Thrombotic thrombocytopenic purpura (TTP) is caused by the persistence of the highly reactive high-molecular-weight multimers of von Willebrand factor (VWF) due to deficiency of the specific VWF-cleaving protease (VWF-CP) ADAMTS13, resulting in microangiopathic disease. The acquired form is caused by autoantibodies against VWF-CP, whereas homozygous or compound heterozygous mutations of ADAMTS13 are responsible for recessively inherited TTP. We investigated 83 children with hemolytic or thrombocytopenic episodes with or without additional neurologic symptoms or renal failure. The presumed diagnosis was chronic idiopathic thrombocytopenic purpura (ITP; n = 50), TTP (n = 8), hemolytic uremic syndrome (HUS; n = 24), and Evans syndrome (n = 1). A severe deficiency of VWF-CP (≤ 5%) was found in all investigated patients with TTP and in none of those with HUS. Additionally, 2 of 50 patients with a prior diagnosis of ITP were deficient for VWF-CP. Antibodies against VWF-CP were found in 4 children. Mutation analysis of the ADAMTS13 gene in the patients deficient in VWF-CP by direct sequencing of all 29 exons identified 8 different mutations, suggesting the hereditary form of TTP in 1 patient with ITP, in the patient with Evans syndrome, and in 5 of the 8 patients with TTP. The phenotype of TTP in childhood can be rather variable. Besides the classical clinical picture, oligosymptomatic forms may occur that can delay the identification of patients at risk.
Introduction
Thrombotic thrombocytopenic purpura (TTP) is a life-threatening microangiopathic disorder. Before the introduction of plasma exchange therapy, the mortality rate was extremely high.1 It is related to hemolytic uremic syndrome (HUS) but can be differentiated from HUS by the more general involvement of many organs compared with the more localized manifestation of HUS in the kidneys, although some overlap of symptoms may occur. It was first described by Moschcowitz in 1924,2 who presented the case of a previously healthy 16-year-old girl with fever, pallor, petechiae, acute renal failure, and paralysis who died 1 week after falling ill. Postmortem histology revealed hyaline platelet thrombi in almost every organ. Reports on similar cases with a congenital onset or familial manifestation, compared with first manifestation in adolescence and adulthood, indicated the presence of an inherited and an acquired form of the disease, respectively.3-9 Although the beneficial effect of plasma infusions suggested a deficiency of an essential plasma factor in the patients,4,6 the nature of the disease-causing factor remained obscure until Moake et al detected unusually large von Willebrand factor (VWF) multimers (ULVWFMs) in the plasma of 4 patients during their remission from TTP.10Relapses of TTP were accompanied by a decrease of these ULVWFMs, suggesting consumption by binding to aggregating platelets. The authors proposed a defect in the processing of the very large VWF multimers after synthesis and secretion by endothelial cells, making the patient susceptible to repeated relapses. This processing defect was identified as deficiency of a VWF-specific metalloprotease,11,12which cleaves VWF at a defined position in the VWF A2 domain between Tyr1605 and Met1606.13 A partial amino acid sequence was determined independently by 3 groups in 2001.14-16 Based on this information, a VWF-cleaving protease (VWF-CP) was identified as a member of the ADAMTS family of metalloproteases. The cDNA sequence was determined,17 and a database search revealed the structure of the gene on chromosome 9q34. Parallel to the biochemical approach, the responsible gene locus was mapped to 9q34 by linkage analysis and identification of specific mutations inADAMTS13.18 Review of the literature reveals very similar courses of congenital TTP in different patients over the years. The reported cases display the full spectrum of symptoms, though not always concurrent at the same time. Occasionally, thrombocytopenia can be the only symptom, but the full-blown clinical picture seems to manifest in later childhood or after puberty and to correlate with certain episodes of infection, stress, alcohol consumption, or pregnancy.3-9 Therefore, especially in early childhood, oligosymptomatic forms may be more frequent, complicating the differential diagnosis. To assess the clinical picture of VWF-CP deficiency in cases of chronic relapsing thrombocytopenia and hemolytic anemia, we determined VWF-CP activity in children with the presumed diagnosis of ITP, TTP, HUS, and Evans syndrome and carried out mutation analysis of the ADAMTS13 gene to establish the molecular background and the spectrum of mutations in our population.
Patients, materials, and methods
Patients
We investigated 83 children with hemolytic or thrombocytopenic episodes with or without additional neurologic symptoms or renal failure. The presumed diagnosis was chronic immune thrombocytopenia (ITP; n = 50), TTP (n = 8), HUS, (n = 24), and Evans syndrome (n = 1). The clinical course of our patients with TTP was rather variable, ranging from isolated thrombocytopenia, at least at some occasions, to the full pentad of symptoms2 and to death of disease (Table 3). Among the 24 patients with HUS, 7 had atypical forms; in 2 children HUS was nonenteropathic, and 1 of these 2 suffered from a relapse after renal transplantation. Two other patients had a relapsing course, and 1 of them had an affected sibling. Three additional patients with enteropathic symptoms also had affected siblings.
All patients or their parents were informed about the nature of this study and their consent was obtained according to the Declaration of Helsinki. Approval for this investigation was obtained from the institutional review board of the Children's University Hospital Hamburg-Eppendorf.
Assay of VWF-CP by the collagen-binding method
The activity of the VWF-CP was determined essentially as described.19 The only modification is the use of recombinant VWF (rVWF) as substrate.20 The rVWF preparation is a fully multimerized molecule and, due to the lack of VWF-CP activity in the cell culture medium in an uncleaved state, displays no triplet pattern. To avoid proteolysis by other proteases it was treated with Pefabloc SC (Roche, Mannheim, Germany) and EDTA (ethylenediaminetetraacetic acid) in final concentrations of 2 and 15 mM, respectively. Thereafter, it was dialyzed against 4% polyethylene glycol 20 000 (Merck, Darmstadt, Germany) and 5 mM Tris (tris(hydroxymethyl)aminomethane), pH 8.0.20This preparation with an rVWF concentration of 1.0 to 1.6 U/mL was stored in aliquots at −80°C. After thawing it was equilibrated with 1.5 M urea and used as the substrate for the VWF-CP. Plasma of a patient with severe von Willebrand disease (VWD) type 3 and normal VWF-CP served as a control. Digestion of rVWF (final concentrations, 0.6-0.9 U/mL) by VWF-CP and the collagen-binding assay of the digested samples was performed as described.19 VWF multimer analysis was carried out in sodium dodecyl sulfate (SDS) agarose gels combined with immunoblotting and luminescent visualization.21 The luminescent blot was stored on electronic media by means of photo imaging (FluorChem 8000; Alpha Innotech, San Leandro, CA).
Inhibitor assay
Detection of inhibitory activity in the sample was carried out by using a screening test, by mixing pooled normal plasma and patient's plasma (1:1), and by incubating for 30 minutes at 37°C. Thereafter the mixture was diluted 1:10 in Tris/urea, pH 8.0, and processed further as the other plasma samples (see “Assay of VWF-CP by the collagen-binding method”).
Molecular studies
DNA was isolated from peripheral nucleated blood cells by standard methods. Polymerase chain reactions (PCRs) were carried out in the T-Gradient and Trio Thermal Cycler, respectively (Biometra, Göttingen, Germany). Direct sequencing was done by means of the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit on an ABI Prism 310 or an ABI 377 (ABI, Foster City, CA) using either PCR primers or additional internal primers (Table1). Sequencing was carried out on both strands. Mutations were confirmed by an independent method such as the restriction enzyme digest (Table 2). If not otherwise stated, in the case of using kits, we followed the instructions of the provider.
| Exon . | Sense . | Antisense . | AT °C . |
|---|---|---|---|
| 7 | ggg tcg acc cgg gtc gga aaa ctc gct ggc | aga gag agg tgg cgc tgg cgg ccc agc ggg | 68 |
| 10 + 11 | ggg act ctc tgt gtg tgt tgg g | tga acc aca gat ggt cac ttt ggc tcc | 62 |
| 12 | gtg cca tgt agt ctc cca gtg aca aca cc | cca gag cct gaa cca ctt tgc cca gtg c | 65 |
| 15 | caa ttt ttc ccg acc agc taa gat cag ctc | act tgc ggg tga tgt cag aag tga ggg | 64 |
| 16 | ctg tgg ctc ctt aga gga ggg ctg ggg | cgc tga atg aat aaa tga acg aat cag gg | 64 |
| 20 | gca gct ggg cta tac ctt ccc ctg gg | aat ggg tgc tcc tcg ttc tcc cag cc | 68 |
| 21 | gga tcg ctg ggt cct cag agg agg ccc | gaa cga ggc aag atg caa gtg gac agg | 65 |
| 22 | gag gtg tcc agt gag cct ggg ctg cag tcc | ggc atg gag cca agg ctc agg tct cc | 67 |
| 26 | cct cct ggt ctc ctt cct cag ctt gg | tgc cag gaa tgg ggc atg cag cgt c | 67 |
| 27 | ctt gaa ctc ggc tca gtc tac cct g | ttc cat ctc agt cag ttc cct ccg | 62 |
| 28 | acg gct ccc tgg ctg tga gtt gtc c | ggg tcc cct agc cct gca cgc cc | 68 |
| 29 | ccg tga gtg cta att att act tgt gg | ggg ttt tca agc cag gtc tgg aca gc | 61 |
| Exon . | Sense . | Antisense . | AT °C . |
|---|---|---|---|
| 7 | ggg tcg acc cgg gtc gga aaa ctc gct ggc | aga gag agg tgg cgc tgg cgg ccc agc ggg | 68 |
| 10 + 11 | ggg act ctc tgt gtg tgt tgg g | tga acc aca gat ggt cac ttt ggc tcc | 62 |
| 12 | gtg cca tgt agt ctc cca gtg aca aca cc | cca gag cct gaa cca ctt tgc cca gtg c | 65 |
| 15 | caa ttt ttc ccg acc agc taa gat cag ctc | act tgc ggg tga tgt cag aag tga ggg | 64 |
| 16 | ctg tgg ctc ctt aga gga ggg ctg ggg | cgc tga atg aat aaa tga acg aat cag gg | 64 |
| 20 | gca gct ggg cta tac ctt ccc ctg gg | aat ggg tgc tcc tcg ttc tcc cag cc | 68 |
| 21 | gga tcg ctg ggt cct cag agg agg ccc | gaa cga ggc aag atg caa gtg gac agg | 65 |
| 22 | gag gtg tcc agt gag cct ggg ctg cag tcc | ggc atg gag cca agg ctc agg tct cc | 67 |
| 26 | cct cct ggt ctc ctt cct cag ctt gg | tgc cag gaa tgg ggc atg cag cgt c | 67 |
| 27 | ctt gaa ctc ggc tca gtc tac cct g | ttc cat ctc agt cag ttc cct ccg | 62 |
| 28 | acg gct ccc tgg ctg tga gtt gtc c | ggg tcc cct agc cct gca cgc cc | 68 |
| 29 | ccg tga gtg cta att att act tgt gg | ggg ttt tca agc cag gtc tgg aca gc | 61 |
Published primers were used for the remaining exons.18 Sense and antisense primers are both given in 5′-3′ direction. Screening for identified mutations among relatives of patients and 50 normal controls (100 alleles) was done either directly by polyacrylamide gel electrophoresis (PAGE) of PCR products in the case of a dinucleotide deletion, or by PAGE after restriction enzyme digest (Table 2). AT indicates annealing temperature.
Confirmation of mutations by restriction enzyme digest
| Mutation . | Exon . | RE . | +/− . | Primer . |
|---|---|---|---|---|
| Leu232Gln | 7 | CfrI | + | See Table 1 exon 7 |
| Ser263Cys | 7 | Eco47I | − | See Table 1exon 7 |
| Pro353Leu | 9 | MboII | + | See Levy et al18 |
| Trp390Xaa | 10 | BseNI | − | Sn: gca gca gtg cat ggg cgc tgg tct aac t |
| Asn: ggt acc tgg ggt tgt tgc act gcc gcc | ||||
| 2549–2550delAT | 20 | NA | See Table 1 exon 20 | |
| Arg910Xaa | 21 | HphI | + | Sn: gag aag gct ccc tcc cca tgg ggc ag |
| Asn: ccc agg agc atc ccg ggg ccc aca cct | ||||
| Arg1034Xaa | 24 | MaeI | − | See Levy et al.18 |
| 4143insA | 29 | MboII | + | Sn: cca tgg gca gca ggt gct cta ctg gga gtg a |
| Asn: cgg tac cca tga ttg gag ggt cca gta c |
| Mutation . | Exon . | RE . | +/− . | Primer . |
|---|---|---|---|---|
| Leu232Gln | 7 | CfrI | + | See Table 1 exon 7 |
| Ser263Cys | 7 | Eco47I | − | See Table 1exon 7 |
| Pro353Leu | 9 | MboII | + | See Levy et al18 |
| Trp390Xaa | 10 | BseNI | − | Sn: gca gca gtg cat ggg cgc tgg tct aac t |
| Asn: ggt acc tgg ggt tgt tgc act gcc gcc | ||||
| 2549–2550delAT | 20 | NA | See Table 1 exon 20 | |
| Arg910Xaa | 21 | HphI | + | Sn: gag aag gct ccc tcc cca tgg ggc ag |
| Asn: ccc agg agc atc ccg ggg ccc aca cct | ||||
| Arg1034Xaa | 24 | MaeI | − | See Levy et al.18 |
| 4143insA | 29 | MboII | + | Sn: cca tgg gca gca ggt gct cta ctg gga gtg a |
| Asn: cgg tac cca tga ttg gag ggt cca gta c |
2549–2550delAT was confirmed by electrophoresis and heteroduplex generation only. Boldface characters refer to mismatch nucleotides in primers used to create a diagnostic PCR product. Primers are given in the 5′-3′ direction.
RE indicates restriction enzyme; +/−, mutation either creates (+) or eliminates (−) a restriction site; Sn, sense; Asn, antisense; and NA, not available.
Mutation screening of the VWF-A2 domain
Besides VWF-CP deficiency itself, TTP could theoretically also be caused by an intrinsic resistance of VWF to proteolysis. However, naturally occurring respective VWF mutations have not been reported to date. We analyzed the VWF A2 domain of all patients with HUS and of 2 patients with TTP for whom no VWF-CP data were available, for possible VWF cleavage site mutations by PCR amplification of VWF exon 28 and direct sequencing as reported previously.20
Mutation screening of ADAMTS13
All coding exons and flanking intron sequences of theADAMTS13 gene were amplified by PCR using either published primers18 or primers chosen from the published sequence (GenBank accession numbers: AL158826, AC002325; Table 1). All PCR products were sequenced directly using an ABI 310 or an ABI 377 automatic sequencer. In the case of a dinucleotide deletion and a single nucleotide insertion, respectively, PCR products were also sequenced after cloning.
Results
VWF-CP
Data obtained from the VWF-CP assay are illustrated in Figure1. Among the 24 patients with HUS, none had VWF-CP activities below 62% (range, 62%-185%; median, 103%) Among the patients with the presumed diagnosis of ITP, 2 had VWF-CP deficiency with activities less than 2% of normal. However, because both also had mild Coombs-negative hemolytic anemia, the former diagnosis was changed to possible TTP. All other ITP patients had values of 33% or higher (range, 33%-175%; median, 59%) All tested patients with the diagnosis of TTP had VWF-CP values of 5% or less and, except one, even below the limit of detection at 2% at least at one or more occasions (Figure 1).
VWF-CP activity in patients with TTP, HUS, and ITP.
VWF-CP activity is severely decreased in patients with TTP, and more than 60% and more than 30% of normal in patients with HUS and ITP, respectively. VWF-CP activities in TTP patients refer to nadir values of each individual. Their VWF-CP ranges are given in Table3.
VWF-CP activity in patients with TTP, HUS, and ITP.
VWF-CP activity is severely decreased in patients with TTP, and more than 60% and more than 30% of normal in patients with HUS and ITP, respectively. VWF-CP activities in TTP patients refer to nadir values of each individual. Their VWF-CP ranges are given in Table3.
The VWF collagen-binding assay is relatively easy to perform and is faster than VWF multimer analysis for determination of VWF-CP activity. However, the action of VWF-CP on VWF is better visualized by the specific VWF cleavage pattern. VWF-CP causes loss of large VWF multimers and generates the typical triplet structure of plasma VWF oligomers. (Actually, a quintuplet structure with inner subbands is present that can be visualized by high-resolution gels.)22This triplet structure is absent in native TTP plasma, in untreated rVWF, and, accordingly, after incubation of rVWF with VWF-CP–deficient plasma. The appearance of pronounced triplets after mixing either VWF-CP–deficient plasma or rVWF with plasma from a patient deficient in VWF but with normal VWF-CP illustrates the specific action of the protease on VWF (Figure 2).
VWF-CP–specific proteolytic action on VWF demonstrated by VWF multimer analysis.
NP indicates normal plasma with typical triplet structure (filled arrows); TTP-P, native plasma of a patient with active TTP (note the absence of a defined triplet structure compared with normal plasma); TTP-P + VWF-CP, plasma of a patient with TTP mixed with control plasma of a patient with VWF deficiency but normal protease (note the appearance of a pronounced triplet structure besides loss of large multimers); rVWF, untreated recombinant VWF; rVWF + VWF-CP, proteolyzed rVWF incubated with plasma of a patient with VWD type 3 (empty arrows refer to inner subbands that can also be demonstrated by high-resolution SDS agarose gel electrophoresis of normal plasma VWF); rVWF + TTP-P, rVWF incubated with plasma of a patient with TTP.
VWF-CP–specific proteolytic action on VWF demonstrated by VWF multimer analysis.
NP indicates normal plasma with typical triplet structure (filled arrows); TTP-P, native plasma of a patient with active TTP (note the absence of a defined triplet structure compared with normal plasma); TTP-P + VWF-CP, plasma of a patient with TTP mixed with control plasma of a patient with VWF deficiency but normal protease (note the appearance of a pronounced triplet structure besides loss of large multimers); rVWF, untreated recombinant VWF; rVWF + VWF-CP, proteolyzed rVWF incubated with plasma of a patient with VWD type 3 (empty arrows refer to inner subbands that can also be demonstrated by high-resolution SDS agarose gel electrophoresis of normal plasma VWF); rVWF + TTP-P, rVWF incubated with plasma of a patient with TTP.
Molecular studies
VWF A2 domain.
In the patients with HUS and in 2 patients with TTP for whom no VWF-CP data were available, we analyzed VWF exon 28 that includes the sequence encoding the VWF A2 domain with the specific VWF cleavage site. No mutations in this region were detected that could possibly provide resistance against the action of VWF-CP.
ADAMTS13 mutations.
The complete coding sequence of ADAMTS13 and flanking intron sequences were analyzed by direct sequencing of PCR products for mutations that could cause VWF-CP deficiency. Altogether, 8 different mutations were identified on 14 disease alleles in 7 patients (Table3; Figure3). One patient had the former diagnosis of ITP, another had the diagnosis of Evans syndrome, and 5 patients had a diagnosis of TTP. No mutations were detected in 4 TTP patients with VWF-CP inhibitors.
Clinical and laboratory parameters of patients with TTP and their available relatives
| Individual . | Age at first symptom manifestation . | VWF-CP, % . | Inhibitor present . | Mutation 1 . | Mutation 2 . | Clinical manifestations . |
|---|---|---|---|---|---|---|
| 01 I-1 (f) | — | 37 | No | Ser263Cys | WT | — |
| 01 I-2 (m) | — | 55 | No | 4143insA | WT | — |
| 01 II-1 (p)3-150 | Congenital | < 2-5 | No | Ser263Cys | 4143insA | Mild |
| 01 II-2 (b) | — | NA | No | Ser263Cys | WT | — |
| 02 I-1 (p)3-150 | 11 mo | < 2 | Var | WT | WT | Mild |
| 03 I-1 (p)3-150 | 2 y | NA | NA | Pro353Leu | Arg910Xaa | DOD |
| 04 I-1 (f) | — | NA | NA | 4143insA | WT | — |
| 04 I-2 (m) | — | 24 | NA | Arg1034Xaa | WT | — |
| 04 II-1 (p)3-150 | Congenital | < 2 | NA | 4143insA | Arg1034Xaa | Severe |
| 04 II-2 (b) | — | 23 | NA | Arg1034Xaa | WT | — |
| 05 I-1 (p)3-150 | Congenital | 2-6 | No | Leu232Gln | Leu232Gln | Severe |
| 06 I-1 (p)3-150 | 9 y | 5-56 | Var | WT | WT | Mild |
| 07 I-1 (p)3-150 | 11 y | < 2-100 | Var | WT | WT | Mild |
| 08 I-1 (f) | — | NA | NA | Trp390Xaa | WT | — |
| 08 I-2 (m) | — | NA | NA | 2549–2550delAT | WT | — |
| 08 II-1 (p)3-150 | Congenital | NA | NA | Trp390Xaa | 2549–2550delAT | Severe |
| 09 I-1 (p)3-150 | 6 y | < 2-71 | Yes | WT | WT | Mild |
| 10 I-1 (f) | — | 21 | No | 4143insA | WT | — |
| 10 I-2 (m) | — | 23 | No | Pro353Leu | WT | — |
| 10 II-1 (p)3-150 | 4 y | < 2-6 | No | Pro353Leu | 4143insA | Mild |
| 10 II-2 (s) | — | 24 | No | 4143insA | WT | — |
| 11 I-1 (f) | — | ≈ 50 | No | Arg910Xaa | WT | — |
| 11 I-2 (m) | — | ≈ 50 | No | 4143insA | WT | — |
| 11 II-1 (p)3-150 | 3 y | < 2% | No | Arg910Xaa | 4143insA | Severe |
| Individual . | Age at first symptom manifestation . | VWF-CP, % . | Inhibitor present . | Mutation 1 . | Mutation 2 . | Clinical manifestations . |
|---|---|---|---|---|---|---|
| 01 I-1 (f) | — | 37 | No | Ser263Cys | WT | — |
| 01 I-2 (m) | — | 55 | No | 4143insA | WT | — |
| 01 II-1 (p)3-150 | Congenital | < 2-5 | No | Ser263Cys | 4143insA | Mild |
| 01 II-2 (b) | — | NA | No | Ser263Cys | WT | — |
| 02 I-1 (p)3-150 | 11 mo | < 2 | Var | WT | WT | Mild |
| 03 I-1 (p)3-150 | 2 y | NA | NA | Pro353Leu | Arg910Xaa | DOD |
| 04 I-1 (f) | — | NA | NA | 4143insA | WT | — |
| 04 I-2 (m) | — | 24 | NA | Arg1034Xaa | WT | — |
| 04 II-1 (p)3-150 | Congenital | < 2 | NA | 4143insA | Arg1034Xaa | Severe |
| 04 II-2 (b) | — | 23 | NA | Arg1034Xaa | WT | — |
| 05 I-1 (p)3-150 | Congenital | 2-6 | No | Leu232Gln | Leu232Gln | Severe |
| 06 I-1 (p)3-150 | 9 y | 5-56 | Var | WT | WT | Mild |
| 07 I-1 (p)3-150 | 11 y | < 2-100 | Var | WT | WT | Mild |
| 08 I-1 (f) | — | NA | NA | Trp390Xaa | WT | — |
| 08 I-2 (m) | — | NA | NA | 2549–2550delAT | WT | — |
| 08 II-1 (p)3-150 | Congenital | NA | NA | Trp390Xaa | 2549–2550delAT | Severe |
| 09 I-1 (p)3-150 | 6 y | < 2-71 | Yes | WT | WT | Mild |
| 10 I-1 (f) | — | 21 | No | 4143insA | WT | — |
| 10 I-2 (m) | — | 23 | No | Pro353Leu | WT | — |
| 10 II-1 (p)3-150 | 4 y | < 2-6 | No | Pro353Leu | 4143insA | Mild |
| 10 II-2 (s) | — | 24 | No | 4143insA | WT | — |
| 11 I-1 (f) | — | ≈ 50 | No | Arg910Xaa | WT | — |
| 11 I-2 (m) | — | ≈ 50 | No | 4143insA | WT | — |
| 11 II-1 (p)3-150 | 3 y | < 2% | No | Arg910Xaa | 4143insA | Severe |
f indicates father, m, mother, b, brother, and s, sister of the patient, respectively; WT, wild type; NA, not available for study; Var, variable; DOD, dead of disease; —, not applicable.
Patient data (p).
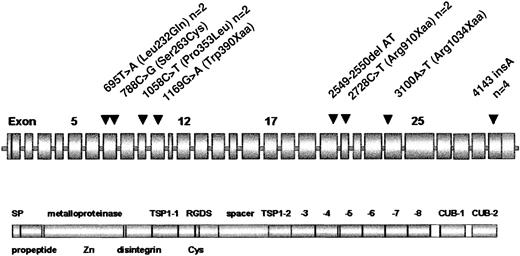
Location of ADAMTS13 mutations.
ADAMTS13 domain structure according to Zheng et al.17 The position of mutations is marked by arrowheads. Frequencies more than 1 are given after the designation.
Location of ADAMTS13 mutations.
ADAMTS13 domain structure according to Zheng et al.17 The position of mutations is marked by arrowheads. Frequencies more than 1 are given after the designation.
Five different molecular defects were truncating mutations, suggesting a causative nature, 3 were candidate missense mutations. Only 1 patient was homozygous, whereas the remaining 6 patients were compound heterozygotes for 2 mutations each. One mutation (4143insA) is particularly frequent and was found in 4 unrelated German patients. One missense mutation (1058C>T; Pro353Leu) and one nonsense mutation, (2728C>T; Arg910Xaa), respectively, were identified in 2 patients each, whereas 6 mutations were confined to single families.
The 695T>A in a Turkish patient (05 I-1) is located in exon 7 and predicts the exchange of leucine 232 to glutamine (Leu232Gln) in the metalloproteinase domain. Homozygosity for the mutation and an intragenic haplotype suggests consanguinity. The clinical course of this patient must be considered as severe due to renal infarction and to neurologic sequelae following stroke, and the need for continuous replacement therapy by fresh-frozen plasma (FFP).
The 788C>G is located in exon 7 and predicts the exchange of serine 263 to cysteine (Ser263Cys) in the metalloproteinase domain. It is found in patient 01 II-1 in combination with 4143insA, a truncating single base insertion in exon 29. He inherited Ser263Cys from his father and 4143insA from his mother (Table 3). To date the only symptoms were mild hemolysis and thrombocytopenia.
The 1058C>T is located in exon 9 and predicts the exchange of proline 353 to leucine (Pro353Leu) in the disintegrin domain. This mutation was found in 2 unrelated patients, both living in the same region in Northern Germany at the Denmark border. Patient 03 I-1 was also compound heterozygous for the nonsense mutation 2728C>T (Arg910Xaa). She was misdiagnosed with Evans syndrome and died from her disease at the age of 3 years. The correct diagnosis was made from archived DNA, 8 years after her death. Patient 10 II-1, with a clinically benign course and VWF-CP residual activity between less than 2% and 6% of normal, had 4143insA as second mutation, which he inherited from his father (Table 3).
The 1169G>A in exon 10 (Trp390Xaa) and an additional truncating mutation, a dinucleotide deletion, in exon 20 (2549-2550delAT) were identified in patient 08 II-1. Trp390Xaa was inherited from the father and 2549-2550delAT from the patient's mother (Table 3). The patient had a severe clinical course with stroke episodes and she is mentally retarded. She is on continuous replacement therapy with FFP. VWF-CP values were not available.
The 3100A>T in exon 24 (Arg1034Xaa) and 4143insA were detected in patient 04 II-1. She had severe clinical symptoms including neurologic and renal involvement. Her VWF-CP was less than 2% of normal. She is on continuous replacement therapy with a virus-inactivated commercial plasma preparation. 4143insA was inherited from the father, and Arg1034Xaa from the patient's mother (Table 3). The patient's unaffected brother is heterozygous for Arg1034Xaa.
In all investigated cases, asymptomatic family members who were heterozygous for one of the mutations displayed VWF-CP values corresponding to the heterozygous state and were clinically not affected (Table 3). By screening of 100 normal alleles none of the mutations was detected in the controls.
Discussion
Although reliable incidence data are not available, TTP is a rare disease in childhood. Our study, however, points to the possibility of underdiagnosis in the past. In our cohort, 3 patients were misdiagnosed as having Evans syndrome and ITP, respectively. It is of interest that the child with TTP reported by Schulman also had a previous diagnosis of ITP.4 These cases illustrate that oligosymptomatic forms may occur with the possibility of misdiagnosis. However, most probably even these patients may eventually suffer from severe TTP episodes when precipitating factors trigger the pathophysiologic cascade that has recently been elucidated. To identify such patients and to differentiate between the acquired and the hereditary form is of crucial importance for immediate care and adequate therapy in acute phases of the disease. Today this can be reliably done by assessing the VWF-CP activity, by screening for VWF-CP inhibitors, and recently, by performing mutation analysis of the ADAMTS13 gene.
All mutations reported to date were confined to single families and most of them were missense mutations.18 None of theADAMTS13 gene mutations that we identified in our patients has been described before. In contrast to the first report,18 we found a higher rate of truncating mutations. One mutation (4143insA) is particularly prevalent in our population and was found in 4 compound heterozygous patients. Only 3 mutations were missense mutations that can only be regarded as candidates, because, to date, expression studies are pending. However, one of them (Leu232Gln) was found in homozygous form, and another one (Pro353Leu) was found in 2 unrelated patients. Furthermore, none of the 8 different mutations was detected among 100 alleles of healthy controls.
A correlation between phenotype and genotype of congenital TTP is hard to establish. The patient's history suggests that precipitating factors such as infections have more impact on the severity of TTP episodes than the genotype. Due to the complex mutation spectrum in congenital TTP, proof of a genotype-phenotype correlation would require a large number of patients.
Although the final events in TTP and childhood HUS are comparable, the initiating triggers are different. Not a single patient among our 24 with HUS displayed VWF-CP values below 62% of normal, not even those with “atypical” HUS. This agrees with the reports of others,23,24 although contradictory observations were also published.25 However, in conclusion, it seems that severe VWF-CP deficiency with values below 5% of normal is indicative of TTP rather than HUS.
Although theoretically possible, we could not detect mutations of VWF that would provide resistance against the proteolytic action of VWF-CP in the patients with HUS. VWF-CP resistance does not seem to contribute significantly to the pathomechanism of HUS.
Two different forms of childhood TTP were observed among our patients. Four patients had the acquired form, most probably caused by autoantibodies against the protease. The inconsistent detection of VWF-CP inhibitors even during longer periods of severe VWF-CP deficiency in these patients complicates the correct classification, which is crucial for the adequate treatment. Possibly, the Bethesda method for the identification of VWF-CP antibodies is not sensitive enough to detect low titer, though effective, inhibitors.
Patients with an inhibitor need a more aggressive therapy, including plasma exchange and immunosuppression, whereas patients with the hereditary form can be treated by VWF-CP replacement therapy alone. Due to the long half-life of VWF-CP of 2 to 3 days26 and the clinical efficacy even at low VWF-CP values, plasma infusion every 2 weeks seems sufficient. This is also confirmed by the clinical response to such prophylaxis in some of our patients.27 28
In all of our patients without an inhibitor we identifiedADAMTS13 gene mutations on both alleles, which is in accordance with the autosomal recessive inheritance of inborn VWF-CP deficiency, whereas no mutation was found in patients with a significant inhibitor detected at least at one time point. Although exact data are lacking, it seems that childhood TTP is more often caused by hereditary VWF-CP deficiency, which would agree with the lower incidence of autoimmune disease in children compared with adults. Given the impact of the correct classification of TTP with respect to therapy and genetic counseling, we propose molecular genetic testing in children with severe VWF-CP deficiency.
Prepublished online as Blood First Edition Paper, October 17, 2002; DOI 10.1182/blood-2002-08-2399.
R.S. and U.B. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Note
A second report on ADAMTS13 gene mutations has been published during revision of this manuscript.29
Author notes
Reinhard Schneppenheim, University Hospital Hamburg-Eppendorf, Department of Pediatric Hematology and Oncology, Martinistr 52, D-20246 Hamburg, Germany; e-mail:schneppenheim@uke.uni-hamburg.de.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal