Mutations of the AML1 gene are frequent molecular abnormalities in minimally differentiated acute myeloblastic leukemia (M0 AML), a rare type of AML. In this retrospective multicenter study, morphologic, immunophenotypical, cytogenetic, and molecular features of 59 de novo M0 AML cases were analyzed and correlated to AML1 mutations. Point mutations ofAML1 gene were observed in 16 cases (27%). They were correlated with higher white blood cell (WBC) count (P = .001), greater marrow blast involvement (P = .03), higher incidence of immunoglobulin H/T-cell receptor (IgH/TCR) gene rearrangement (P < .0001), and with a borderline significant lower incidence of complex karyotypes. In the 59 patients, FLT3 mutations were the only significant prognostic factors associated with short survival.
Introduction
Minimally differentiated acute myeloblastic leukemia (AML), now classified as M0 AML, is a rare type of AML associated with poor prognosis.1,2 Recently, we and others reported in M0 AML a high frequency of mutations of AML1gene, a gene that plays a pivotal role in myeloid differentiation.3-6 In this multicenter cooperative work, we analyzed clinical and biologic characteristics of a large series of M0 AML and compared, in particular, patients with and withoutAML1 mutation.
Study design
Patient population
Fifty-nine patients diagnosed as de novo M0 AML between 1993 and 2000 in the hematology departments of 9 French university hospitals (Besançon, Bordeaux, Dijon, Lille, Marseille, Nantes, Paris, Reims, and Toulouse) were included. All cases were centrally reviewed by members of Groupe Français d'Hématologie Cellulaire (GFHC) for cytology, cytochemistry, and immunophenotyping and by members of Groupe Français de Cytogénétique Hématologique (GFCH) for cytogenetic findings.
Patients with older age and/or poor clinical condition received supportive care only or moderate chemotherapy (Table1). Other patients received anthracycline-AraC induction chemotherapy, based on European or French multicenter protocols for AML (acute Leukaemia French Association [ALFA], Bordeaux Grenoble Montpellier Toulouse [BGMT], European Organisation for Research and Treatment of Cancer [EORTC], and Groupe ouest des Leucemies et Autres Maladies du Sang [GOELAMS] cooperative groups).7-10 Patients who achieved complete remission (CR) received consolidation with high-dose AraC-based chemotherapy and autologous or allogeneic stem cell transplantation, depending on age, donor availability, and trial design.
Biological and clinical data
| Patient . | Sex . | Age, y . | Karyotype . | FLT3 duplication . | Rearrangement . | AML1 gene mutation . | AML1 gene alteration . | Intensive Treatment . | Induction regimen . | CR achievement . | CR duration, mo . | Survival, mo . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IgH . | TCR . | First allele . | Second allele . | |||||||||||
| 1 | M | 45 | Not done | No | DJ | No | Mutated | C114ins | F131del | Yes | AraC-anthracycline | Yes | 6+ | 6+ |
| 2 | M | 38 | 46,XY [20] | No | DJ | No | Mutated | R135G | del | Yes | AraC-anthracycline | Yes | 20+ | 20+ |
| 3 | F | 64 | 46,XX [19] | No | No | No | Mutated | D171G | del | Yes | AraC-anthracycline | Yes | 56+ | 56+ |
| 4 | M | 86 | Not done | Yes | No | No | Mutated | R139 ter | del | No | Supportive care | No | — | 2 |
| 5 | F | 45 | 46,XX,inv (3)(q21q26) [1]/45,XX,id. −7 [4]/ 46,XX [8] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 2 |
| 6 | F | 43 | 45,XX,inv (3)(q21q26), −7 [16] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 1 |
| 7 | F | 46 | 46,XX [20] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 28+ | 28+ |
| 8 | M | 23 | 46,XY [20] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 10 | 11 |
| 9 | F | 49 | Not done | No | FR3+ FR1+ | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | <1 |
| 10 | F | 35 | 45-46,XX −X[3],del(5)(q15q33),−7, add(7)(p22),+17,−20, add(20)(p13)[2]+mar1,+r[cp44]/ 44-46, XX-X[3],idem,del(3)(q21)[3], ?der(3)t(3:?)[3], −del(5) (q15q33)[4],−6,−8[4],−14[6], +add(16)(q22)[6],−17[2], −17[4],add(17)(p13)[3), +m2[7], +m3[5][cp]7 | No | No | VG1JP1/2-VD1-VG11JP1/2 | Mutated | S140 ins | Splice site ex4/int4 | No | Supportive care | No | — | <1 |
| 11 | F | 80 | Not done | Not done | Not done | Not done | Germ line | — | — | No | Supportive care | No | — | <1 |
| 12 | M | 80 | 46,XY [20] | Not done | Not done | Not done | Germ line | — | — | No | Supportive care | No | — | 1 |
| 13 | M | 48 | 45,XX, −1, add(I)(q?10),−5, −7,der(7)t(7;?)(p?;?), −12, −17, i(17)(q10), + 4 mar, +dmin [17]/ 46, XY [1] ishder(1) wcp1 +, wcp7+, mar1der(1)del(1)(q22) (wcp1+;wcp5+),mar2 der(5) (wcp5+), mar3 der(5) (?:5?;7p14− qter)(wcp5+;wcp7+) | No | No | No | Germ line | Yes | AraCanthracycline-VP 16 | No | — | <1 | ||
| 14 | M | 78 | 47,XY,del(20)(q13.1),+ mar [5]/ 47,id, der(13;13)(q10;q10) [16] | No | No | No | Mutated | R49S | del(38 bp) | No | Supportive care | No | — | <1 |
| 15 | M | 28 | 46,XY, ins(10;11)(p14;q14q23).ish der(10)(CALM+,AF10+)[15] / 46,XY [7] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline-VP16 | Yes | 12+ | 12+ |
| 16 | M | 78 | Not done | No | DJ | No | Mutated | N119del | del | Yes | AraC-anthracycline | No | — | <1 |
| 17 | M | 78 | Not done | No | No | No | Mutated | R135M | G138C | No | Supportive care | No | — | <1 |
| 18 | M | 61 | 47,XY,i(21)(q10),+mar[14]/47, XY,idem,del(3)(p11), der(11)add(11)(p15)add (11)(q25), −18, +min.ish der(11)(MLL+)[2] /46,XY [1] | No | No | No | Germ line | Yes | AraC-anthracycline | No | — | 1 | ||
| 19 | M | 64 | 47,XY,−21,+der(21)(q2?)×2 .ish der(21)(AML1 × 3)[16] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 1 |
| 20 | M | 29 | 79,XXYY,−2,−3,−5,−6,−7, −9,−10,−11,15,−15, −16,−17,−18,−21 [4]/ 46,XY [25] | Yes | No | No | Mutated | R139 ter | del | Yes | AraC-anthracycline | Yes | 7.5 | 10 |
| 21 | M | 76 | 45,XY,−21.ish (21)(wcp21 +)×1[12]/ 46,XY [21] | No | No | No | Germ line | — | — | No | Supportive care | No | — | <1 |
| 22 | M | 53 | 48,XY, del (3)(p2?), +del (3)(p2?),+19 [13]/ 46,XY[18] | No | No | VG1J1J2 | Germ line | — | — | Yes | AraC-anthracycline | Yes | 7 | 10 |
| 23 | M | 77 | Not done | No | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 24 | M | 86 | Not done | No | FR3+ FR1+ | No | Mutated | R139 ter | del | No | Supportive care | No | — | <1 |
| 25 | M | 81 | Not done | No | No | VG9J1J2 | Mutated | G138 ins | del | No | Supportive care | No | — | 1 |
| 26 | M | 38 | 46,XY, ins(10;?)(q23;?)[3]/ 46, XY [32] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 2 |
| 27 | M | 36 | 46,XY, t(6;12)(q25;q11), t(8;9)(q24;q33), t(11;16)(q13;q13) [29]/46, XY [2] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 7 | 8 |
| 28 | M | 64 | 46, XY [29] | Yes | FR3+ FR1+ | No | Mutated | R174Q | Ins. splice site exon 4 | Yes | AraC-anthracycline | No | — | 1 |
| 29 | F | 84 | 46, XX [30] | No | No | No | Germ line | — | — | No | Supportive care | No | — | <1 |
| 30 | M | 82 | 46, XY [31] | No | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 31 | M | 47 | Not done | Yes | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 32 | F | 73 | 44,XX, −4,−5,−7,−17,−19, add(22),+3mar .ish der(5)(5pter−5q?::22::19) (wcp5+,wcp19+,wcp22+), der(7)t(4;7)(q21;q22)(wcp4+, wcp7+),der[19]t(4;19)(q21;?) (wcp4+,19+),add(22).ish dup(22)(q?)(wcp22+) [12] | No | No | No | Germ line | — | — | No | Supportive care | No | — | <1 |
| 33 | F | 88 | 46, XX [21] | Yes | DJ | No | Mutated | V137ins | del | No | Supportive care | No | — | 1 |
| 34 | F | 16 | 50,XX,+2,+6,+20,+22[3]/ 50,idem,−6, +add(6)(q26).ish add(6)(wcp6+,wcp 11−)[11] / 46,XX [6] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 47 | 48 |
| 35 | F | 74 | 46,XX [22] | No | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 36 | M | 31 | 47,XY, dup(14)(q3?)+mar .ish mar (wcp10+) [11] / 46, XY [10] | No | No | VG1JP1/2 | Germ line | — | — | Yes | AraC-anthracycline | Yes | 26 | 26 |
| 37 | M | 64 | Not done | No | FR1+ FR3+ | VG1JP1/2-VG1J1J2 9J1J2 | Mutated | R177ter | del | No | Supportive care | No | — | 1 |
| 38 | F | 34 | 46,XX, t(1,9)(q31;p21).ish t(1;9;11)(q31;p21−22;q23−24) (wcp1+,wcp9+,wcp11+) (MLL+X3) [21]/ 46,XX [2] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 94+ | 94+ |
| 39 | M | 50 | 91,XXYY,−5,−11,+der(5;11) (p10;p10)[29] /46, XY [1] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 11 | 20 |
| 40 | F | 62 | 46,XX [24] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 5 |
| 41 | H | 57 | 46,XY,t(3;12)(q26;p13).ish der(3)(ETV6+)der(12)(ETV6+) [15]/ 46,XY [7] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 13 | 15 |
| 42 | H | 49 | 46,XY,i(7)(q10) [20] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 7 | 7 |
| 43 | F | 17 | 89,XXXX, −5, −7, +13, −16,−21,−22, +mar inc[8]/ 46, XX [1] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 8 | 9 |
| 44 | F | 52 | 46,XX [20] | No | No | No | Mutated | Splice site int3/ex4 | del | Yes | AraC-anthracycline | No | — | 3 |
| 45 | F | 76 | Not done | No | No | No | Germ line | — | — | No | Supportive care | No | — | <1 |
| 46 | F | 87 | Not done | No | No | No | Germ line | — | — | No | Supportive care | No | — | <1 |
| 47 | M | 63 | 47,XY, +8.nuc ish (D8z2×3)(86%)[16]/ 46, XY [4] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | <1 |
| 48 | F | 15 | 46,XX [18] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 1+ | 1+ |
| 49 | M | 67 | Not done | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 2 |
| 50 | M | 38 | 46,XY [20] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 1 |
| 51 | F | 59 | Not done | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 49 | 51 |
| 52 | M | 65 | 46,XY [30] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 12 | 13 |
| 53 | M | 35 | 47,XY, +1, del (5)(q12q34), del (11)(q22.1q23.2) [7] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 103+ | 103+ |
| 54 | M | 70 | Not done | No | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 55 | F | 73 | Not done | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 28 | 50 |
| 56 | M | 47 | 46,XY [15] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 15+ | 15+ |
| 57 | M | 60 | 46,XY, t(4;13)(q21;q21) [19]/ 46, id. del (7)(q2?2q3?4) [1]/46, id. del (5)(q15q35) [1] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 48 | 50 |
| 58 | M | 82 | Not done | No | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 59 | M | 79 | Not done | No | FR3+ | No | Mutated | R135ins | R174G | No | Supportive care | No | — | <1 |
| Patient . | Sex . | Age, y . | Karyotype . | FLT3 duplication . | Rearrangement . | AML1 gene mutation . | AML1 gene alteration . | Intensive Treatment . | Induction regimen . | CR achievement . | CR duration, mo . | Survival, mo . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IgH . | TCR . | First allele . | Second allele . | |||||||||||
| 1 | M | 45 | Not done | No | DJ | No | Mutated | C114ins | F131del | Yes | AraC-anthracycline | Yes | 6+ | 6+ |
| 2 | M | 38 | 46,XY [20] | No | DJ | No | Mutated | R135G | del | Yes | AraC-anthracycline | Yes | 20+ | 20+ |
| 3 | F | 64 | 46,XX [19] | No | No | No | Mutated | D171G | del | Yes | AraC-anthracycline | Yes | 56+ | 56+ |
| 4 | M | 86 | Not done | Yes | No | No | Mutated | R139 ter | del | No | Supportive care | No | — | 2 |
| 5 | F | 45 | 46,XX,inv (3)(q21q26) [1]/45,XX,id. −7 [4]/ 46,XX [8] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 2 |
| 6 | F | 43 | 45,XX,inv (3)(q21q26), −7 [16] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 1 |
| 7 | F | 46 | 46,XX [20] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 28+ | 28+ |
| 8 | M | 23 | 46,XY [20] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 10 | 11 |
| 9 | F | 49 | Not done | No | FR3+ FR1+ | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | <1 |
| 10 | F | 35 | 45-46,XX −X[3],del(5)(q15q33),−7, add(7)(p22),+17,−20, add(20)(p13)[2]+mar1,+r[cp44]/ 44-46, XX-X[3],idem,del(3)(q21)[3], ?der(3)t(3:?)[3], −del(5) (q15q33)[4],−6,−8[4],−14[6], +add(16)(q22)[6],−17[2], −17[4],add(17)(p13)[3), +m2[7], +m3[5][cp]7 | No | No | VG1JP1/2-VD1-VG11JP1/2 | Mutated | S140 ins | Splice site ex4/int4 | No | Supportive care | No | — | <1 |
| 11 | F | 80 | Not done | Not done | Not done | Not done | Germ line | — | — | No | Supportive care | No | — | <1 |
| 12 | M | 80 | 46,XY [20] | Not done | Not done | Not done | Germ line | — | — | No | Supportive care | No | — | 1 |
| 13 | M | 48 | 45,XX, −1, add(I)(q?10),−5, −7,der(7)t(7;?)(p?;?), −12, −17, i(17)(q10), + 4 mar, +dmin [17]/ 46, XY [1] ishder(1) wcp1 +, wcp7+, mar1der(1)del(1)(q22) (wcp1+;wcp5+),mar2 der(5) (wcp5+), mar3 der(5) (?:5?;7p14− qter)(wcp5+;wcp7+) | No | No | No | Germ line | Yes | AraCanthracycline-VP 16 | No | — | <1 | ||
| 14 | M | 78 | 47,XY,del(20)(q13.1),+ mar [5]/ 47,id, der(13;13)(q10;q10) [16] | No | No | No | Mutated | R49S | del(38 bp) | No | Supportive care | No | — | <1 |
| 15 | M | 28 | 46,XY, ins(10;11)(p14;q14q23).ish der(10)(CALM+,AF10+)[15] / 46,XY [7] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline-VP16 | Yes | 12+ | 12+ |
| 16 | M | 78 | Not done | No | DJ | No | Mutated | N119del | del | Yes | AraC-anthracycline | No | — | <1 |
| 17 | M | 78 | Not done | No | No | No | Mutated | R135M | G138C | No | Supportive care | No | — | <1 |
| 18 | M | 61 | 47,XY,i(21)(q10),+mar[14]/47, XY,idem,del(3)(p11), der(11)add(11)(p15)add (11)(q25), −18, +min.ish der(11)(MLL+)[2] /46,XY [1] | No | No | No | Germ line | Yes | AraC-anthracycline | No | — | 1 | ||
| 19 | M | 64 | 47,XY,−21,+der(21)(q2?)×2 .ish der(21)(AML1 × 3)[16] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 1 |
| 20 | M | 29 | 79,XXYY,−2,−3,−5,−6,−7, −9,−10,−11,15,−15, −16,−17,−18,−21 [4]/ 46,XY [25] | Yes | No | No | Mutated | R139 ter | del | Yes | AraC-anthracycline | Yes | 7.5 | 10 |
| 21 | M | 76 | 45,XY,−21.ish (21)(wcp21 +)×1[12]/ 46,XY [21] | No | No | No | Germ line | — | — | No | Supportive care | No | — | <1 |
| 22 | M | 53 | 48,XY, del (3)(p2?), +del (3)(p2?),+19 [13]/ 46,XY[18] | No | No | VG1J1J2 | Germ line | — | — | Yes | AraC-anthracycline | Yes | 7 | 10 |
| 23 | M | 77 | Not done | No | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 24 | M | 86 | Not done | No | FR3+ FR1+ | No | Mutated | R139 ter | del | No | Supportive care | No | — | <1 |
| 25 | M | 81 | Not done | No | No | VG9J1J2 | Mutated | G138 ins | del | No | Supportive care | No | — | 1 |
| 26 | M | 38 | 46,XY, ins(10;?)(q23;?)[3]/ 46, XY [32] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 2 |
| 27 | M | 36 | 46,XY, t(6;12)(q25;q11), t(8;9)(q24;q33), t(11;16)(q13;q13) [29]/46, XY [2] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 7 | 8 |
| 28 | M | 64 | 46, XY [29] | Yes | FR3+ FR1+ | No | Mutated | R174Q | Ins. splice site exon 4 | Yes | AraC-anthracycline | No | — | 1 |
| 29 | F | 84 | 46, XX [30] | No | No | No | Germ line | — | — | No | Supportive care | No | — | <1 |
| 30 | M | 82 | 46, XY [31] | No | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 31 | M | 47 | Not done | Yes | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 32 | F | 73 | 44,XX, −4,−5,−7,−17,−19, add(22),+3mar .ish der(5)(5pter−5q?::22::19) (wcp5+,wcp19+,wcp22+), der(7)t(4;7)(q21;q22)(wcp4+, wcp7+),der[19]t(4;19)(q21;?) (wcp4+,19+),add(22).ish dup(22)(q?)(wcp22+) [12] | No | No | No | Germ line | — | — | No | Supportive care | No | — | <1 |
| 33 | F | 88 | 46, XX [21] | Yes | DJ | No | Mutated | V137ins | del | No | Supportive care | No | — | 1 |
| 34 | F | 16 | 50,XX,+2,+6,+20,+22[3]/ 50,idem,−6, +add(6)(q26).ish add(6)(wcp6+,wcp 11−)[11] / 46,XX [6] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 47 | 48 |
| 35 | F | 74 | 46,XX [22] | No | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 36 | M | 31 | 47,XY, dup(14)(q3?)+mar .ish mar (wcp10+) [11] / 46, XY [10] | No | No | VG1JP1/2 | Germ line | — | — | Yes | AraC-anthracycline | Yes | 26 | 26 |
| 37 | M | 64 | Not done | No | FR1+ FR3+ | VG1JP1/2-VG1J1J2 9J1J2 | Mutated | R177ter | del | No | Supportive care | No | — | 1 |
| 38 | F | 34 | 46,XX, t(1,9)(q31;p21).ish t(1;9;11)(q31;p21−22;q23−24) (wcp1+,wcp9+,wcp11+) (MLL+X3) [21]/ 46,XX [2] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 94+ | 94+ |
| 39 | M | 50 | 91,XXYY,−5,−11,+der(5;11) (p10;p10)[29] /46, XY [1] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 11 | 20 |
| 40 | F | 62 | 46,XX [24] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 5 |
| 41 | H | 57 | 46,XY,t(3;12)(q26;p13).ish der(3)(ETV6+)der(12)(ETV6+) [15]/ 46,XY [7] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 13 | 15 |
| 42 | H | 49 | 46,XY,i(7)(q10) [20] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 7 | 7 |
| 43 | F | 17 | 89,XXXX, −5, −7, +13, −16,−21,−22, +mar inc[8]/ 46, XX [1] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 8 | 9 |
| 44 | F | 52 | 46,XX [20] | No | No | No | Mutated | Splice site int3/ex4 | del | Yes | AraC-anthracycline | No | — | 3 |
| 45 | F | 76 | Not done | No | No | No | Germ line | — | — | No | Supportive care | No | — | <1 |
| 46 | F | 87 | Not done | No | No | No | Germ line | — | — | No | Supportive care | No | — | <1 |
| 47 | M | 63 | 47,XY, +8.nuc ish (D8z2×3)(86%)[16]/ 46, XY [4] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | <1 |
| 48 | F | 15 | 46,XX [18] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 1+ | 1+ |
| 49 | M | 67 | Not done | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 2 |
| 50 | M | 38 | 46,XY [20] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | No | — | 1 |
| 51 | F | 59 | Not done | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 49 | 51 |
| 52 | M | 65 | 46,XY [30] | Yes | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 12 | 13 |
| 53 | M | 35 | 47,XY, +1, del (5)(q12q34), del (11)(q22.1q23.2) [7] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 103+ | 103+ |
| 54 | M | 70 | Not done | No | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 55 | F | 73 | Not done | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 28 | 50 |
| 56 | M | 47 | 46,XY [15] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 15+ | 15+ |
| 57 | M | 60 | 46,XY, t(4;13)(q21;q21) [19]/ 46, id. del (7)(q2?2q3?4) [1]/46, id. del (5)(q15q35) [1] | No | No | No | Germ line | — | — | Yes | AraC-anthracycline | Yes | 48 | 50 |
| 58 | M | 82 | Not done | No | No | No | Germ line | — | — | No | Supportive care | No | — | 1 |
| 59 | M | 79 | Not done | No | FR3+ | No | Mutated | R135ins | R174G | No | Supportive care | No | — | <1 |
Morphologic studies
Bone marrow and peripheral blood smears were stained by May-Grünwald-Giemsa. Myeloperoxidase, naphthol AS-D-acetate esterases with and without fluoride inhibition, or naphthyl acetate butyrate esterases cytochemical reactions were performed.
Immunophenotypic studies
Immunophenotyping was performed by flow cytometry. Membrane expression of CD2, 3, 5, 7, 10, 19, 20, 22, 24, 79a, 34, 13, 33, 117, 14, 15, 41, 61, 65, and HLA-DR was tested. For CD3, CD13, CD22, and myeloperoxidase (MPO), intracytoplasmic antigen expression was also tested. Diagnostic criteria of M0 AML were (1) less than 3% myeloperoxidase-positive blasts; (2) expression of at least one of the following myeloid markers: CD13, CD33, or MPO; and (3) no expression of lymphoid markers except CD4 or CD7 (European Group for the Immunological Characterization of Leukemias [EGIL] criteria).11
Cytogenetic and fluorescence in situ hybridization (FISH) studies
For conventional cytogenetic analysis, chromosomes were identified by R-bands by heating using Giemsa (RHG) and/or G-bands with trypsin using Giemsa (GTG) banding, and abnormalities were described according to International System for Cytogenetic Nomenclature (ISCN).12 FISH was performed according to standard methods and manufacturer's instructions by using whole chromosomes or locus-specific probe mixed lineage lymphoma (MLL), AML1 (Vysis, Downers Grove, IL), and yeast artificial chromosomes (YACs) probes for ALL1 fused gene from chromosome 10 (AF10), clathrin assembly lymphoid-myeloid leukemia (CALM), and ETS variant gene 6 (ETV6) regions obtained from the CENTRE d'Étude du Polymorphisme humain (CEPH; Paris, France) (815c7, 814d9, 936e2 YAC clones).
Molecular studies
Detection of AML1 mutations was made on DNA and/or cDNA from bone marrow cells, as previously reported.3,6Study of FLT3 duplication, IgH (FR1, FR3, DJ), TCR-γ (Vg 1→Vg 11; J1J2; Jγ1/2; JP1/2; JP) and TCR-δ (Vd2Dδ3; Vd1Jδ1) gene rearrangements were performed according to Kiyoi et al,13 Landman-Parker et al,14 Davi et al,15 Delabesse et al,16 and Cave et al,17 respectively.
Results and discussion
Clinical and biologic characteristics and outcome
Clinical and hematologic findings are summarized in Table2. They confirmed the association between M0 AML and older age, high WBC counts, CD7 expression, and high incidence of cytogenetic abnormalities.18-21 Median age of the 59 patients was 62 years. No patient was younger than 15 years, confirming the very low incidence of M0 AML in children. The morphology of blast cells was that of small to medium-sized blasts with high nucleocytoplasmic ratio and agranular basophilic cytoplasm and less often that of monocytoid-shaped blast, as previously reported.22 Morphologic and cytochemical data were not sufficient, and immunophenotypical studies were required in all cases for accurate diagnosis of M0 AML.
Pretreatment characteristics of M0 AML patients according to AML1 gene mutations
| . | Overall population . | Presence of AML1 gene mutation . | Absence ofAML1 gene mutation . | P . |
|---|---|---|---|---|
| N | 59 | 16 | 43 | |
| Mean age, y (range) | 62 (15-88) | 65 (29-88) | 56 (15-87) | .1146 |
| Females, % | 34 | 25 | 37 | .48 |
| Mean WBC count, g/L (range) | 52.7 (1.1-309) | 115 (1.4-309) | 30 (1.1-208) | .001 |
| Mean platelet count, giga/L (range) | 91.3 (4-287) | 83 (8-223) | 94 (4-287) | .71 |
| Mean Hb, g/dL (range) | 8.8 (3.4-14.8) | 8.46 (3.9-14.8) | 9 (3.4-14.7) | .27 |
| Mean circulating blasts, % (range) | 67.3 (1-100) | 77 (12-100) | 63.5 (1-100) | .08 |
| Mean marrow blasts, % (range) | 84.4 (36-100) | 91.4 (75-100) | 82 (36-100) | .032 |
| WBC more than 50 g/L, % pts | 35 | 80 | 19 | < .0001 |
| Dysgranulopoiesis, % pts | 22 | 30 | 19 | .437 |
| Dyserythropoiesis, % pts | 16 | 9 | 19 | .65 |
| Dysmegakaryopoiesis, % pts | 5 | 0 | 8 | 1 |
| FLT3 duplication, % pts | 22 | 28 | 23 | 1 |
| IgH orTCR gene rearrangement, % pts | 22 | 58 | 7 | < .0001 |
| Abnormal karyotypes, % | 32.5 | 12.5 | 37.5 | .166 |
| AML1 mutation, % | 27 | — | — | — |
| CD13 expression, % | 85 | 100 | 77 | .0965 |
| CD33 expression, % | 61 | 41 | 70 | .036 |
| CD7 expression, % | 35 | 15 | 40 | .176 |
| CD34 expression, % | 86 | 93 | 83 | .666 |
| DR expression, % | 83 | 100 | 77 | .049 |
| CR achievement in patients treated intensively, % | 62 | 68 | 62.5 | .76 |
| Median CR duration, mo | 12 | 10 | 17.5 | .69 |
| . | Overall population . | Presence of AML1 gene mutation . | Absence ofAML1 gene mutation . | P . |
|---|---|---|---|---|
| N | 59 | 16 | 43 | |
| Mean age, y (range) | 62 (15-88) | 65 (29-88) | 56 (15-87) | .1146 |
| Females, % | 34 | 25 | 37 | .48 |
| Mean WBC count, g/L (range) | 52.7 (1.1-309) | 115 (1.4-309) | 30 (1.1-208) | .001 |
| Mean platelet count, giga/L (range) | 91.3 (4-287) | 83 (8-223) | 94 (4-287) | .71 |
| Mean Hb, g/dL (range) | 8.8 (3.4-14.8) | 8.46 (3.9-14.8) | 9 (3.4-14.7) | .27 |
| Mean circulating blasts, % (range) | 67.3 (1-100) | 77 (12-100) | 63.5 (1-100) | .08 |
| Mean marrow blasts, % (range) | 84.4 (36-100) | 91.4 (75-100) | 82 (36-100) | .032 |
| WBC more than 50 g/L, % pts | 35 | 80 | 19 | < .0001 |
| Dysgranulopoiesis, % pts | 22 | 30 | 19 | .437 |
| Dyserythropoiesis, % pts | 16 | 9 | 19 | .65 |
| Dysmegakaryopoiesis, % pts | 5 | 0 | 8 | 1 |
| FLT3 duplication, % pts | 22 | 28 | 23 | 1 |
| IgH orTCR gene rearrangement, % pts | 22 | 58 | 7 | < .0001 |
| Abnormal karyotypes, % | 32.5 | 12.5 | 37.5 | .166 |
| AML1 mutation, % | 27 | — | — | — |
| CD13 expression, % | 85 | 100 | 77 | .0965 |
| CD33 expression, % | 61 | 41 | 70 | .036 |
| CD7 expression, % | 35 | 15 | 40 | .176 |
| CD34 expression, % | 86 | 93 | 83 | .666 |
| DR expression, % | 83 | 100 | 77 | .049 |
| CR achievement in patients treated intensively, % | 62 | 68 | 62.5 | .76 |
| Median CR duration, mo | 12 | 10 | 17.5 | .69 |
Hb indicates hemoglobin; WBC, white blood cell; pts, patients; —, not applicable.
Abnormal karyotype was found in 24 of 40 available cases and was complex (≥ 3 abnormalities) in 54% of them (Table 1). Aberrations involved preferentially chromosome 5, 7, 3, and 11. This high incidence of complex karyotypes with unbalanced changes such as −5/del(5q), −7, del(7q), and 3q abnormalities is usual in AML, occurring in elderly patients, and could partially explain the poor prognosis observed in our cohort. Abnormalities of chromosome 13 were found in only 2 cases, contrasting with a previous report that linked those abnormalities to M0 AML.20 22
Molecular and immunophenotypic findings are summarized in Table 1 and Figure 1. FLT3 duplication was found in 13 patients (22%) as compared with 16% of M0 AML in the recent series of Thiede et al.23IgH or TCR gene rearrangement was found in 9 (15%) and 5 (8%), respectively, of the patients and were not correlated to lymphoid marker expression. One patient had simultaneous IgH and TCRγ rearrangements.
Antigenic expression profile.
The percentage of patients positive for the expression for each antigen for the whole series is compared with that of patients withAML1 gene mutation and without AML1 gene mutation. Gray bars indicate whole series of patients; black bars, patients with AML1 mutations; and white bars, patients without AML1 mutations.
Antigenic expression profile.
The percentage of patients positive for the expression for each antigen for the whole series is compared with that of patients withAML1 gene mutation and without AML1 gene mutation. Gray bars indicate whole series of patients; black bars, patients with AML1 mutations; and white bars, patients without AML1 mutations.
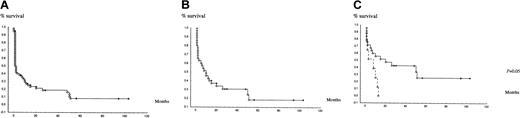
Three patients, aged 35, 47, and 64 years, died before onset of treatment. Nineteen patients received moderate single-agent chemotherapy or supportive care only, because of older age and/or poor clinical condition (18 of them were older than 70 years). Intensive chemotherapy was administered to the 37 remaining patients, of whom 23 (62%) achieved CR. Median CR duration was 12 months. Median survival of intensively treated patients was 10 months and 20 months in patients who achieved CR. Median survival of the whole cohort was 1 month (Figure 2A). Those results confirmed the poor prognosis of M0 AML, because of (besides frequent chromosomal rearrangements) the older age of many patients, who could not receive intensive chemotherapy.2,19 24
Survival of patients.
(A) Kaplan-Meier overall survival of the whole series of patients. (B) Kaplan-Meier survival of patients intensively treated. (C) Kaplan-Meier survival according to FLT3 duplication in patients intensively treated: patient without FLT3 duplication, patients with FLT3 duplication, (P = .05 between the 2 groups).
Survival of patients.
(A) Kaplan-Meier overall survival of the whole series of patients. (B) Kaplan-Meier survival of patients intensively treated. (C) Kaplan-Meier survival according to FLT3 duplication in patients intensively treated: patient without FLT3 duplication, patients with FLT3 duplication, (P = .05 between the 2 groups).
The poor prognosis of FLT3 duplication in other AMLs as a whole is now well documented.25 Here, in patients with M0 AML treated intensively, FLT3 duplication was the only prognostic factor with a median survival of 9 months in patients with FLT3 duplication versus 16 months in patients without FLT3 duplication (P = .05) (Figure 2C).
AML1 mutation in the M0 AML population
AML1 mutation was found in 16 patients (27%) (Table2), an incidence similar to that observed in previous reports.3-6 All mutations involved theRUNT domain, were missense (n = 7) or stop codon mutation (n = 15), and were biallelic (except one case), therefore probably inactivating the AML1 protein. Those characteristics are similar to our preliminary results and show the strong correlation between lack of AML1 function and M0 AML subtype.26
In our M0 AML series, no differences were found between patients with or without AML1 mutations for age, sex, platelet count, hemoglobin value, myelodysplastic features, response to chemotherapy, and survival (Table 2). However, patients with AML1 mutation showed significantly higher leukocyte counts and higher marrow blast percentage, suggesting that AML1 mutations are associated with greater cell proliferation, lower incidence of CD33, and higher incidence of HLA-DR expression and higher frequency ofIgH or TCR gene rearrangement. There was also a trend for lower incidence of chromosomal abnormalities and complex chromosomal abnormalities in our M0 AML cases with AML1mutation. The higher incidence of IgH/TCR gene rearrangement in mutated cases could be related to AML1 loss of function. Indeed,AML1 has been reported to act as a transcriptional repressor by recruitment of transducinlike enhancer of split (TLE)/Groucho proteins.27 One of the target genes of this repression is TCR enhancer.28,29 AML1-mutated blast cells did not show expression of lymphoid markers, indicating absence of lymphoid differentiation, which could be linked to abnormal variable diversity joining (VDJ) recombination in myeloid-committed cells.30 However, we cannot exclude thatAML1 mutation occurred only in very immature cells not committed to lymphoid or myeloid differentiation, in whichTCR or immunoglobulin gene recombination would be a normal event.
Prepublished online as Blood First Edition Paper, October 10, 2002; DOI 10.1182/blood-2002-05-1474.
Supported by the Centre Hospitalier Universitaire of Lille (PHRC 1997), The Ligue Nationale contre le cancer (Comité du Nord et de l'Aisne), and the Fondation de France (Comité Leucémie).
P.F. and C.P. have equally contributed as last authors to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Claude Preudhomme, Unité INSERM 524 and Laboratoire d'Hématologie A, 1 place de Verdun, Hôpital Calmette, 59037 Lille, France; e-mail:cpreudhomme@chru-lille.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal