Chronic myelogenous leukemia (CML) is characterized by the presence of a Bcr-Abl fusion protein with deregulated tyrosine kinase activity that is required for maintaining the malignant phenotype. Imatinib, a selective inhibitor ofBcr-Abl, induces major cytogenetic remission (MCR) or complete cytogenetic remission (CCR) in the majority of patients with CML in first chronic phase. However, thorough re-evaluation of cytogenetics in a cohort of patients in MCR or CCR demonstrated clonal karyotypic abnormalities in more than 10% of cases, some of which were clinically associated with a myelodysplastic syndrome (MDS). Further analysis identified previous exposure to cytarabine and idarubicin as significant risk factors for the subsequent occurrence of abnormalities in Philadelphia chromosome–negative (Ph−) cells. To investigate if cytogenetically normal but clonal hematopoiesis might be present in other patients in cytogenetic remission, we studied X-chromosome inactivation as a marker of clonality by polymerase chain reaction analysis of the human androgen receptor (HUMARA). We find that imatinib restores a polyclonal pattern in most patients in CCR and MCR. Nonetheless, our results are consistent with the notion that targeted therapy of CML with imatinib favors the manifestation of Ph− clonal disorders in some patients. They indicate that patients on imatinib should be followed with conventional cytogenetics, even after induction of CCR.
Introduction
Almost all patients with chronic myeloid leukemia (CML) carry a t(9;22)(q34;q11) reciprocal translocation that results in the formation of a BCR-ABL fusion gene.1 The chimeric Bcr-Abl protein derived from the fusion has deregulated protein tyrosine kinase (PTK) activity, which is sine qua non for cellular transformation.2 Current thinking holds that the initialBCR-ABL translocation occurs in a hematopoietic stem cell that, by virtue of the Bcr-Abl protein, acquires a proliferative advantage over normal hematopoiesis. There is evidence that BCR-ABL is both necessary and sufficient to induce CML. Most notably, lethally irradiated mice receiving transplants of syngeneic bone marrow infected with a BCR-ABL retrovirus develop a CML-like myeloproliferative syndrome (MDS).3
The fact that Philadelphia chromosome–negative (Ph−) progenitor cells can be mobilized even in patients with long-standing CML clearly indicates that residual Ph− hematopoiesis coexists with the malignant clone.4 Certain therapeutic interventions, such as interferon α (IFN-α) and autologous stem cell transplantation, are capable of reversing the growth advantage of the Ph+ cell clone and enable Ph−hematopoiesis to recover at least transiently.5,6It is generally thought that this Ph− hematopoiesis is normal, a notion based on 2 observations. First, the Ph−cells have, as a rule, a normal karyotype. Second, the Ph−cells are polyclonal in most cases studied thus far,7,8 By contrast, studies in Epstein-Barr virus (EBV)–transformed Ph− B-lymphoblastoid cell lines established from patients with CML showed skewing toward the type of glucose-6-phosphate dehydrogenase (G-6PD) isoenzyme of the CML clone in a subset of patients with CML9,10; this implies that, in these cases, the Ph translocation arose in an already clonal population. Cytogenetic abnormalities9 as well as an increased propensity to radiation-induced chromosomal translocations11 have also been described in such lines. Furthermore, clonal chromosomal abnormalities are occasionally seen in Ph− metaphases from cytogenetic responders to IFN-α.12 It was speculated that IFN-α may either induce these abnormalities or favor the growth of abnormal clones that had been present prior to IFN-α therapy. If the latter were true, this would clearly question the assumption that Ph−hematopoiesis in IFN-α responders is generally normal.
Treatment with imatinib (formerly STI571) induces major cytogenetic remissions (MCRs) or complete cytogenetic remissions (CCRs) in more than 40% of patients resistant or intolerant to IFN-α.13 As a rule, these patients are in complete hematologic remission (CHR). Unexpectedly, we detected clonal chromosomal abnormalities in Ph− cells from several patients in MCR or CCR to imatinib. This prompted us to perform a systematic study of clonality in a larger cohort of female patients and to thoroughly re-evaluate conventional cytogenetics in a series of male and female patients in CCR or MCR. We show that polyclonal hematopoiesis is present in most women in CCR and MCR to imatinib. However, we found clonal chromosomal abnormalities in 8 of 48 patients and evidence for an MDS in an additional patient.BCR-ABL–targeted therapy of CML may thus favor the manifestation of Ph− hematologic disorders.
Patients, materials, and methods
Patients
Bone marrow (BM) or peripheral blood (PB) samples for clonality analysis were obtained from 23 female patients with CML in CCR or MCR to imatinib. PB or BM from 5 patients without cytogenetic response, 1 patient with active Ph− CML (termed patients in nonresponse [NR]), and from 12 healthy women were analyzed as controls. Cytogenetics was re-evaluated in additional 5 female and 20 male patients in CCR or MCR to imatinib. The analysis included all patients enrolled in successive STI571 protocols (102, 109, 110, 106, 113, 114, 115) at the Department of Hematology, University of Leipzig, Germany, between August 1999 and October 2001, who achieved CCR and MCR and whose cytogenetics had been done in Leipzig. Two patients (nos. 14 and 15; Table 1) were treated at the Department of Haematology, Western General Hospital, Edinburgh, United Kingdom. All cytogenetic and morphologic studies were performed as part of the STI571 protocols, and informed consent was obtained in all cases.
Characteristics of female patients and results of HUMARA-PCR
| Patient no. . | Age, y . | Disease duration, mo . | Previous therapy for CML . | Phase at imatinib start . | Imatinib therapy, mo . | Ph+, % . | Karyotype . | RCI CD3+ . | RCI CD33+ . | RCI CD33+/CD3+/− . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 37 | 23 | IFN, Ara-C | CP1 | 23 | 0 | 46XX[20] | 1.43 | 2.78 | 1.94 |
| 2 | 57 | 1 | HU | CP 1, initial | 9 | 0 | 46XX[20] | 1.04 | 1.28 | 1.23 |
| 3 | 58 | 6 | IFN | CP1 | 19 | 0 | 46XX[20] | 2.94 | 6.22 | 2.13 |
| 4 | 32 | 1 | HU | CP 1, initial | 11 | 0 | 46XX[20] | 1.02 | 0.89 | 0.89 |
| 5 | 25 | 4 | HU | CP 1, initial | 16 | 0 | 46XX[20] | 2.28 | 2.70 | 1.19 |
| 6 | 57 | 30 | HU, IFN | CP1 | 19 | 0 | 46XX[20] | 0.49 | 0.48 | 0.98 |
| 7 | 62 | 19 | HU, IFN | CP1 | 20 | 0 | 46XX[20] | 0.60 | 0.69 | 1.14 |
| 8 | 50 | 28 | HU, IFN, Ida, Ara-C | CP1 | 21 | 0 | 46XX[20] | 1.72 | 1.40 | 0.82 |
| 9 | 60 | 83 | IFN, HU, Ara-C, Ida, Bu, ABMT | CP1 | 6 | 0 | 46XX[20] | 1.86 | 3.16 | 1.69 |
| 10-R | 40 | 1 | HU | CP1, initial | 3 | 0 | 46XX[20] | 0.90 | 1.09 | 1.21 |
| 10-I | 40 | 1 | HU | CP 1, initial | 2 | 100 | 46,XX,t(9;22)[25] | 1.51 | 4.59 | 0.33 |
| 11 | 51 | 11 | HU, IFN | CP1 | 4 | 0 | 46XX[20] | 0.89 | 0.94 | 1.05 |
| 12 | 36 | 1 | HU | CP 1, initial | 11 | 0 | 46XX[20] | 0.95 | 1.83 | 1.92 |
| 13 | 73 | 39 | HU, IFN | CP1 | 1 | 0 | 0/100 Ph+(IP-FISH) | 0.95 | 1.83 | 1.92 |
| 14 | 58 | 32 | IFN | CP1 | 16 | 0 | 46XX[20] | 1.70 | 2.20 | 1.30 |
| 15 | 45 | 9 | HU, IFN, Ara-C | AP | 16 | 2 | 46XX[29]/45XX,−7[28]/46XX, t(9;22)[1] | 0.97* | 1.86* | 1.92* |
| 16 | 36 | 52 | HU, IFN | CP1 | 22 | 10 | 46XX[18]/46XX,t(9;22)[2] | 1.26 | 3.71 | 2.94 |
| 17 | 28 | 18 | IFN | CP1 | 9 | 10 | 46XX[18]/46XX,t(9;22)[2] | 1.28 | 0.88 | 0.69 |
| 18 | 29 | 10 | HU | CP1 | 6 | 15 | 46XX[17]/46XX,t(9;22)[3] | 0.59 | 0.59 | 1.00 |
| 19 | 67 | 29 | IFN | CP1 | 17 | 32 | 47,XX,+8[11]/46XX,t(9;22)[8]/ 46XX[6] | 0.59 | 0.48 | 0.81 |
| 20 | 48 | 34 | HU, IFN | CP1 | 14 | 24 | 46XX[14];46XX,t(9;22)[6] | 1.13 | 2.48 | 2.17 |
| 21 | 58 | 85 | HU, Ara-C, IFN | CP1 | 19 | 35 | 46XX[13];46XX,t(9;22)[7] | 1.65 | 1.30 | 0.79 |
| 22 | 29 | 89 | IFN, HU, dendritic cells | CP1 | 19 | 96 | 46,XX,t(9;22)[24]/46,XX[1] | 1.24 | 0.41 | 0.33 |
| 23 | 43 | 91 | HU, IFN, thioguanine | CP1 | 23 | 92 | 46,XX,t(9;22)[23]/46,XX[2] | 1.17 | 0.03 | 0.03 |
| 24 | 62 | 56 | HU | CP1 | 24 | 100 | 46,XX,t(9;22)[25] | 1.17 | 1.55 | 1.32 |
| 25 | 64 | 16 | IFN, HU | CP1 | NA | NA | Ph−CML | 1.05 | >100 | >100 |
| 26 | 50 | 157 | HU, IFN, Ara-C, Ida | AP (previous myeloid BC) | 4 | 100 | Multiaberrant | 0.65 | >100 | >100 |
| 27 | 69 | 63 | IFN, HU | CP1 | 19 | 0 | 46XX[20] | NI | NI | NA |
| 28 | 62 | 28 | HU, Ara-C, IFN, 6-mercaptopurine | CP1 | 13 | 0 | 46XX[20] | NI | NI | NA |
| 29 | 52 | 14 | IFN | CP1 | 5 | 0 | 46XX[20] | ND | ND | NA |
| 30 | 69 | 44 | IFN | CP1 | 14 | 5 | 46XX[19];46XX,t(9;22)[1] | ND | ND | NA |
| 31 | 74 | 7 | IFN, HU | BC | 11 | 0 | 46XX[20] | ND | ND | NA |
| 32 | 55 | 20 | IFN, HU | CP1 | 11 | 15 | 46XX[17];46XX,t(9;22)[3] | ND | ND | NA |
| 33 | 47 | 22 | Ida, Ara-C, IFN | CP1 | 19 | 4 | 46,XX,ider(20)(q10)t(20;21) (q22;q22)[24]/46,XX, t(6;9;22)[1] | ND | ND | NA |
| Patient no. . | Age, y . | Disease duration, mo . | Previous therapy for CML . | Phase at imatinib start . | Imatinib therapy, mo . | Ph+, % . | Karyotype . | RCI CD3+ . | RCI CD33+ . | RCI CD33+/CD3+/− . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 37 | 23 | IFN, Ara-C | CP1 | 23 | 0 | 46XX[20] | 1.43 | 2.78 | 1.94 |
| 2 | 57 | 1 | HU | CP 1, initial | 9 | 0 | 46XX[20] | 1.04 | 1.28 | 1.23 |
| 3 | 58 | 6 | IFN | CP1 | 19 | 0 | 46XX[20] | 2.94 | 6.22 | 2.13 |
| 4 | 32 | 1 | HU | CP 1, initial | 11 | 0 | 46XX[20] | 1.02 | 0.89 | 0.89 |
| 5 | 25 | 4 | HU | CP 1, initial | 16 | 0 | 46XX[20] | 2.28 | 2.70 | 1.19 |
| 6 | 57 | 30 | HU, IFN | CP1 | 19 | 0 | 46XX[20] | 0.49 | 0.48 | 0.98 |
| 7 | 62 | 19 | HU, IFN | CP1 | 20 | 0 | 46XX[20] | 0.60 | 0.69 | 1.14 |
| 8 | 50 | 28 | HU, IFN, Ida, Ara-C | CP1 | 21 | 0 | 46XX[20] | 1.72 | 1.40 | 0.82 |
| 9 | 60 | 83 | IFN, HU, Ara-C, Ida, Bu, ABMT | CP1 | 6 | 0 | 46XX[20] | 1.86 | 3.16 | 1.69 |
| 10-R | 40 | 1 | HU | CP1, initial | 3 | 0 | 46XX[20] | 0.90 | 1.09 | 1.21 |
| 10-I | 40 | 1 | HU | CP 1, initial | 2 | 100 | 46,XX,t(9;22)[25] | 1.51 | 4.59 | 0.33 |
| 11 | 51 | 11 | HU, IFN | CP1 | 4 | 0 | 46XX[20] | 0.89 | 0.94 | 1.05 |
| 12 | 36 | 1 | HU | CP 1, initial | 11 | 0 | 46XX[20] | 0.95 | 1.83 | 1.92 |
| 13 | 73 | 39 | HU, IFN | CP1 | 1 | 0 | 0/100 Ph+(IP-FISH) | 0.95 | 1.83 | 1.92 |
| 14 | 58 | 32 | IFN | CP1 | 16 | 0 | 46XX[20] | 1.70 | 2.20 | 1.30 |
| 15 | 45 | 9 | HU, IFN, Ara-C | AP | 16 | 2 | 46XX[29]/45XX,−7[28]/46XX, t(9;22)[1] | 0.97* | 1.86* | 1.92* |
| 16 | 36 | 52 | HU, IFN | CP1 | 22 | 10 | 46XX[18]/46XX,t(9;22)[2] | 1.26 | 3.71 | 2.94 |
| 17 | 28 | 18 | IFN | CP1 | 9 | 10 | 46XX[18]/46XX,t(9;22)[2] | 1.28 | 0.88 | 0.69 |
| 18 | 29 | 10 | HU | CP1 | 6 | 15 | 46XX[17]/46XX,t(9;22)[3] | 0.59 | 0.59 | 1.00 |
| 19 | 67 | 29 | IFN | CP1 | 17 | 32 | 47,XX,+8[11]/46XX,t(9;22)[8]/ 46XX[6] | 0.59 | 0.48 | 0.81 |
| 20 | 48 | 34 | HU, IFN | CP1 | 14 | 24 | 46XX[14];46XX,t(9;22)[6] | 1.13 | 2.48 | 2.17 |
| 21 | 58 | 85 | HU, Ara-C, IFN | CP1 | 19 | 35 | 46XX[13];46XX,t(9;22)[7] | 1.65 | 1.30 | 0.79 |
| 22 | 29 | 89 | IFN, HU, dendritic cells | CP1 | 19 | 96 | 46,XX,t(9;22)[24]/46,XX[1] | 1.24 | 0.41 | 0.33 |
| 23 | 43 | 91 | HU, IFN, thioguanine | CP1 | 23 | 92 | 46,XX,t(9;22)[23]/46,XX[2] | 1.17 | 0.03 | 0.03 |
| 24 | 62 | 56 | HU | CP1 | 24 | 100 | 46,XX,t(9;22)[25] | 1.17 | 1.55 | 1.32 |
| 25 | 64 | 16 | IFN, HU | CP1 | NA | NA | Ph−CML | 1.05 | >100 | >100 |
| 26 | 50 | 157 | HU, IFN, Ara-C, Ida | AP (previous myeloid BC) | 4 | 100 | Multiaberrant | 0.65 | >100 | >100 |
| 27 | 69 | 63 | IFN, HU | CP1 | 19 | 0 | 46XX[20] | NI | NI | NA |
| 28 | 62 | 28 | HU, Ara-C, IFN, 6-mercaptopurine | CP1 | 13 | 0 | 46XX[20] | NI | NI | NA |
| 29 | 52 | 14 | IFN | CP1 | 5 | 0 | 46XX[20] | ND | ND | NA |
| 30 | 69 | 44 | IFN | CP1 | 14 | 5 | 46XX[19];46XX,t(9;22)[1] | ND | ND | NA |
| 31 | 74 | 7 | IFN, HU | BC | 11 | 0 | 46XX[20] | ND | ND | NA |
| 32 | 55 | 20 | IFN, HU | CP1 | 11 | 15 | 46XX[17];46XX,t(9;22)[3] | ND | ND | NA |
| 33 | 47 | 22 | Ida, Ara-C, IFN | CP1 | 19 | 4 | 46,XX,ider(20)(q10)t(20;21) (q22;q22)[24]/46,XX, t(6;9;22)[1] | ND | ND | NA |
Patients nos. 15, 19, and 33 had abnormalities in Ph− cells.
Ara-C indicates cytarabine; CP1 chronic phase; Ida, idamycin; Bu, busulfan; IP-FISH, interphase–fluorescence in situ hybridization; AP, accelerated phase; BC, blast crisis; NI, not informative; NA, not applicable; and ND, not done.
Analysis was done from peripheral blood, which showed 28% WBCs with monosomy 7 by FISH.
Cell separation
White blood cells (WBCs) were labeled with anti-CD3 (for selection of T cells) and anti-CD33 (for selection of myeloid cells) monoclonal antibodies (Beckton-Dickinson, Heidelberg, Germany) or isotypic controls, respectively. Between 5 × 104 and 105 CD3+ and CD33+ cells were sorted on a Calibur cell sorter (Beckton-Dickinson) and stored at −70°C until used.
Clonality analysis of HUMARA-PCR
For DNA extraction the cell pellet was lysed in 40 μL lysis buffer (500 mM Tris [tris(hydroxymethyl)aminomethane]–HCl,10 mM NaCl, 20 mM EDTA [ethylenediaminetetraacetic acid],1% sodium dodecyl sulfate [SDS], pH 8.9). Next, 40 μL phosphate-buffered saline (PBS) and 4 μL proteinase K (Qiagen, Hilden, Germany) were added. The samples were incubated at 56°C for 4 to 6 hours, then proteinase K was inactivated by 10 minutes of incubation at 95°C. The debris was pelleted by 10 minutes centrifugation at 13 000 rpm and the supernatant transferred to a fresh Eppendorf tube. After adding 135 μL isopropanol, the DNA was precipitated by 10 minutes centrifugation at 15 000 rpm, washed once with 70% ethanol, and then dissolved in 50 μL deionized water.
DNA (12 μL) was digested with 10 U HpaII (Gibco, Karlsruhe, Germany) overnight. A control reaction was set up under the same conditions but without restriction enzyme. The human androgen receptor (HUMARA) gene was amplified from 5 μL of each sample in a 25-μL reaction containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2, 200 nM each primer, and 0.2 μLTaq polymerase (Platinum, Gibco). Primers were 5′-CTCTACGATGGGCTTGGGGAGAAC-3′ and 6-carboxy-flourescein–labeled 5′-TCCAGAATCTGTTCCAGAGCGTGC-3′, respectively. Amplification conditions were 5 minutes at 95°C, 30 seconds at 85°C followed by 27 to 30 cycles of denaturation at 95°C (1 minute), annealing at 68°C (30 seconds), and extension at 72°C. Then, 7 μL polymerase chain reaction (PCR) product was analyzed on an ABI Prism 377 (Applied Biosystems, Foster City, CA). “Blanks” were included at the levels of DNA extraction and PCR to monitor for PCR contamination.
X-chromosome inactivation was quantified as described by Delabesse et al.14 Briefly, the peak areas for both alleles were determined. The relative corrected index (RCI) was then calculated for CD3+ and CD33+ cells as the ratio of the 2 undigested alleles divided by the ratio of the 2HpaII-digested alleles. Nonrandom X-chromosome inactivation in a given population was assumed if the RCI was more than 3:1 or less than 1:3.15 To control for constitutional skewing,16 the ratio RCICD33/RCICD3 was calculated. A ratio more than 2 or less than 0.5 was considered as evidence of clonality.17 For statistical analysis, all values less than 1 were inverted.
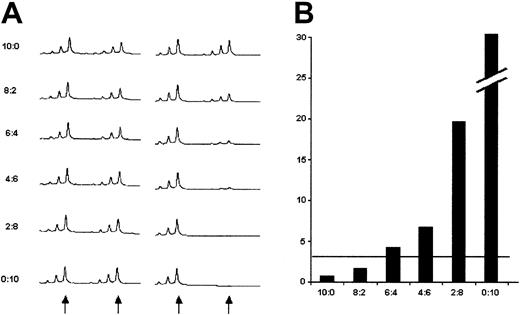
To assess the sensitivity of the method to detect a clonal population in a background of polyclonal cells, (polyclonal) CD3+cells and (monoclonal) CD33+ cells from a CML patient with 100% Ph+ metaphases were mixed at graded concentrations, and the cell mix subjected to HUMARA-PCR (Figure1). An RCI more than 3, indicative of monoclonality, was obtained if the mix contained between 30% and 40% of CD33+ cells. Thus, monoclonal subpopulations that are smaller than approximately 30% are not reliably detectable.
HUMARA-PCR performed on a mix of FACS-sorted CD3+ and CD33+ from a patient with CML and 100% Ph+ metaphases.
(A) Undigested DNA (left panel) versus HpaII-digested DNA (right panel). The arrows indicate the positions of the 2 alleles. The smaller peaks represent stutter bands that were not included in the calculations. The proportion of CD33+ cells was increased from 0% (10:0) to 100% (0:10). (B) RCI values for the respective mixes. The horizontal line indicates an RCI of 3, the limit indicating unbalanced X inactivation (see “Patients, materials, and methods”).
HUMARA-PCR performed on a mix of FACS-sorted CD3+ and CD33+ from a patient with CML and 100% Ph+ metaphases.
(A) Undigested DNA (left panel) versus HpaII-digested DNA (right panel). The arrows indicate the positions of the 2 alleles. The smaller peaks represent stutter bands that were not included in the calculations. The proportion of CD33+ cells was increased from 0% (10:0) to 100% (0:10). (B) RCI values for the respective mixes. The horizontal line indicates an RCI of 3, the limit indicating unbalanced X inactivation (see “Patients, materials, and methods”).
FISH and classical cytogenetics
Interphase and metaphase preparations were done according to standard protocols. Probes used were LSI ETO/AML1 (for detection of t(8;21)(q22;q22)), LSI BCR/ABL ES (for detection of t(9;22)(q34;q11)), LSI21 (locus-specific probe corresponding to 21q22.13-21q22.2), CEP 8 (centromer probe for chromosome 8), CEPX/Y (centromer probe system for X and Y chromosomes), WCP 3 (whole chromosome paint for chromosome 3), all from Vysis (Downers Grove, IL). In addition, whole chromosome paints for chromosomes 20 and 21 were purchased from Metasystems (Altlussheim, Germany). Hybridization was done with the Hybrite semiautomated system (Vysis) and generally followed the recommendations of the manufacturer. In 2 selected cases, cells sorted by fluorescence-activated cell sorting (FACS) were analyzed by fluorescence in situ hybridization (FISH). R-banding technique was used for classical cytogenetics.
Statistical analysis
All statistics was done with the SPSS software package (SPSS, Chicago, IL). Comparison of noncategorical variables was done by 2-tailed Mann-Whitney U test. Categorical variables were compared by 2-sided Fisher exact test.
Results
HUMARA-PCR
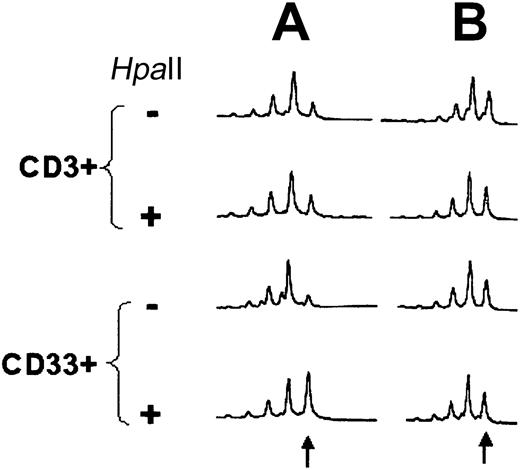
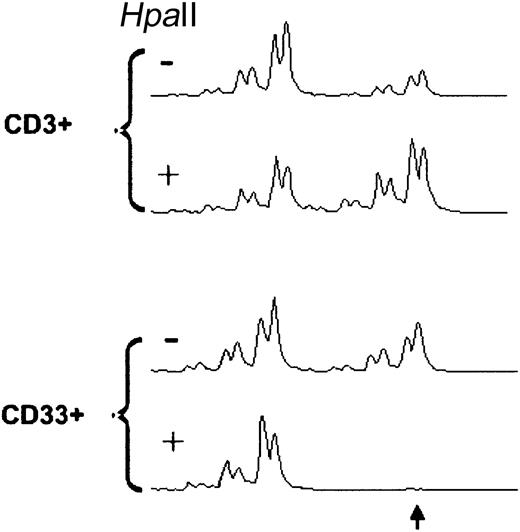
Fourteen females with CML in CCR (nos. 1-9, 10-R, 11-14), 7 females in MCR (10%-35% Ph+ metaphases, nos. 15-21), 6 females with more than 90% Ph+ metaphases (including 1 patient with Ph− CML, nos.10-I, 22-26; Table 1) and 10 healthy volunteers (Table 2) were informative for clonality analysis by HUMARA-PCR (37 of 40 tested = 92.5%). One patient was studied prior (10-I) to imatinib and after induction of CCR (10-R). Unbalanced X inactivation in CD33+ cells as defined by an RCI more than 3:1 or less than 1:3 was found in 2 of 14 CML patients (14.3%) in CCR and 1 of 7 patients in MCR (14.3%), in 4 of 6 patients (66.7%) with CML in NR, and in 1 of 10 healthy volunteers (10%). RCI values were not significantly different between healthy individuals and patients in CCR (P = .538) and MCR (P = .435). In contrast, there was a significant difference between healthy volunteers and patients in NR (P = .009), between patients in CCR and NR (P = .017), and between patients in MCR and NR (P =.045). Applying the criterion of RCICD33/RCICD3 ratio less than 0.5 or more than 2, evidence of clonality was found in 1 of 14 patients in CCR (6.7%), 2 of 7 (28.6%) in MCR, 5 of 6 in NR (83.3%), and 1 of 10 (10%) of healthy volunteers. The differences were significant for CCR and MCR versus NR (P = .002 and P = .015, respectively) and healthy individuals versus NR (P = .003), but not for CCR and MCR versus healthy individuals (P = .747 and P = .204, respectively) and CCR versus MCR (P = .279). In one patient, analysis of samples prior to imatinib (no. 10-I) and at the time of CCR (no. 10-R) was possible and demonstrated normalization of the RCI of the CD33+ cells (Figure2). Of note, at the time of the analysis, there was cytogenetic evidence of clonal evolution in Ph−metaphases in patients nos. 15, 19, and 33, and histologic signs of a myeloproliferative disease in patient no. 1 (see “Cytogenetics and FISH”). In patient 33, HUMARA-PCR was performed on PB 6 months after the cytogenetic analysis and demonstrated clonality of the CD33+ cells (Figure 3). At the time this sample was obtained, BCR-ABL was undetectable by FISH, but 78% of WBCs were positive for t(20;21).
HUMARA-PCR in healthy women
| Patient no. . | RCI CD3+ . | RCI CD33+ . | RCI CD33+/CD3+ . |
|---|---|---|---|
| 1 | 1.86 | 2.31 | 1.25 |
| 2 | 1.50 | 1.63 | 1.09 |
| 3 | 0.26 | 0.33 | 1.27 |
| 4 | 0.99 | 1.07 | 1.09 |
| 5 | 1.13 | 1.21 | 1.06 |
| 6 | 1.07 | 1.89 | 1.75 |
| 7 | 0.68 | 0.71 | 1.05 |
| 8 | 2.18 | 1.25 | 0.58 |
| 9 | 1.57 | 1.36 | 0.87 |
| 10 | 0.90 | 2.00 | 2.22 |
| Patient no. . | RCI CD3+ . | RCI CD33+ . | RCI CD33+/CD3+ . |
|---|---|---|---|
| 1 | 1.86 | 2.31 | 1.25 |
| 2 | 1.50 | 1.63 | 1.09 |
| 3 | 0.26 | 0.33 | 1.27 |
| 4 | 0.99 | 1.07 | 1.09 |
| 5 | 1.13 | 1.21 | 1.06 |
| 6 | 1.07 | 1.89 | 1.75 |
| 7 | 0.68 | 0.71 | 1.05 |
| 8 | 2.18 | 1.25 | 0.58 |
| 9 | 1.57 | 1.36 | 0.87 |
| 10 | 0.90 | 2.00 | 2.22 |
Results of HUMARA-PCR in patient no. 10.
HUMARA-PCR (A) at the time of diagnosis (10-I) and (B) after induction of CCR by imatinib (10-R). HpaII indicatesHpaII-digested DNA. The arrow indicates the position of the larger allele. In this case, the 2 alleles differed only by 1 triplet.
Results of HUMARA-PCR in patient no. 10.
HUMARA-PCR (A) at the time of diagnosis (10-I) and (B) after induction of CCR by imatinib (10-R). HpaII indicatesHpaII-digested DNA. The arrow indicates the position of the larger allele. In this case, the 2 alleles differed only by 1 triplet.
HUMARA-PCR in CD33+ from patient no. 33 at 6 months after induction of MCR by imatinib.
HpaII indicates HpaII digestion of DNA. FISH of CD33+ WBCs failed but 77% of total WBCs carried t(20;21), whereas BCR-ABL was not detected. Note that the larger allele (arrow) is lacking from the HpaII-digested CD33+ cells but not from the CD3+ cells, consistent with a monoclonal CD33+ population.
HUMARA-PCR in CD33+ from patient no. 33 at 6 months after induction of MCR by imatinib.
HpaII indicates HpaII digestion of DNA. FISH of CD33+ WBCs failed but 77% of total WBCs carried t(20;21), whereas BCR-ABL was not detected. Note that the larger allele (arrow) is lacking from the HpaII-digested CD33+ cells but not from the CD3+ cells, consistent with a monoclonal CD33+ population.
Cytogenetics and FISH
Cytogenetic preparations were re-evaluated in 28 women (Table 1; patients nos. 1-21, 27-33) and 20 men (Table3) in CCR or MCR induced by imatinib. In 8 patients (nos. 15, 19, 33, 34, 47, 51, 52, 53), nonrandom chromosomal abnormalities were detected (Table4).
Characteristics of male patients
| Patient no. . | Age, y . | Disease duration, mo . | Previous therapy for CML . | Phase at imatinib start . | Imatinib therapy, mo . | Ph+, % . | Karyotype . |
|---|---|---|---|---|---|---|---|
| 34 | 50 | 18 | IFN, Ara-C, Ida | CP1 | 11 | 0 | 46XY[4]/46,XY,t(3;21)(q27?;q22)[15] |
| 35 | 41 | 1 | HU | CP 1, initial | 11 | 0 | 46XY[18] |
| 36 | 63 | 1 | None | CP 1, initial | 6 | 0 | 46XY[20] |
| 37 | 48 | 2 | HU | CP1, initial | 16 | 0 | 46XY[20] |
| 38 | 59 | 27 | IFN, HU | CP1 | 12 | 0 | 46XY[20] |
| 39 | 61 | 36 | HU, IFN, Ara-C, Ida, Bu, ABMT | CP1 | 6 | 0 | 46XY[20] |
| 40 | 50 | 48 | HU, IFN, Ara-C, Ida, Bu, ABMT | CP1 | 11 | 0 | 46XY[20] |
| 41 | 66 | 4 | None | CP 1, initial | 8 | 0 | 46XY[20] |
| 42 | 36 | 6 | HU | CP 1, initial | 16 | 0 | 46XY[20] |
| 43 | 46 | 3 | HU | CP 1, initial | 6 | 0 | 46XY[20] |
| 44 | 72 | 45 | HU | CP1 | 22 | 0 | 46XY[20] |
| 45 | 39 | 31 | HU, IFN, Ara-C, Ida, Bu, ABMT | CP2 | 21 | 0 | 46XY[20] |
| 46 | 49 | 18 | HU, IFN, Ara-C | AP | 8 | 0 | 46XY[20] |
| 47 | 47 | 69 | HU, IFN, Ara-C, Ida, Bu, ABMT | CP1 | 16 | 0 | 46,XY[13]/46,XY,del(20q)[9] |
| 48 | 33 | 6 | HU | CP 1, initial | 16 | 5 | 46XY[19];46XY,t(9;22)[1] |
| 49 | 73 | 32 | HU, IFN, Bu, 6-mercaptopurine | CP1 | 8 | 10 | 46XY[18];46XY,t(9;22)[2] |
| 50 | 41 | 55 | HU, IFN | CP1 | 22 | 5 | 46XY[19];46XY,t(9;22)[1] |
| 51 | 72 | 14 | HU | AP | 10 | 11 | 46,XY[6]/45,X-Y[2]/46,XY,t(9;22)[1] |
| 52 | 55 | 35 | IFN, HU | CP1 | 27 | 8 | 46,XY[14]/45,X-Y[9]/46,XY,t(9;22)[2] |
| 53 | 37 | 58 | HU, IFN, Ida, Ara-C, Bu, ABMT, 2 Gy TBI, fludarabine, MUD allograft | MCR | 12 | 8 | 6,XY[18]/46,XY,t(7;17)(q11;q21)[5]/ 46,XY,t(9;22)[2] |
| Patient no. . | Age, y . | Disease duration, mo . | Previous therapy for CML . | Phase at imatinib start . | Imatinib therapy, mo . | Ph+, % . | Karyotype . |
|---|---|---|---|---|---|---|---|
| 34 | 50 | 18 | IFN, Ara-C, Ida | CP1 | 11 | 0 | 46XY[4]/46,XY,t(3;21)(q27?;q22)[15] |
| 35 | 41 | 1 | HU | CP 1, initial | 11 | 0 | 46XY[18] |
| 36 | 63 | 1 | None | CP 1, initial | 6 | 0 | 46XY[20] |
| 37 | 48 | 2 | HU | CP1, initial | 16 | 0 | 46XY[20] |
| 38 | 59 | 27 | IFN, HU | CP1 | 12 | 0 | 46XY[20] |
| 39 | 61 | 36 | HU, IFN, Ara-C, Ida, Bu, ABMT | CP1 | 6 | 0 | 46XY[20] |
| 40 | 50 | 48 | HU, IFN, Ara-C, Ida, Bu, ABMT | CP1 | 11 | 0 | 46XY[20] |
| 41 | 66 | 4 | None | CP 1, initial | 8 | 0 | 46XY[20] |
| 42 | 36 | 6 | HU | CP 1, initial | 16 | 0 | 46XY[20] |
| 43 | 46 | 3 | HU | CP 1, initial | 6 | 0 | 46XY[20] |
| 44 | 72 | 45 | HU | CP1 | 22 | 0 | 46XY[20] |
| 45 | 39 | 31 | HU, IFN, Ara-C, Ida, Bu, ABMT | CP2 | 21 | 0 | 46XY[20] |
| 46 | 49 | 18 | HU, IFN, Ara-C | AP | 8 | 0 | 46XY[20] |
| 47 | 47 | 69 | HU, IFN, Ara-C, Ida, Bu, ABMT | CP1 | 16 | 0 | 46,XY[13]/46,XY,del(20q)[9] |
| 48 | 33 | 6 | HU | CP 1, initial | 16 | 5 | 46XY[19];46XY,t(9;22)[1] |
| 49 | 73 | 32 | HU, IFN, Bu, 6-mercaptopurine | CP1 | 8 | 10 | 46XY[18];46XY,t(9;22)[2] |
| 50 | 41 | 55 | HU, IFN | CP1 | 22 | 5 | 46XY[19];46XY,t(9;22)[1] |
| 51 | 72 | 14 | HU | AP | 10 | 11 | 46,XY[6]/45,X-Y[2]/46,XY,t(9;22)[1] |
| 52 | 55 | 35 | IFN, HU | CP1 | 27 | 8 | 46,XY[14]/45,X-Y[9]/46,XY,t(9;22)[2] |
| 53 | 37 | 58 | HU, IFN, Ida, Ara-C, Bu, ABMT, 2 Gy TBI, fludarabine, MUD allograft | MCR | 12 | 8 | 6,XY[18]/46,XY,t(7;17)(q11;q21)[5]/ 46,XY,t(9;22)[2] |
Patients 34, 47, 51, 52, and 53 had abnormalities in Ph− cells; the karyotype reflects the most recent evaluation.
MUD indicates matched unrelated donor; and TBI, total body irradiation. Other abbreviations are explained in the text and Table 1.
Patients with karyotypic abnormalities in Ph−cells
| Patient no. . | Age at diagnosis, y . | Sex . | Treatment prior to imatinib . | Best response prior to imatinib . | Disease duration prior to imatinib, mo . | Disease phase at starting imatinib . | Best response to imatinib . | Time to detection of abnormality, mo . | MDS . | Survival from diagnosis, mo . | Initial karyotype . | Karyotype at best response prior to imatinib . | Karyotype prior to imatinib . | First karyotype with abnormalities in Ph− cells . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 43 | F | IFN, HU | PHR | 10 | AP | CCR | 15 | Yes | Allograft 26 | 46XX t(9;22)[20] | 46XX,t(9;22),t(12;16)[33] | 46XX,t(9;22),t(12;16)[20] | 46,XX[29]/45XX,−7[28]/ 46,XX,t(9,22),t(12,16)[1] |
| 19 | 68 | F | IFN, HU | mCR | 29 | mCR | MCR | 12 | No | 54+ | 46XX t(9;22)[25] | ND | 30/50 BCR-ABL (HMF); 0/50 +8 (HMF) | 46XX[11]/46,XX, t(9;22)[3]/47,XX,+8[8] |
| 33 | 46 | F | Ara-C, HU, Ida, IFN | CHR | 23 | CP | CCR | 12 | No | 47+ | 46,XX,t(6;9;22) (q22;q34;q11) [25] | 46,XY,t(6;9;22)[25] | 46,XX,t(6;9;22)[25] | 46,XX,t(6;9;22)[13]/ 46,XX[8]/46,XX,ider(20) (q10)t(20;21)(q22;q22)[4] |
| 34 | 48 | M | Ida, Ara-C, HU, IFN | CCR | 17 | CP | CCR | 9 | Yes | 43+ | 46XX t(9;22)[17] | ND | 46,XX,t(9;22)[25]; 0/35 t(3;21) (HMF); 1/300 t(3;21) (IPF) | 46XY[4]/46,XY, t(3;21)(q27?;q22)[15] |
| 47 | 40 | M | Ida, Ara-C, HU, IFN, Bu (ABMT) | MCR (leukapheresis product) | 67 | CP | CCR | 3 | No | 96+ | ND | 46,XY,t(9;22)[5]/46,XY[24] | 46,XY,t(9;22)[25]; 0/50 del(20) (HMF); 11/500 del(20) (IPF) | 46,XY[9]/46,XY, t(9;22)[6]/46,XY,del(20q)[5] |
| 51 | 70 | M | HU | PHR | 14 | AP | MCR | 6 | No | 31+ | ND | ND | 46,XY,t(9,22)[24]/47, XY,t(9,22),+der (22)[1];0/100-Y(IPF) | 46,XY[6]/45,X-Y[2]/ 46,XY,t(9;22)[1] |
| 52 | 56 | M | IFN, HU | PHR | 36 | CP | CCR | 6 | No | 68+ | 46XX t(9;22)[30] | ND | 46,XY,t(9;22)[25]; 0/500-Y(IPF) | 46,XY[11]/45,X-Y[4] |
| 53 | 36 | M | Ida, Ara-C, HU, IFN, Bu (ABMT), MUD allograft4-150 | CCR4-151 | 52 | MCR | CCR | 12 | No | 76+ | 46,XY,t(9;22) | ND | 46,XY[25];0/25 del(20) (HMF); 9/300 del(20) (IPF) | 6,XY[18]/46,XY,t(7;17) (q11;q21)/del(20q) [5]/46,XY,t(9;22)[2] |
| Patient no. . | Age at diagnosis, y . | Sex . | Treatment prior to imatinib . | Best response prior to imatinib . | Disease duration prior to imatinib, mo . | Disease phase at starting imatinib . | Best response to imatinib . | Time to detection of abnormality, mo . | MDS . | Survival from diagnosis, mo . | Initial karyotype . | Karyotype at best response prior to imatinib . | Karyotype prior to imatinib . | First karyotype with abnormalities in Ph− cells . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 43 | F | IFN, HU | PHR | 10 | AP | CCR | 15 | Yes | Allograft 26 | 46XX t(9;22)[20] | 46XX,t(9;22),t(12;16)[33] | 46XX,t(9;22),t(12;16)[20] | 46,XX[29]/45XX,−7[28]/ 46,XX,t(9,22),t(12,16)[1] |
| 19 | 68 | F | IFN, HU | mCR | 29 | mCR | MCR | 12 | No | 54+ | 46XX t(9;22)[25] | ND | 30/50 BCR-ABL (HMF); 0/50 +8 (HMF) | 46XX[11]/46,XX, t(9;22)[3]/47,XX,+8[8] |
| 33 | 46 | F | Ara-C, HU, Ida, IFN | CHR | 23 | CP | CCR | 12 | No | 47+ | 46,XX,t(6;9;22) (q22;q34;q11) [25] | 46,XY,t(6;9;22)[25] | 46,XX,t(6;9;22)[25] | 46,XX,t(6;9;22)[13]/ 46,XX[8]/46,XX,ider(20) (q10)t(20;21)(q22;q22)[4] |
| 34 | 48 | M | Ida, Ara-C, HU, IFN | CCR | 17 | CP | CCR | 9 | Yes | 43+ | 46XX t(9;22)[17] | ND | 46,XX,t(9;22)[25]; 0/35 t(3;21) (HMF); 1/300 t(3;21) (IPF) | 46XY[4]/46,XY, t(3;21)(q27?;q22)[15] |
| 47 | 40 | M | Ida, Ara-C, HU, IFN, Bu (ABMT) | MCR (leukapheresis product) | 67 | CP | CCR | 3 | No | 96+ | ND | 46,XY,t(9;22)[5]/46,XY[24] | 46,XY,t(9;22)[25]; 0/50 del(20) (HMF); 11/500 del(20) (IPF) | 46,XY[9]/46,XY, t(9;22)[6]/46,XY,del(20q)[5] |
| 51 | 70 | M | HU | PHR | 14 | AP | MCR | 6 | No | 31+ | ND | ND | 46,XY,t(9,22)[24]/47, XY,t(9,22),+der (22)[1];0/100-Y(IPF) | 46,XY[6]/45,X-Y[2]/ 46,XY,t(9;22)[1] |
| 52 | 56 | M | IFN, HU | PHR | 36 | CP | CCR | 6 | No | 68+ | 46XX t(9;22)[30] | ND | 46,XY,t(9;22)[25]; 0/500-Y(IPF) | 46,XY[11]/45,X-Y[4] |
| 53 | 36 | M | Ida, Ara-C, HU, IFN, Bu (ABMT), MUD allograft4-150 | CCR4-151 | 52 | MCR | CCR | 12 | No | 76+ | 46,XY,t(9;22) | ND | 46,XY[25];0/25 del(20) (HMF); 9/300 del(20) (IPF) | 6,XY[18]/46,XY,t(7;17) (q11;q21)/del(20q) [5]/46,XY,t(9;22)[2] |
PHR indicates partial hematologic remission; mCR, minor cytogenetic remission; and HMF, hypermetaphase FISH. Other abbreviations are explained in Table 1 and the text.
2 Gy TBI/fludarabine.
Graft rejection and autologous reconstitution in CCR.
The karyotypic abnormalities included one case each of trisomy 8, monosomy 7, t(3;21)(q27?;q22) (Figure 4), del(20q), ider20(q10)t(20;21)(q22;q22), one multiaberrant karyotype including del(20q) and t(7;17)(q11;q21), and 2 cases with loss of the Y chromosome. In all patients, archive material was analyzed to see if the abnormalities were detectable at any time prior to imatinib treatment. Patients no. 19 and no. 34 had achieved cytogenetic remission with IFN-α and after stem cell mobilization, respectively, and thus Ph− cells were available for study. However, the respective abnormalities were not detected (in patient no. 34, a total of 900 interphases was analyzed by FISH).
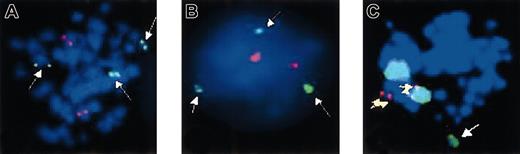
FISH analysis of patient no. 35 after 11 months on imatinib.
(A) Metaphase and (B) interphase preparations were hybridized with anAML1-ETO translocation probe. The 3 green signals (white arrows) correspond to chromosome 21q22 and indicate a break in this region in one chromosome 21; the 2 red signals correspond to the normal chromosome 8p22. (C) Metaphases were hybridized with a chromosome 3 paint (large green signals) and a locus-specific probe corresponding to chromosome 21q22.13-q22.2 (small red signals). There is colocalization of both probes (yellow arrows). Note the additional green signal corresponding to the translocated part of chromosome 3 (white arrow).
FISH analysis of patient no. 35 after 11 months on imatinib.
(A) Metaphase and (B) interphase preparations were hybridized with anAML1-ETO translocation probe. The 3 green signals (white arrows) correspond to chromosome 21q22 and indicate a break in this region in one chromosome 21; the 2 red signals correspond to the normal chromosome 8p22. (C) Metaphases were hybridized with a chromosome 3 paint (large green signals) and a locus-specific probe corresponding to chromosome 21q22.13-q22.2 (small red signals). There is colocalization of both probes (yellow arrows). Note the additional green signal corresponding to the translocated part of chromosome 3 (white arrow).
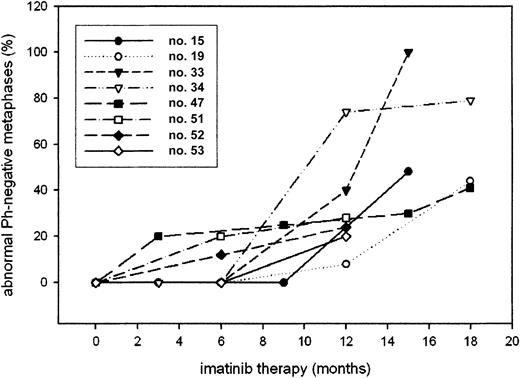
In patients no. 47 and no. 53, FISH detected 2.2% and 3% interphases with a signal constellation typical of del(20) prior to iamtinib therapy; however, both values are lower than the laboratory cutoff (3.7%). In 3 patients (nos. 19, 34, and 53), the abnormalities developed after a period of CCR or MCR, without evidence of abnormal Ph− cells. Cohybridization studies were done in the 2 patients with loss of Y and failed to detect cells with a BCR-ABL signal and −Y. In patient no. 19, constitutional trisomy 8 mosaicism could be excluded because FACS-sorted CD3+(T) cells were negative for trisomy 8 by FISH (1 of 300 cells = 0.3%, below cutoff). In all cases, the proportion of cells carrying the karyotypic abnormality increased over time (Figure5).
Proportion of abnormal Ph− metaphases in the various patients over time.
Patients with and without abnormalities in Ph− cells were analyzed for a number variables (Table5). Factors that were significantly associated with chromosomal changes in Ph− cells were previous treatment with cytarabine or idarubicin, whereas IFN, hydroxyurea (HU) as well as autologous bone marrow transplantation (ABMT) had no impact. Similarly, no relation to disease duration, age, and duration of imatinib treatment or disease phase was found.
Characteristics of patients with and without abnormalities
| Feature . | Patients without abnormalities in Ph−metaphases (n = 40) . | Patients with abnormalities in Ph−metaphases (n = 8) . | P . |
|---|---|---|---|
| Demographics | |||
| Age at diagnosis, y, median (range) | 51 (25-78) | 50 (37-72) | .9765-150 |
| Sex, male/female | 15/25 | 5/3 | .2515-151 |
| Disease duration, mo, median (range) | 20 (1-85) | 22 (9-69) | .2995-150 |
| Treatment prior to imatinib | |||
| IFN, yes/no | 25/15 | 7/1 | .2405-151 |
| HU, yes/no | 32/8 | 5/3 | .3615-151 |
| Ara-C, yes/no | 9/31 | 5/3 | .0375-151 |
| Ida, yes/no | 5/35 | 4/4 | .0315-151 |
| Other, yes/no | 2/38 | 1/8 | .4295-151 |
| ABMT, yes/no | 4/36 | 2/6 | .2585-151 |
| Phase at starting imatinib | |||
| Newly diagnosed, yes/no | 12/28 | 0/8 | .1745-151 |
| CPI/beyond CP1 | 37/3 | 6/2 | .1895-151 |
| Duration of imatinib therapy, mo, median (range) | 11 (1-23) | 16 (10-24) | .3065-150 |
| Feature . | Patients without abnormalities in Ph−metaphases (n = 40) . | Patients with abnormalities in Ph−metaphases (n = 8) . | P . |
|---|---|---|---|
| Demographics | |||
| Age at diagnosis, y, median (range) | 51 (25-78) | 50 (37-72) | .9765-150 |
| Sex, male/female | 15/25 | 5/3 | .2515-151 |
| Disease duration, mo, median (range) | 20 (1-85) | 22 (9-69) | .2995-150 |
| Treatment prior to imatinib | |||
| IFN, yes/no | 25/15 | 7/1 | .2405-151 |
| HU, yes/no | 32/8 | 5/3 | .3615-151 |
| Ara-C, yes/no | 9/31 | 5/3 | .0375-151 |
| Ida, yes/no | 5/35 | 4/4 | .0315-151 |
| Other, yes/no | 2/38 | 1/8 | .4295-151 |
| ABMT, yes/no | 4/36 | 2/6 | .2585-151 |
| Phase at starting imatinib | |||
| Newly diagnosed, yes/no | 12/28 | 0/8 | .1745-151 |
| CPI/beyond CP1 | 37/3 | 6/2 | .1895-151 |
| Duration of imatinib therapy, mo, median (range) | 11 (1-23) | 16 (10-24) | .3065-150 |
Comparison by 2-tailed Mann-Whitney U test.
Comparison by 2-sided Fisher exact test.
The chromosomal abnormalities in Ph− cells were detected at a median of 12 months (range, 3-15 months) after starting imatinib treatment (Table 4). Patients no. 34 and no. 15 developed clinical and morphologic signs of an MDS. Patient no. 34, with t(3;21), developed grade 3 thrombocytopenia that resolved to grade 2 after imatinib had been stopped. His bone marrow showed consistently more than 10% blasts. Patient no. 15, with monosomy 7, required continuous granulocyte colony-stimulating factor (G-CSF) support while on imatinib; her bone marrow exhibited severe dysplasia of granulopoiesis and erythropoiesis. In view of her monosomy 7, she underwent allogeneic stem cell transplantation. In 3 other patients (nos. 33, 47, and 53), imatinib had to be stopped temporarily because of grade 3 neutropenia and grade 2 thrombocytopenia. As of now, all patients are alive.
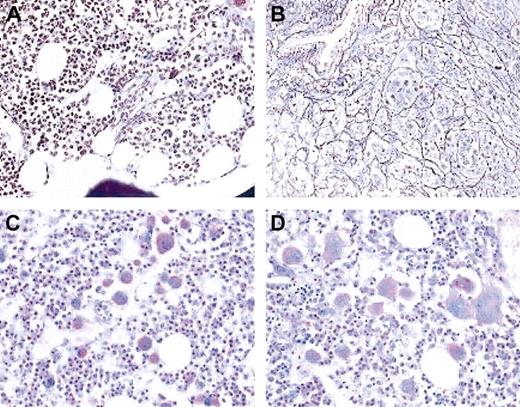
Of note, profound hematologic and morphologic abnormalities were also detected in one woman (patient no. 1) with a normal karyotype. This patient had achieved MCR on IFN-α/cytarabine but had subsequently relapsed in CP, with a platelet count of 635 × 109/L and 100% Ph+ metaphases. Bone marrow histology showed some atypical megakaryocytes as well as an increase in reticulin fibers (Figure 6A,C). Imatinib induced CCR within 3 months, but the platelet count rose to 2440 × 109/L. Bone marrow histology at 6 months demonstrated clusters of atypical megakaryocytic and marked fibrosis (Figure 6B,D). HUMARA-PCR failed to demonstrate clonal myeloid cells. Attempts to analyze platelet clonality at the RNA level18failed because all markers tested were noninformative.
Bone marrow histology.
Bone marrow histology in patient no. 1 prior to imatinib (A,C) and after 6 months on imatinib (B,D). Upper panels are silver stain; lower panels are periodic acid-Schiff stain. Cytogenetic analysis showed 100% Ph+ prior to imatinib and 0% Ph+ at the time of the follow-up biopsy. Note the increase in reticulin fibers and the marked change in megakaryocyte morphology. Magnification × 20 in all panels.
Bone marrow histology.
Bone marrow histology in patient no. 1 prior to imatinib (A,C) and after 6 months on imatinib (B,D). Upper panels are silver stain; lower panels are periodic acid-Schiff stain. Cytogenetic analysis showed 100% Ph+ prior to imatinib and 0% Ph+ at the time of the follow-up biopsy. Note the increase in reticulin fibers and the marked change in megakaryocyte morphology. Magnification × 20 in all panels.
Discussion
We show that therapy with imatinib restores polyclonal hematopoiesis in the majority of patients who achieve CCR. This is indicated by the fact that there is no difference between the RCI values of CD33+ cells from patients in CCR and healthy women. Applying the commonly used criterion of RCI less than 0.33 or more than 3 to diagnose clonality, 18 of 21 patients in CCR or MCR and 9 of 10 healthy women had polyclonal CD33+ cells. In contrast, 4 of 6 patients without cytogenetic response had a clonal pattern in their CD33+ cells. Correcting for constitutional skewing led to similar results. In the one patient, where material was available, a monoclonal pattern was demonstrable prior to imatinib therapy and a polyclonal pattern at the time of CCR. Three considerations should, however, be kept in mind. First, because the analysis of clonality based on X-chromosome inactivation cannot distinguish between a truly monoclonal and a polyclonal population with extreme lyonization,15 it is not capable of diagnosing clonality with absolute certainty. Second, clearly monoclonal populations may occasionally show a polyclonal pattern, as demonstrated by patient no. 24. It has been suggested that in such cases the malignant transformation leads to unstable X-chromosome methylation.19 Third, as shown in our dilution experiment as well as in the patients in MCR, the sensitivity of HUMARA-PCR to detect a clonal subpopulation in a polyclonal background is limited to 30% to 40%. This is likely to explain our failure to detect clonal hematopoiesis in most patients in MCR, and patients nos. 15 and 19, where the percentage of interphases carrying the respective abnormalities is below 30%. However, the example of patient no. 33 (Figure 3) clearly shows that clonality can be diagnosed by HUMARA-PCR if the majority of the cells carry the cytogenetic abnormality. Thus, within these limitations, our data provide compelling evidence that, similar to complete cytogenetic responders to IFN-α7 or ABMT,8 the re-established Ph− hematopoiesis in cytogenetic responders to imatinib is polyclonal. Thus, suppression of the Ph+ clone with a specific targeted therapy allows normal hematopoiesis to recover.
Surprisingly, we found cytogenetic abnormalities in Ph−cells in 8 of 48 patients studied, and clear evidence of a hematologic disorder in another patient with a normal karyotype. Considering only the patients treated in Leipzig, who were selected only on the basis of CCR or MCR to imatinib and availability of material for re-evaluation, the frequency of cytogenetic abnormalities in Ph− cells is 7 of 46 (15.2%). There are several reports on patients treated with IFN-α who developed clonal cytogenetic abnormalities in Ph− metaphases. The most common finding in these cases is trisomy 8,20-23 but other abnormalities such as various deletions have also been reported.12 Because no systematic study has been conducted, it is impossible to estimate the incidence of such abnormalities. However, it is certainly less than the 15.2% observed in our series. Some of the cytogenetic abnormalities detected in our patients, most notably monosomy 7 and t(3;21), are associated with acute myeloid leukemia and MDS.24 Of note, both are also found during progression of CML to blast crisis, although they are not frequent.25 MDS developed in the patient with t(3;21) who has thrombocytopenia and more than 10% bone marrow blasts and in the patient with monosomy 7 who exhibited dysplastic features in red and white cells, without increased blasts. Of note, on imatinib the latter patient required continuous G-CSF support because of neutropenia. Del(20q), present alone or in conjunction with t(7;17) in 2 of our patients, is associated with MDS as well as MDSs.26 Notably, all patients with chromosome 20 abnormalities (nos. 33, 47, 53) had frequent treatment interruptions because of hematologic toxicity, although they did not develop morphologic features indicative of MDS. It should be noted that some degree of mild dysplasia is frequently observed in patients treated with imatinib.27 Isolated loss of the Y chromosome is found in a substantial proportion of healthy older men and not necessarily of significance. However, at least one of our patients (no. 52) shows a progressive increase of −Y metaphases and may eventually reach the proposed 75% cutoff that appears to predict a pathologic state, even in older men.28 Of note, there is also one patient (no. 1) with a normal karyotype who developed excessive thrombocytosis. Because HUMARA-PCR of CD33+ cells failed to detect evidence of clonal hematopoiesis and the patient was not informative for all markers used to study mRNA from platelets, there is no way of proving clonal hematopoiesis. However, clusters of atypical megakaryocytes and excessive reticulin fibers are apparent on histology, consistent with an myeloproliferative syndrome and clearly indicating that the Ph− hematopoiesis is not normal.
Perhaps the most intriguing question refers to the time when the abnormal Ph− clones arise. It has long been speculated that a Ph− pathologic state may predate the acquisition of the Ph chromosome. A mathematical model found the epidemiology of chronic-phase CML most consistent with a 2-step pathogenesis.29 Furthermore, the observation thatBCR-ABL transcripts are detectable in a sizable proportion of healthy individuals raises the question if a first lesion is necessary to set the ground on which BCR-ABL becomes transforming. The most persuasive finding, however, is the observation that the distribution of G-6PD isoenzymes in Ph− B-cell lines derived from CML patients is skewed toward the phenotype of the CML cells, suggesting that the BCR-ABL translocation originates in a population of clonal Ph− cells.9,10 In regard to cytogenetic abnormalities, one would expect to see the additional changes both in Ph+ and Ph− cells. There is, however, no such case in our series and, to our knowledge, only one reported in the literature, where a trisomy 8 was detected both in Ph− and Ph+ cells.22 Moreover, HUMARA-PCR analysis shows polyclonal hematopoiesis in most patients in CCR. Thus, our data do not support the concept of a Ph− monoclonal state predating the acquisition of the BCR-ABL translocation.
The second explanation is that a patient's hematopoiesis as a whole has been subjected to genomic damage, producing multiple abnormal clones, among them one with a t(9;22). This clonal diversity would not become apparent as long as the Ph+ clone is at a proliferative advantage. Application of selective pressure by imatinib would then shift the balance, allowing Ph− clones to predominate. In contrast to imatinib, IFN-α may suppress not only the Ph+ but also other abnormal clones. This latter speculation might derive some support from the fact that in 2 of the 3 cases reported by Fayad et al12 and in the case reported by Ariyama et al20 the abnormalities in Ph−cells developed after IFN-α had been discontinued for 60, 36, and 4 months, respectively. It appears, however, more likely that the damage to a Ph− stem cell occurred at a later stage and as a result of CML therapy. This is supported by the significant association with previous exposure to cytarabine and idarubicin. In line with this notion, the emergence of an MDS with a t(3;21) in Ph−cells after autologous transplantation has been reported.30 Similarly, almost all patients with +8 in Ph− cells that were reported in the literature had been exposed to busulfan.23 Clearly, due to the small number of cases with abnormalities in Ph− cells, our statistical analysis must be interpreted with caution, and the study of a larger cohort is warranted.
The third possibility is that imatinib treatment itself induced or favored the acquisition of the additional abnormalities. Ablis known to interact with a number of proteins involved in the response to DNA damage and in DNA repair such asp73,31,32DNA-PK,33Atm,34,35 andRad51.36 37 Thus, its permanent inhibition by imatinib could contribute to the accumulation of new genetic damage. This may be particularly relevant in a situation where the hematopoiesis must be restored from a limited pool of Ph−stem cells, as in imatinib-induced cytogenetic remissions long after the initial CML diagnosis. It is obvious that these stem cells are under enormous stress to maintain cellular output. A strong argument against a causative role of imatinib is, however, the fact that the phenomenon has not yet been observed in patients treated with imatinib up-front.
Two considerations are important regarding the frequency of abnormalities in Ph− cells. First, the rate of major cytogenetic responses induced by imatinib is unprecedented, particularly in long-standing CML. Thus, it is now possible to investigate the Ph− hematopoiesis in situations where this was not the case before. Second, complete karyotyping of Ph− metaphases during follow-up on IFN-α is not always performed routinely, and subtle aberrations may be missed. This may explain, why such abnormalities were only rarely observed in patients treated with IFN-α.
What are the implications of these findings for the practical management of patients? One immediate conclusion is that patients on imatinib should be monitored by classical cytogenetics, even after achieving CCR. The second point is that the use of cytotoxic agents other than HU may have to be balanced against an increased risk of inducing abnormalities in Ph− cells. This would have implications for the design of combination trials. The third conclusion is that the hematopoiesis in some CML patients suffers from more than just CML. In such individuals, allografting would seem the best option, preferably before clones with dubious prognostic features (such as monosomy 7) have been selected. However, with the limited information available, as yet no therapeutic decisions should be based on our results.
Given the potential clinical implications, a systematic data collection is required. There are plans to establish an international registry at Oregon Health and Science University with the aim of collecting data on patients who develop chromosomal abnormalities in Ph− cells. This will allow gathering the information necessary for adequate counseling of patients about their treatment options.
We thank Ms Gerlinde Patzer for technical assistance.
Prepublished online as Blood First Edition Paper, October 31, 2002; DOI 10.1182/blood-2002-07-2053.
Supported by a grant from IZKF Leipzig, BMB-F, Interdisciplinary Center for Clinical Research at the University of Leipzig 9504 (K.K.) (01KS 9504, project Z3) and (M.D.) (01KS 9504, project D5).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Note added in proof
While this manuscript was in review, Andersen et al reported data on 3 patients with abnormalities in Ph− cells.38Several other cases have been brought to our attention that have been published in abstract form39-41 or will be published soon.27 These reports emphasize the importance of establishing a registry to study this phenomenon.
Author notes
Michael Deininger, OHSU, BMT/Leukemia Center, 3181 SW Sam Jackson Park Rd, Portland, OR 97239; e-mail:deininge@ohsu.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal