investigations using murine models of thalassemia are required.
The different HbF-inducing potencies of the investigated HDAC inhibitors might be due to their different affinities for the respective histone deacetylases associated with the fetal globin genes or other involved target genes. For example TSA exhibits much lower IC50 values for HDAC1 compared with HDAC4, and trapoxin B has a 3000-fold higher IC50 value for HDAC6 compared with HDAC1.29
In addition to affecting chromatin structure by histone hyperacetylation, HDAC inhibitors may induce other biologic responses in cells as well. We have observed that the MAP kinase signal transduction system contributes to the molecular action of butyrate, a compound with HDAC-inhibiting activity, during erythroid differentiation of K562 cells.17 In the present report, we found that activation of p38 MAP kinase is also involved in the HbF-inducing activity of apicidin. P38 belongs to a group of kinases known to be activated by cellular stress such as heat, hyperosmolarity, x-radiation, and heavy metal ions,30,31 and, thus, we were able to show in a previous report that heat shock and hyperosmolarity can induce hemoglobin production in K562 cells.32Additionally, p38 has been shown to be involved in erythropoietin-induced erythroid differentiation of mouse erythroleukemia cells,33 34 demonstrating that HDAC inhibitors and cytokines might share the same signaling pathways with respect to induction of globin gene expression. In contrast to butyrate, apicidin did not affect ERK signaling, but ERK pathway inhibitor UO126 acted synergistically with both butyrate and apicidin on stimulation of hemoglobin production in K562 cells. The molecular link between inhibition of histone deacetylase activity and p38 MAP kinase signaling needs further investigation.
In summary, we have identified the HDAC inhibitor apicidin as a compound with strong HbF-inducing potential at nanomolar to micromolar concentrations. Our data outline the role of HDAC inhibition and p38 MAP kinase signaling as molecular targets for pharmacologic stimulation of HbF production in erythroid cells. Further studies need to investigate the in vivo potential of apicidin in the treatment of β-thalassemia.
Prepublished online as Blood First Edition Paper, October 17, 2002; DOI 10.1182/blood-2002-08-2617.
Supported by a grant from the Deutsche Forschungsgemeinschaft (Wi 1461/3) and by the B. Braun-Stiftung, Melsungen.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Olaf Witt, Laboratory for Hematological and Cancer Research, Children's Hospital, University of Göttingen, Robert-Koch-Str. 40; D-37075 Göttingen, Germany; e-mail:owitt@gwdg.de.


![Fig. 7. Activation of Aγ-globin gene promoter by apicidin in K562 cells. / Cells were transiently transfected with a 1435-bp Aγ-promoter/luciferase reporter gene construct and subsequently treated with 0.5 μM apicidin for the various times indicated. After determination of luciferase activity from cell lysates as described in “Materials and methods,” values were expressed relative to the activity of the 0 [h] value. Hatched bars show results of transfected cells that have been treated with p38 inhibitor SB203580 prior to addition of apicidin. Each experiment was performed 3 times, and standard errors were calculated as indicated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/5/10.1182_blood-2002-08-2617/3/m_h80533885007.jpeg?Expires=1769862251&Signature=nS6a9Cu3bB15HhOaPY1E4mvd1D3hUp5lF6wbZUK576S5cP71dZmriZakYRDboCWSGxde72k~6hS-KG0NfSnNgsU4rccW5InplnDVXkCuG5nE9AetgVApa2IUtM8U~x4mb2qAnvRzKYbcLSiqJfvDFV4fQxPO9GjZVwDcsqEl77sSIO9DyCO-tm8APDwfe2uop6kUSehrbIDpIeJrNNf7z3mBOtdILR6odJw-rL1~O2~rgbL9C6N7c6tMq-b5PUWJvGHRyddfzdZ10f534gMxAvxRzMEbns7cR21~Gcc13kr8vf8Rd72~fl7au7E1TzzjAePo3BIc8qkL9t8poAIL4g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal