Disruption of the physiologic balance between cell proliferation and death is a universal feature of all cancers. In general terms, human B-cell lymphomas can be subdivided into 2 main groups, low- and high-growth fraction lymphomas, according to the mechanisms through which this imbalance is achieved. Most types of low-growth fraction lymphomas are initiated by molecular events resulting in the inhibition of apoptosis, such as translocations affecting BCL2, in follicular lymphoma, or BCL10 and API2/MLT1, in mucosa-associated lymphoid tissue (MALT) lymphomas. This results in cell accumulation as a consequence of prolonged cell survival. In contrast, high-growth fraction lymphomas are characterized by an enhanced proliferative activity, as a result of the deregulation of oncogenes with cell cycle regulatory functions, such asBCL6, in large B-cell lymphoma, or c-myc, in Burkitt lymphoma. Low- and high-growth fraction lymphomas are both able to accumulate other alterations in cell cycle regulation, most frequently involving tumor suppressor genes such asp16INK4a, p53, andp27KIP1. As a consequence, these tumors behave as highly aggressive lymphomas. The simultaneous inactivation of several of these regulators confers increased aggressivity and proliferative advantage to tumoral cells. In this review we discuss our current knowledge of the alterations in each of these pathways, with special emphasis on the deregulation of cell cycle progression, in an attempt to integrate the available information within a global model that describes the contribution of these molecular changes to the genesis and progression of B-cell lymphomas.
Introduction
Although studies in experimental models and analyses of virus-induced cell transformation have shown that cell cycle subversion is a key step in tumorigenesis, the available information on human tumors has only recently reached the critical threshold of knowledge that allows a reasonably clear understanding of the mechanisms of cell cycle inactivation and their contribution to the genesis and progression of human cancer. Here, we have chosen B-cell lymphoproliferative lesions on the understanding that the accumulation of data concerning alterations in specific key genes makes it possible to propose a general model that describes the role of these specific alterations in cell cycle regulators in the initiation and progression of these tumors.
Lymphoma/leukemia is a group of different types of cancer of the lymphoid system, in which numerous entities feature distinctive molecular alterations (Table 1). The most frequent types of lymphoma are collectively denominated B-cell lymphomas (BCLs), a term that encompasses different entities with variable clinical behavior and diverse molecular features. Nevertheless, in general terms, it is possible to segregate BCLs into 2 main groups, defined as low- and high-growth fraction lymphomas that roughly overlap with previous definitions of low- and high-histologic grade.
Chromosomal alterations in cell cycle and apoptosis regulatory genes in B-cell lymphomas
| Lymphoma . | Gene . | Function . | Alteration . | Frequency, % . | Reference . |
|---|---|---|---|---|---|
| FL | BCL2 | Apoptosis inhibition | t(14;18) | 70-90 | Yunis et al1, Tsujimoto et al2, Cleary et al3, Weiss et al4, Yunis et al5, Tsujimoto et al6, Bakhshi et al7, Cleary et al8, and Albinger-Hegyi et al9 |
| MALT | BCL10 | Apoptosis inhibition through NF-κB | t(1;14) | Willis et al10, Zhang et al11 | |
| API2/MLT1 | t(11;18) | 50 | Levine et al12, Dierlamm et al13, Baens et al14 15, Inagaki et al16, and Liu et al17 | ||
| MCL | Cyclin D1 | G1/S cell cycle transition | t(11;14) | 90 | Tsujimoto et al18 19, Bosch et al20, and Vaandrager et al21 |
| B-CLL | ? | ? | 13q14 deletion | 50-60 | Kalachikov et al22 |
| LPL | PAX5 | B-cell differentiation | t(9;14) | 50 | Offit et al23, Iida et al24 |
| LBCL | BCL6 | B-cell activation and differentiation | t(3;N) | 30-40 | Bastard et al25, Offit et al26, and Lo Coco et al27 |
| Inflammation | |||||
| Cell cycle control | |||||
| Apoptosis inhibition | |||||
| BL | c-myc | Differentiation | t(8;14) | 80 | Boxer and Dang28, Cory29, |
| Metabolism | t(8;22) | 10 | Dalla-Favera et al30, and Taub et al31 | ||
| Cell cycle control | t(2;8) | 10 | |||
| Apoptosis induction | |||||
| Adhesion |
| Lymphoma . | Gene . | Function . | Alteration . | Frequency, % . | Reference . |
|---|---|---|---|---|---|
| FL | BCL2 | Apoptosis inhibition | t(14;18) | 70-90 | Yunis et al1, Tsujimoto et al2, Cleary et al3, Weiss et al4, Yunis et al5, Tsujimoto et al6, Bakhshi et al7, Cleary et al8, and Albinger-Hegyi et al9 |
| MALT | BCL10 | Apoptosis inhibition through NF-κB | t(1;14) | Willis et al10, Zhang et al11 | |
| API2/MLT1 | t(11;18) | 50 | Levine et al12, Dierlamm et al13, Baens et al14 15, Inagaki et al16, and Liu et al17 | ||
| MCL | Cyclin D1 | G1/S cell cycle transition | t(11;14) | 90 | Tsujimoto et al18 19, Bosch et al20, and Vaandrager et al21 |
| B-CLL | ? | ? | 13q14 deletion | 50-60 | Kalachikov et al22 |
| LPL | PAX5 | B-cell differentiation | t(9;14) | 50 | Offit et al23, Iida et al24 |
| LBCL | BCL6 | B-cell activation and differentiation | t(3;N) | 30-40 | Bastard et al25, Offit et al26, and Lo Coco et al27 |
| Inflammation | |||||
| Cell cycle control | |||||
| Apoptosis inhibition | |||||
| BL | c-myc | Differentiation | t(8;14) | 80 | Boxer and Dang28, Cory29, |
| Metabolism | t(8;22) | 10 | Dalla-Favera et al30, and Taub et al31 | ||
| Cell cycle control | t(2;8) | 10 | |||
| Apoptosis induction | |||||
| Adhesion |
LPL indicates lymphoplasmacytic lymphoma; ?; unknown.
Low-growth fraction BCLs, including follicular lymphoma (FL), marginal zone lymphomas (MZLs), mantle cell lymphoma (MCL), and B-cell chronic lymphocytic leukemia (B-CLL), are distinguished by a relatively low proliferative index, small cell size, formation of large tumoral masses in lymph nodes, bone marrow or extranodal locations, and a paradoxical combination of advanced clinical stages associated with low clinical aggressivity (for a review see Seng and Peterson32 and Capello and Gaidano33). This clinicopathologic presentation seems to be the final consequence of significant advantages to cell accumulation as a result of alterations in apoptosis regulators rather than in cell cycle control genes. The situation is the opposite of that commonly observed in high-growth fraction BCLs, including large B-cell lymphoma (LBCL) and Burkitt lymphoma (BL), in which an increased proliferative index and larger cell size are associated with more frequently localized clinical stages but a higher clinical aggressivity, as a consequence of alterations in cell cycle regulators such as BCL6 or c-Myc that usually occur in combination with the more common defects in apoptosis.
A fraction of both low- and high-growth fraction lymphomas are characterized by the acquisition of additional alterations in cell cycle control, usually involving cyclin-dependent kinase inhibitors (CKIs), such as p16INK4a, p21CIP1, or p27KIP1. These tumors, irrespective of their histologic grade, show an increased clinical aggressivity and more frequent treatment resistance.
Low-growth fraction lymphomas
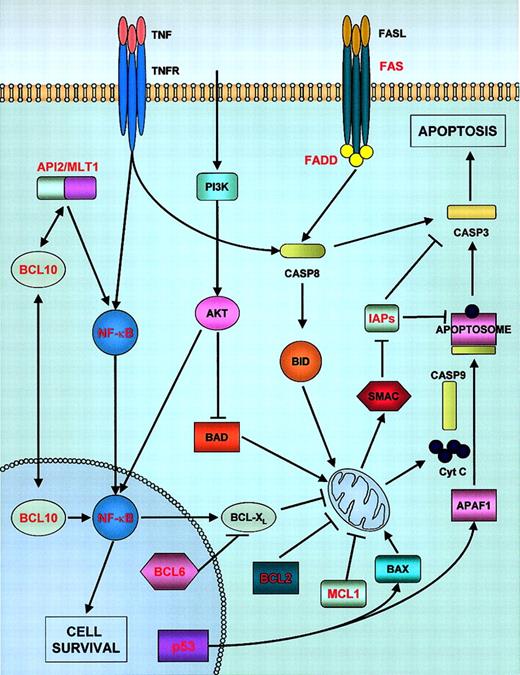
Alterations in genes controlling apoptosis have been recognized in most of these tumors, although the nature of the observed changes seems to be more complex than initially expected. Apoptosis can be initiated in cells by 2 alternative convergent pathways (reviewed in Igney and Krammer34): the “extrinsic” pathway involves death receptors, such as tumor necrosis factor (TNF) receptors or FAS, whose binding to their ligands leads to the activation of caspase-8. The mitochondrial or “intrinsic” pathway is thought to be triggered by the translocation into mitochondria of BCL2 family members, which causes alterations in mitochondrial permeability and the consequent release of cytochrome c, which, in association with apoptotic protease-activating factor-1 (APAF1), activates caspase-9 (Figure 1). Caspases-8 and -9 (initiator caspases) both subsequently activate caspases-3, -6, and -7 (executioner caspases), which, in turn, cleave substrates involved in the regulation of cell death. Alterations in the proteins involved in these pathways could lead to tumor initiation; moreover, the suppression of apoptosis facilitates the accumulation of further oncogenic lesions, eventually leading to uncontrolled proliferation.
Apoptosis pathways and their alterations in lymphomas.
Activation of death receptors by their respective ligands (extrinsic pathway) leads to the activation of caspase-8, which subsequently activates the caspase cascade. Some receptors, such as TNFRα, have the ability to activate either cell survival (through IAP proteins, components of the TNFR complex) or apoptosis. The intrinsic or mitochondrial apoptotic pathway is controlled by BCL2 family members, including proapoptotic BID (activated by caspase-8) or BAX, or antiapoptotic proteins, such as BCL2, MCL1, and BCL-XL. Activation of the mitochondrial pathway involves release of cytochrome c from mitochondria and its association with APAF-1 and procaspase-9 to form the apoptosome. Caspase-9 is activated within this complex. IAP (inhibitor of apoptosis proteins) are, in turn, inhibited by second mitochondria-derived activator of caspase (SMAC). The extrinsic and intrinsic pathways are connected via BID and caspase-3. Some of the receptors also exert prosurvival functions through NF-κB pathway activation. API2/MLT1 fusion proteins and BCL10 also activate NF-κB, inducing cell survival. The phosphatidylinositol 3-kinase (PI3K)/AKT pathway also has a role in cell survival control through NF-κB activation and by inhibiting BAD protein (an inducer of apoptosis). Those proteins known to be involved in lymphomagenesis are marked in red. Casp, Caspase.
Apoptosis pathways and their alterations in lymphomas.
Activation of death receptors by their respective ligands (extrinsic pathway) leads to the activation of caspase-8, which subsequently activates the caspase cascade. Some receptors, such as TNFRα, have the ability to activate either cell survival (through IAP proteins, components of the TNFR complex) or apoptosis. The intrinsic or mitochondrial apoptotic pathway is controlled by BCL2 family members, including proapoptotic BID (activated by caspase-8) or BAX, or antiapoptotic proteins, such as BCL2, MCL1, and BCL-XL. Activation of the mitochondrial pathway involves release of cytochrome c from mitochondria and its association with APAF-1 and procaspase-9 to form the apoptosome. Caspase-9 is activated within this complex. IAP (inhibitor of apoptosis proteins) are, in turn, inhibited by second mitochondria-derived activator of caspase (SMAC). The extrinsic and intrinsic pathways are connected via BID and caspase-3. Some of the receptors also exert prosurvival functions through NF-κB pathway activation. API2/MLT1 fusion proteins and BCL10 also activate NF-κB, inducing cell survival. The phosphatidylinositol 3-kinase (PI3K)/AKT pathway also has a role in cell survival control through NF-κB activation and by inhibiting BAD protein (an inducer of apoptosis). Those proteins known to be involved in lymphomagenesis are marked in red. Casp, Caspase.
BCL2 protein family
The BCL2 gene encodes a 26-kDa protein that inhibits induction of apoptosis through the mitochondrial pathway. BCL2 gives its name to a family of apoptosis regulatory molecules that include antiapoptotic (BCL2, BCL-XL, MCL1…) and proapoptotic (BAX, BAK, BCL-XS, BID, BIK…) proteins; the antiapoptotic or proapoptotic status seems to depend on the relative levels of the members of the 2 subfamilies.
Chromosomal translocations involving the BCL2 gene are the characteristic type of genetic alteration of follicular lymphoma. The translocation t(14;18)(q32;q21), which is present in the large majority of FL cases (70%-90%), places BCL2 under the control of the immunoglobulin heavy chain (IgH) Eμenhancer, leading to high expression of the BCL2 protein.1-7 Chromosomal breakpoints occur in 3′-noncoding regions of BCL2 locus and cluster in 3 regions: the major breakpoint region (MBR) (50%-70% of translocations), the intermediate cluster region (icr), and the minor cluster region (mcr).8,9 Generally, close associations have been found between the presence of translocation and the expression of BCL2 protein, although in some cases BCL2 expression seems to be independent of translocations.35-37 The absence of BCL2 expression in some FL cases suggests that tumoral cells may acquire apoptosis inhibition through mechanisms other than that of BCL2 overexpression; for instance, an important role has been postulated for BCL-XL.38 In fact, BCL2 overexpression alone is apparently not sufficient to induce a malignant phenotype, because clonal BCL2-Ig rearrangement products are detectable in normal B cells.39 Moreover, in transgenic mice mimicking t(14;18), BCL2 overexpression results in prolonged B-cell survival without an increase in cell cycling; as a consequence, these mice are susceptible to developing autoimmune diseases and follicular hyperplasia40-43 that progresses to malignant large-cell lymphoma only after a long latency and through the acquisition of secondary alterations.42 Therefore, other oncogenic events in addition to BCL2 deregulation appear to be necessary in the genesis of FL. As a final step, when alterations in key cell cycle regulators, such as p53, p16INK4a, or c-myc, are added to the initial antiapoptotic lesions, FL undergoes progression to more aggressive variants.44-48
A paradigm of a neoplasm caused by a failure in the mechanisms regulating apoptosis without an increase in cell proliferation is provided by B-cell chronic lymphocytic leukemia. Data obtained by different experimental approaches has demonstrated that the nondividing cells that constitute the hallmark of this entity carry on an increased expression of the antiapoptotic proteins BCL2 (in virtually all cases) and MCL1 (in approximately one half of the cases)49-52 and show down-regulation of the proapoptotic gene BID, as has been observed in expression profiling experiments.50
NF-κB survival pathway
The term NF-κB (nuclear factor κB) refers to dimeric transcription factors belonging to the REL family (for a review see Karin and Lin53). Studies in mice defective in NF-κB activation suggest a role for this family in adaptive immunity, inflammation, and lymphoid organ development.54 Another function assigned to these transcription factors is the inhibition of apoptosis: NF-κB induces the expression of antiapoptotic factors such as the inhibitor of apoptosis proteins (IAP), c-FLIP, and TRAF1-2, as well as members of the BCL2 family such as BCL-XL andA1.53
Mammals express 5 REL proteins, encoded by the genes RELA,RELB, c-REL, NFKB1, andNFKB2, whose activity is regulated by specific inhibitors (IκB). Because of their important function in the promotion of cell survival, alterations in these proteins may play a key role in the development of some lymphomas and leukemias. The human c-RELproto-oncogene (on 2p12-16) is amplified in 23% of LBCLs,55,56 especially in extranodal tumors, and exclusively in the germinal center B-cell–like subgroup as defined by expression profiling.57 c-REL is also amplified in primary mediastinal B-cell lymphoma,58 and gains in the region containing this gene are the most frequent chromosomal imbalance in classical Hodgkin lymphoma (HL).59,60 The 10q24 region, where NFKB2 is located, is occasionally rearranged in cases of B-CLL and multiple myeloma (MM).61,62BCL3encodes an IκB protein and is the target of t(14;19), a rare translocation found in B-CLL and other B-cell neoplasms.63High levels of nuclear NF-κB activity have been found in unstimulated B-CLL cells in comparison with that detected in nonmalignant human B cells; CD40 binding to its ligand further increased NF-κB activity and prolonged B-CLL cell survival in vitro.64
The IAP gene family encodes a group of proteins involved in the suppression of programmed cell death, in addition to other cellular functions. Their ability to suppress apoptosis is mediated, at least in part, by their ability to block caspase activity. Some IAPs (XIAP, c-IAP1, c-IAP2) bind and directly inhibit certain caspases (-3, -7, and -9)65,66 and can act as E3-ligases, catalyzing the ubiquitination of the caspases they bind.67 68 IAPs may also have caspase-independent functions, such as cell cycle regulation.
In mucosa-associated lymphoid tissue (MALT) lymphomas, the most common translocation is t(11;18)(q21;q21), which occurs in 30% to 50% of these lymphomas.12-17 The genes involved in this translocation are API2 (c-IAP2) on 11q21, a member of the IAP family with caspase-inhibitory functions, andMLT1 (on 18q21), encoding the human paracaspase (a protein with certain sequence similarity to caspases whose precise function is not fully understood).69 The translocation results in a chimeric transcript encoding an API2/MLT1 fusion protein, whose action may lead to increased inhibition of apoptosis and, therefore, confer a survival advantage on tumoral cells.70
BCL10, an apoptosis-regulatory molecule containing an amino-terminal caspase-recruitment domain, is overexpressed as a result of t(1;14)(p22;q32), a translocation that is also recurrently detected in a group of MALT lymphomas.10,11 In normal cells, BCL10 weakly promotes apoptosis, induces NF-κB activation, and suppresses transformation in culture.71-74 In normal lymphoid tissue, BCL10 is detected in the cytoplasm; however, the protein partially localizes to the nucleus in a fraction of MALT lymphomas with or without the translocation (although BCL10 levels are higher in the presence of t(1;14)).75,76BCL10 mutations occur at low frequency and often result in truncated proteins that are unable to induce apoptosis but are sometimes able to activate NF-κB.10,11,76,77 Abnormal nuclear expression of BCL10 and the t(11;18) translocation tend to appear together, and their joint occurrence is associated with advanced MALT lymphomas.17,77 Further points of association between t(11;18) and BCL10 are that both BCL10 and API2/MLT1 fusion products (although not API2 or MLT1 on their own) activate NF-κB.10,11,74 Moreover, BCL10 interacts with MLT1 and some of the API2/MLT1 products, synergizing the activation of NF-κB. These observations suggest that BCL10 and API2/MLT1 operate in a common mechanism, involving NF-κB–mediated inhibition of apoptosis, in the promotion of MALT lymphomas.75 76
Death receptors: FAS
FAS (CD95, Apo-1) is a death receptor belonging to the TNF-receptor family. The binding of FAS to its physiologic ligand (FASL) leads to the assembly of a death-inducing signaling complex, which consists of trimerized FAS, Fas-associated via death domain (FADD), and procaspase-8. When recruited to this complex, caspase-8 is activated by autocleavage.
FAS-induced apoptosis mediates the process of negative selection of B cells within the germinal center. Additionally, it has been suggested that FAS acts as a tumor suppressor gene. Approximately 20% of BCLs derived from germinal center or postgerminal center B cells (FL, MALT, LBCL, MM, HL) have somatic mutations in the FASgene,78-82 which are especially frequent in extranodal lymphomas. Most of these mutations are deleterious and mainly affect the death domain, giving rise to mutant proteins that may behave as dominant-negative forms.
In addition, germline mutations of FAS are associated with BCLs in both mice and humans. The mouse strain MRL/lpr-lpr, harboring an inactivating mutation in the FAS gene, develops lymphadenopathy and is prone to autoimmunity and the development of BCL.83 Germline mutations of FAS in humans lead to the autoimmune lymphoproliferative syndrome, which is characterized by symptoms that are similar to those in MRL/lpr-lpr mice and by a high predisposition to the development of BCL.84
FAS mutations have not been detected in BCLs derived from immature or pregerminal center B cells (B-cell acute lymphoblastic leukemia [ALL], MCL, B-CLL).79 However, the FAS apoptotic pathway might be disrupted in these lymphomas through different mechanisms. B-CLL cells show down-regulation ofFAS and are resistant to FAS-induced apoptosis85,86; this may contribute to the extended in vivo survival of B-CLL cells. Similarly, MCL cells express low or null levels of FAS and high levels of CD40, thereby favoring the CD40 cell survival pathway, and exogenous FAS ligand has no effect on MCL cells.87
Other alterations observed in low-growth fraction lymphomas
The hallmark of mantle cell lymphoma is t(11;14)(q13;q32); this translocation juxtaposes the cyclin D1 gene with anIgH enhancer,18,19 resulting in up-regulation of cyclin D1. Virtually 100% of MCLs overexpress cyclin D1, in most cases as a consequence of a t(11;14).20,21 Despite the role of cyclin D1 as a positive regulator of G1-phase progression, cyclin D1 expression in MCL is not related to proliferative activity; moreover, overexpression of cyclin D1 in Eμ-cyclin D1 mice is not sufficient to promote malignant transformation, although it strongly cooperates with other oncogenes such as mycalleles.88 These observations imply that additional alterations in MCL development are needed, although the precise nature of these molecular changes has not been completely defined. Gene expression profiling in MCL has suggested the existence of a broad spectrum of alterations in pathways regulating apoptosis, including the mitochondrial apoptotic pathway (up-regulation of BCL2, down-regulation of cytochrome C1 and caspase-9) and death-receptor signaling pathways (down-regulation ofFADD, DAXX, and RAIDD).89A different study found down-regulation of FAS and resistance to FAS-induced apoptosis in MCL cells (see “Death receptors: FAS”).87 At the same time, several genes stimulating cell cycle progression were found to be up-regulated (CDK4, MDM2, SKP2, myc,E2F5/DP2, some growth factors and their receptors).89 Ataxia telangiectasia mutated gene(ATM), a kinase involved in p53 activation in response to DNA damage, is inactivated by mutation in a significant proportion of MCLs.90 A subset of MCL has been shown to accumulate alterations in p16INK4a and p53, leading to the generation of blastoid variants characterized by higher proliferative activity and more aggressive clinical behavior (see “Alterations in the Rb pathway in lymphomas”).
A subset of MM also features the t(11;14) translocation and displays overexpression of cyclin D1.91 In addition, cyclin D1 expression is frequent in hairy cell leukemia (HCL), although it is often weaker than it is in MCL, and it appears to be unrelated to translocations or amplification of the cyclin D1gene.92
Alterations in the ATM gene (loss of heterozygosity [LOH] and mutations, ∼15% each) have also been described in a group of B-CLL cases and are associated with impaired p53 function and resistance to the induction of apoptosis93-97;p53 itself is mutated in a similar percentage of B-CLL tumors.98 p53 dysfunction because of p53 orATM mutation is an adverse prognostic factor in B-CLL99 and is independent of known predictors of poor outcome (CD38 expression and absence of IgVHmutations100). Interestingly, p53 dysfunction occurs exclusively in the subset of B-CLL with unmutatedIgVH.99
PAX5, a member of the paired-box gene family, encodes a B-cell–specific transcription factor that regulates B-cell genes (CD19, Ig genes) and plays a key role in B-lymphoid differentiation. In addition, PAX5 stimulates B-cell proliferation and is able to transcriptionally repress p53in vitro.101PAX5 is juxtaposed with theIgH locus by the t(9;14)(p13;q32) translocation, which is characteristic of a subset of LPLs23,24 and has rarely been detected in other BCLs such as splenic marginal zone lymphoma (SMZL).102 The inappropriately increased expression of PAX5 in terminally differentiated B cells, in which this gene is normally down-regulated, may contribute to the pathogenesis of LPL.
High-growth fraction lymphomas
In the case of lymphomas with a high-growth fraction, the most common chromosomal translocations seem to affect genes involved in proliferation control such as BCL6, in LBCL, or c-myc, in BL.
Contribution of BCL6 to lymphomagenesis
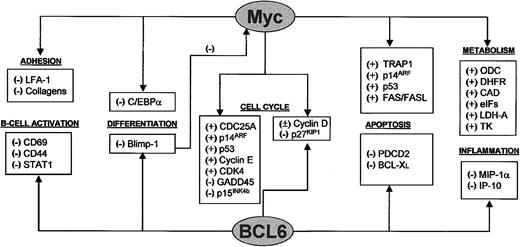
BCL6 acts as a multifunctional regulator involved in cell cycle control and other critical cell functions such as lymphocyte differentiation and immune response (Figure2). It was initially identified because of its involvement in chromosomal translocations affecting 3q27, present in a group of FLs and LBCLs.103 AlthoughBCL6 mRNA can be detected in different cell types,104,105 the protein seems to be expressed at higher levels in lymphocytes. Therefore, the oncogenic properties ofBCL6 have mainly been explored in the lymphoid system, where it is expressed by B and T cells in the germinal center, isolated cells in the lymph node interfollicular area, and cortical thymocytes.106-108 Knockout mice studies have demonstrated that BCL6 is required for germinal center formation and T-cell–dependent antigen responses.109 The selective expression of BCL6 by germinal center B cells has led to its use as a surrogate marker for malignancies derived from the germinal center; thus, BCL6 is expressed in those tumors assumed to arise from follicular center cells, such as FL (almost 100%), BL (100%), a majority of LBCLs (> 80%), and nodular lymphocyte predominance–Hodgkin lymphoma (NLP-HL) (> 80%).106-108In general terms, BCL6 expression is a finding associated with cells with a high proliferation index. Thus, BCL6 is not expressed by low-growth fraction lymphomas, with the exception of FL, in which BCL6 is detected in proliferating cells and down-regulated in the interfollicular compartment, where the growth fraction is also diminished. A similar exception is provided by NLP-HL, in which proliferating tumoral cells are distinguished by BCL6 expression. In general terms, the pattern of expression of BCL6 in normal and tumoral lymphoid tissue is the opposite of that of p27KIP1; thus, it seems that BCL6, probably as a consequence of its ability to down-regulate p27KIP1 expression,110 is a key marker in B cells leading to proliferation (Figure3).
c-Myc and BCL6 are involved in regulation of multiple cell functions and are interconnected.
c-Myc and BCL6 are involved in the regulation of multiple pathways. Some processes, such as cell cycle control, differentiation, and apoptosis, are regulated by both c-Myc and BCL6, although through different targets. c-Myc also regulates the processes of cell adhesion, cell growth, and metabolism. BCL6 also plays a role in B-cell activation and inflammation. Interconnections between the 2 proteins are provided by cell cycle regulation through cyclin D and p27KIP1 and by Blimp-1, a transcriptional repressor of c-myc that is, in turn, repressed by BCL6.
c-Myc and BCL6 are involved in regulation of multiple cell functions and are interconnected.
c-Myc and BCL6 are involved in the regulation of multiple pathways. Some processes, such as cell cycle control, differentiation, and apoptosis, are regulated by both c-Myc and BCL6, although through different targets. c-Myc also regulates the processes of cell adhesion, cell growth, and metabolism. BCL6 also plays a role in B-cell activation and inflammation. Interconnections between the 2 proteins are provided by cell cycle regulation through cyclin D and p27KIP1 and by Blimp-1, a transcriptional repressor of c-myc that is, in turn, repressed by BCL6.
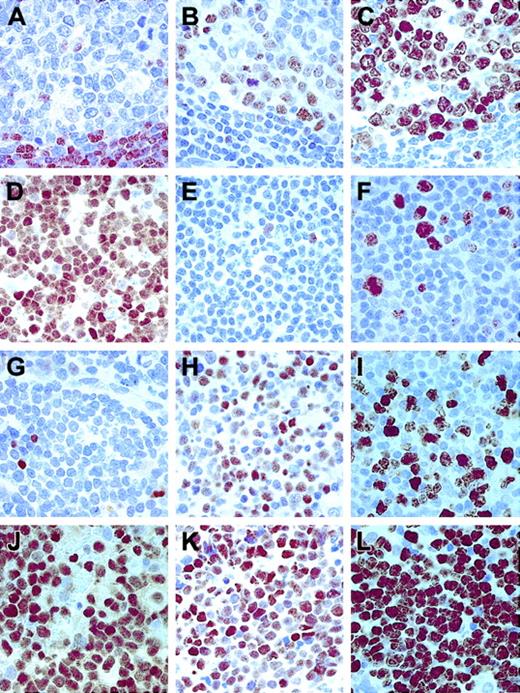
Different patterns of expression of Ki67, p27KIP1, and BCL6 in lymphomas.
(A) p27KIP1 staining in nontumoral lymphoid tissue (a reactive tonsil). (B) BCL6 and (C) Ki67 in the same sample. (D) A B-CLL case with a high level of p27KIP1 expression. (E) Absence of BCL6 in the same sample and Ki67 (F), in this case, indicating the low-growth fraction of this tumor. (G) A DLBCL case showing absence of p27KIP1 in tumoral cells, with high levels of BCL6 (H) and Ki67 (I) expression. (J) An aggressive DLBCL case showing p27KIP1 overexpression associated with high levels of BCL6 (K) and Ki67 (L). Original magnifications, × 400.
Different patterns of expression of Ki67, p27KIP1, and BCL6 in lymphomas.
(A) p27KIP1 staining in nontumoral lymphoid tissue (a reactive tonsil). (B) BCL6 and (C) Ki67 in the same sample. (D) A B-CLL case with a high level of p27KIP1 expression. (E) Absence of BCL6 in the same sample and Ki67 (F), in this case, indicating the low-growth fraction of this tumor. (G) A DLBCL case showing absence of p27KIP1 in tumoral cells, with high levels of BCL6 (H) and Ki67 (I) expression. (J) An aggressive DLBCL case showing p27KIP1 overexpression associated with high levels of BCL6 (K) and Ki67 (L). Original magnifications, × 400.
BCL6 rearrangements (chromosomal translocations involving the BCL6 5′-noncoding region in 3q27) occur at a frequency of 20% to 40% in large B-cell lymphoma and 6% to 14% in FL.25-27 In contrast, rearrangements of BCL6(as well as those affecting BCL2 or c-myc) are very rare in primary mediastinal B-cell lymphoma, which suggests that this tumor is a separate type of LBCL arising from different pathogenetic mechanisms.111 These translocations contribute to BCL6 deregulation; however, we know that the level ofBCL6 expression seems to depend on the partner involved in the translocation, because tumors in which BCL6 is translocated to an Ig locus express higher levels ofBCL6 mRNA than those with non-Ig/BCL6translocation, and also that overexpression of BCL6 is often independent of chromosomal translocations.112
In this respect, it has recently been claimed that somatic mutations involving the 5′-noncoding region of the BCL6 gene can contribute to the deregulation of BCL6expression.113 In LBCL, somatic mutations in the major mutational cluster have been described as playing a role in the regulation of BCL6 expression. Mutations within the 423 to 443 hotspot in the first intron of BCL6 are associated with increased protein levels, suggesting that this region may include a regulatory element whose mutation may modify the binding of hypothetical BCL6-regulating transcription factors.
The BCL6 gene encodes a sequence-specific transcriptional repressor.114-117 Expression profiling of B-cell lines in which BCL6 was positively or negatively modulated revealed that BCL6 represses a group of genes involved in B-cell activation and terminal differentiation (such as Blimp-1) and cell cycle control (cyclin D2, p27KIP1), and it has been hypothesized that this indirectly induces c-mycexpression.110 Therefore, the lack of germinal center formation, as observed in BCL6 knockout mice,109 could be at least partially a consequence of the absence of the proliferative signal that is dependent on BCL6.
Blimp-1 (B-lymphocyte–induced maturation protein 1) is a transcriptional repressor with a key role in the differentiation of B cells into plasma cells.118 One of the targets of Blimp-1 repression is c-myc.119 In vitro studies have shown that inhibition of BCL6 function induces Blimp-1 and down-regulates c-myc expression, causing cell cycle arrest in G1.110 Conversely, it might be hypothesized that overexpression of BCL6 because of translocations or somatic mutations could indirectly activate c-myc through repression of Blimp-1 by BCL6, although this has not been formally demonstrated in human tumors.
The altered expression of BCL6 could also repress genes involved in apoptosis, thereby favoring neoplastic cell growth. One candidate target is PDCD2,120 previously found to be associated with programmed cell death in thymocytes. However, the role of BCL6 in apoptosis is controversial because the report cited earlier, and others, suggest that BCL6 may be an inhibitor of apoptosis,121,122 whereas other studies suggest that it induces cell death. For example, it has been proposed that AFX (a forkhead transcription factor) activates apoptosis by induction ofBCL6, which in turn repressesBCL-XL.123 This dual role of BCL6 in the regulation of apoptosis could be dependent on factors such as cell type or level of expression.
BCL6 has also been identified as a potent inhibitor of senescence in murine embryo fibroblasts and primary B cells, making cells unresponsive to negative proliferation control by the p19ARF-p53 pathway through a mechanism mediated by up-regulation of cyclin D1.124 This finding may be potentially relevant, but we need to bear in mind that cyclin D1 expression in B cells is mainly restricted to MCL.
The prognostic significance of 3q27 translocation and BCL6 expression has been analyzed in different series. BCL6 rearrangement has been described as an independent marker of favorable clinical outcome in LBCL.26 The partner in BCL6translocation is predictive of survival, because non-Ig/BCL6translocations are an indicator of poor prognosis in LBCL compared withIg/BCL6.125 Most studies coincide in showing that a high level of BCL6 expression, as determined by real-time polymerase chain reaction (PCR) or immunohistochemistry, is a favorable prognostic marker. These observations may be linked by the finding that non-Ig/BCL6 translocations lead to much lowerBCL6 mRNA levels than do Ig/BCL6translocations.112 Mutations in the 423 to 443 cluster, which are associated with high BCL6 levels, are also associated with a better clinical outcome.113
The increased expression of BCL6 was one of the features that made it possible to define a subset of germinal center B-cell–like LBCLs, characterized by lower aggressivity, increased frequency of c-REL amplification, and translocations involving the BCL2 gene.57,126,127 Thus, Rosenwald et al57 have recently identified 3 different LBCL subgroups (germinal center B-cell–like, activated B-cell–like, and type 3 LBCL) through the use of gene expression profiling. The different clinical behavior of the germinal center B-cell–like group and the fact that some molecular alterations appear to occur exclusively in this concrete subset may support the idea that this LBCL subgroup actually represents a distinct entity with specific mechanisms of transformation.
The c-Myc transcription factor
c-Myc, a helix-loop-helix leucine zipper transcription factor, has been proposed as being involved in multiple cellular functions such as cell cycle regulation, apoptosis, cell growth, metabolism, cell adhesion, and differentiation. However, much of the information available is still controversial, and, to some extent, the precise involvement of c-Myc in these functions might be cell type- and context-dependent (reviewed in Eisenman,128 Boxer and Dang,28 Dang,129 and Amati et al130). The ability of c-Myc to bind to specific DNA sequences (E-boxes) and activate transcription requires heterodimerization with Max. Max also heterodimerizes with other Myc family proteins, such as Mad or Mxi1; Mad-Max dimers mediate transcriptional repression in E-boxes and might antagonize the activity of Myc-Max, at least in some promoters.131,132 Myc-Max complexes can also exert repressing activity on initiator (Inr) elements, in conjunction with Miz-1.133
Myc promotes proliferation through the induction of genes involved in cell cycle control such as CDK4, Id2,CDC25A, and cyclins D1, D2, A,and E. Additionally, Myc suppresses growth-arresting genes such as p15INK4b,p21CIP1, GADD45, orGAS1.128-130,133-135 p27KIP1 is negatively regulated by Myc at different levels, including transcriptional repression,136 the increase of its degradation, and its sequestration by CDK4/cyclin D2 (Figure 2). c-Myc also induces cellular immortalization, a function that may be mediated by induction of the catalytic subunit of telomerase.137
Overexpression of c-Myc in primary cells results in cell cycle arrest or apoptosis, in contrast to what happens in immortalized cells. High levels of expression of c-Myc result in induction of p19ARF, which sequesters MDM2, leading to activation of p53-mediated arrest or apoptosis.138 Other targets that contribute to Myc-mediated apoptosis may be FAS,FASL, and BAX.28 The ability of Myc to induce apoptosis explains why, in transgenic mice, tumors overexpressing Myc frequently select for alterations that neutralize its apoptotic program through inactivation of the p19ARF/p53 pathway139 or BCL2 overexpression.140
The c-myc gene is subject to complex transcriptional and posttranscriptional regulation. Four promoters have been identified, although one of them (P2) usually contributes to 80% to 90% of c-myc RNA in normal cells.28 Expression of c-myc is positively regulated by multiple factors, such as BCL6,110 epidermal growth factor (EGF), platelet-derived growth factor (PDGF), and interleukin 7 (IL-7),141 and negatively controlled by Blimp-1,119 transforming growth factor β (TGFβ), or p21CIP1. The c-Myc protein has a short half-life and is degraded via the ubiquitin/26S proteasome system. Two phosphorylation sites (Thr58 and Ser62) seem particularly relevant to the control of c-Myc degradation. Ras controls the phosphorylation of these residues through the extracellular signal-related kinase (ERK) and AKT pathways.142
The c-myc gene is translocated to the Ig loci in virtually all Burkitt lymphomas.28-31 The most frequent translocation, c-myc/IgH (t(8;14)(q24;q32)), is detected in about 80% of BLs. Each of the variant translocations, t(2;8)(p12:q24) and t(8;22)(q24;q11), involving IgK and IgL, respectively, occur at a frequency of approximately 10%. These or other translocations involving c-myc have also been detected in LBCL (6%)143 and other hematologic malignancies, such as T-cell ALL and MM. These translocations are believed to place c-myc under the control of an intronic and/or 3′Ig enhancer, leading to a deregulated expression from the translocated allele (the normal allele remains silent). This expression could be explained by a combination of events, including a switch from P2 to P1 promoters and the release of the block on transcription elongation. NF-κB is an important positive regulator of the expression of the translocated allele of c-myc in BL cells.144
Most translocated c-myc alleles in BL are subjected to somatic hypermutation.145 Mutations tend to cluster in either the exon 1-intron 1 region, where they may cancel the block on transcription elongation, or the transactivation domain (Thr58 is a hot spot), possibly interfering with phosphorylation and thereby contributing to the stabilization of the protein. Similarly distributed somatic mutations were detected in LBCL (32%).146
Deregulation of c-myc by alternative mechanisms, such as amplification, occurs in diverse types of tumors and has been implicated in the pathogenesis of these diseases; in LBCL c-myc amplification has been found in up to 16% of cases.56 Increased expression or activation of upstream c-myc regulators (NF-κB, BCL6, Blimp-1) may be another cause of alterations in c-Myc expression and function.110,119 144
Alterations in c-myc seem to constitute one of the primary events in malignant transformation in BL, but they could also be a secondary event in tumor progression. In fact, translocations of c-myc have been observed in transformed FL.47,48 According to experimental models, overexpression of Myc alone may not be sufficient to induce cellular transformation.Eμ-myc mice develop clonal B-cell malignancies (not comparable with BL, however) that usually accumulate alterations in the p19ARF/p53 pathway or BCL2, thereby allowing the cancellation of Myc-induced apoptosis.139 This mimics the situation observed in samples of progressed FL and aggressive B-cell lymphoma with double c-myc and BCL2translocation, in which simultaneous BCL2 and c-Myc expression gives a critical advantage to the tumoral cells.47 48
There are few conclusive studies regarding the relationship between c-Myc and clinical or biologic variables in lymphomas, and there is some variability in the findings among these reports concerning protein levels, cell distribution, and subcellular localization. Nevertheless, these analyses seem to coincide in showing that c-Myc overexpression is common in aggressive lymphomas and appears to be related to advanced clinical stage, high International Prognostic Index, or shorter overall survival.147-149
Highly aggressive lymphomas
Both low- and high-growth fraction lymphomas can evolve to highly aggressive tumors; the process takes place through the acquisition of alterations in the major tumor suppressor pathways, most frequently affecting CKI. In fact, inactivation of CKI is a marker of highly aggressive lymphomas, irrespective of their histologic grade. CKI inactivation takes place in a concerted manner to confer a proliferative advantage to tumoral cells and, at the same time, determines increased treatment resistance. The G1/S transition of the cell cycle is regulated by 3 main pathways that might be altered in virtually 100% of aggressive tumors and that are represented by the proteins p16INK4a-cyclin D-CDK4-Rb, p14ARF-MDM2-p53-p21CIP1, and p27KIP1-cyclin E-CDK2 (reviewed in Sherr150).
Alterations in the Rb pathway in lymphomas
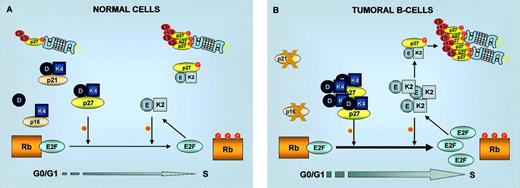
In normal cells, the transition through the restriction point is negatively regulated by the retinoblastoma (Rb) protein (Figure4A). Rb binds to transcription factors (E2Fs) whose activity is necessary for the expression of S-phase genes, and it assembles a transcriptionally repressing complex that keeps E2F-responsive promoters inactive. Under conditions of mitogenic stimulation, accumulation of D-type cyclins allows the formation of active CDK4/6-cyclin D complexes, which phosphorylate and inactivate Rb, thus promoting E2F-mediated transcription and subsequent progression through the cell cycle. An additional level of regulation is provided by the INK4 (inhibitors of cdk4) family of CKI (p16INK4a, p15INK4b, p18INK4c, p19INK4d). These 4 homologous proteins inhibit the kinase activity of CDK4/6 by preventing its interaction with the activating cyclins D. The integrity of this regulatory cascade, termed the “Rb pathway” (INK4-CDK4,6/cyclin D-Rb-E2F), appears to be compromised in most human cancers. This inactivation can be achieved by an increased activity of CDK-cyclin complexes and/or by inactivation of the tumor suppressor genes INK4a or Rb.
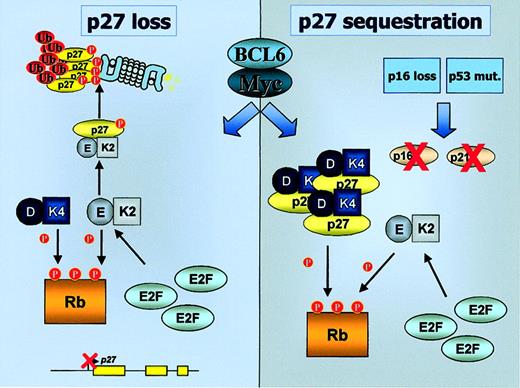
G1/S cell cycle transition in normal and tumoral cells.
G1/S transition in normal cells is regulated by Rb phosphorylation by cyclin D/CDK4 and cyclin E/CDK2 complexes. In tumoral B cells, inactivation of p53 leading to absence of p21CIP1 or loss of p16INK4a both lead to increased Rb phosphorylation and consequent release of E2F transcription factors. Inactivation of p27KIP1 by increase in its degradation or by sequestration in cyclin D/CDK4 complexes also stimulates Rb phosphorylation by cyclin E/CDK2. K2, CDK2; K4, CDK4; E, cyclin E; D, cyclin D; Ub, ubiquitin.
G1/S cell cycle transition in normal and tumoral cells.
G1/S transition in normal cells is regulated by Rb phosphorylation by cyclin D/CDK4 and cyclin E/CDK2 complexes. In tumoral B cells, inactivation of p53 leading to absence of p21CIP1 or loss of p16INK4a both lead to increased Rb phosphorylation and consequent release of E2F transcription factors. Inactivation of p27KIP1 by increase in its degradation or by sequestration in cyclin D/CDK4 complexes also stimulates Rb phosphorylation by cyclin E/CDK2. K2, CDK2; K4, CDK4; E, cyclin E; D, cyclin D; Ub, ubiquitin.
The distribution of Rb protein in reactive lymphoid tissue follows a pattern that is parallel to that of Ki67 (a proliferation marker), with preferential expression in most germinal center B cells and cortical thymocytes.151 This normal regulation of Rb in relation to proliferative activity is frequently preserved in different types of lymphoma, in which genetic alterations of the Rb gene appear to be relatively rare.152-154 Levels of Rb are low in low-growth fraction BCLs, whereas they are higher in most high-growth fraction tumors.151,155 NLP-HL also displays this direct correlation between Rb and proliferation,156 and in blastoid MCL the expression and phosphorylation level of Rb are higher than in classical types.152 A deviation from this pattern, consisting of partial or total loss of Rb expression in highly proliferative tumors, was detected in a group of high-growth fraction non-Hodgkin lymphomas (NHLs)151,157 and classical HL156; this may be, at least in part, a consequence of genetic alterations, which have been found at low frequencies in different lymphoid malignancies. Loss of Rb was found to be an adverse prognostic factor in LBCL, in which there was a correspondence between high Rb protein levels and extended overall survival time,158 and in HL, in which Rb loss was related to failure to achieve complete remission159; monoallelic deletions of Rb predict poor outcome in MM.160Notably, the Rb-like p130 protein is mutated in most cases of endemic BL.161
Of the 3 D-type cyclins (D1, D2, D3), cyclin D3 is the most ubiquitously expressed in lymphoid tissue. It is detected mainly in proliferating lymphocytes, either in germinal centers or interfollicular areas.162,163 Several observations point to a possible oncogenic role of cyclin D3 in lymphomagenesis. Immunohistochemical analysis of aggressive BCL revealed that a significant number of these tumors overexpressed cyclin D3, a finding that was more frequent in BL.162 Recent work points to an association between high levels of cyclin D3 expression and poor clinical outcome in LBCL.164 It is interesting to note that, in addition to the direct activation of CDK4/6, cyclin D3 appears to play another role in the promotion of cell proliferation, ie, the indirect activation of CDK2–cyclin E achieved by the sequestration of p27KIP1 (see “p27KIP1 alterations in lymphomas”). Recent data suggest that cyclin D3 overexpression may have a genetic basis in some cases, because t(6;14)(p21;q32) translocations involving cyclin D3 and IgH genes have been detected in several hematologic malignancies (myeloma, plasma cell leukemia, LBCL, SMZL) and result in the deregulated expression of cyclin D3.165 In contrast, the level of cyclin D2 expression is low in normal and tumoral lymphoid tissue (except in B-CLL and LPL in which it is often overexpressed166), and cyclin D1 is virtually absent167 (with the exception of MCL, HCL, and a subset of MM; see “Other alterations observed in low-growth fraction lymphomas”).
Molecular studies concerning the status of CDK4/6 in lymphomas are scarce; amplification of the CDK4 gene has been identified in 11% of LBCLs,56 and translocations affecting CDK6 (t(7;14) or t(2;7)) have been described in splenic lymphoma with villous lymphocytes.168
Despite the fact that the INK4 proteins share structural and functional homology, only p16INK4a has gained credence as a bona fide tumor suppressor gene in vivo. Thep16INK4a and p15INK4bgenes are located in 9p21, which is, after p53, the most frequently altered locus in human cancer and which also encodesp14ARF; in contrast,p18INK4c and p19INK4d are very rarely altered in human tumors, including lymphomas.169 170
In reactive lymphoid tissue, p16INK4a protein is detected in all compartments, with a slight increase in the most proliferative ones (germinal centers, interfollicular areas) and a decrease in the mantle zone, which mainly consists of resting lymphocytes.171 Low-growth fraction BCLs display normal p16INK4a expression by small- and medium-sized tumoral cells. There is a dramatic difference in high-growth fraction BCLs, more than 50% of which exhibit partial or total loss of expression of p16INK4a protein by tumoral cells. This finding is often associated with alterations in the p16INK4a gene (Table 2). Thus, althoughp16INK4a alterations are rare in low-growth fraction BCLs (< 5%), genetic or epigenetic inactivation ofp16INK4a occurs in 30% to 50% of high-growth fraction BCLs,173,178,179,183,184 predominantly by 9p21 deletion/LOH or promoter hypermethylation (the most frequent mechanism in LBCL); point mutations of p16INK4a are infrequent. It has been shown that p15INK4b is either frequently codeleted with p16INK4a or hypermethylated at variable frequency and thatp15INK4b hypermethylation can occur independently of that ofp16INK4a.172,181,185 Inactivation of p16INK4a may also play a role in the development of HL, because many HL cases show selective loss of p16INK4aexpression by Reed-Sternberg (RS) cells; hypermethylation of thep16INK4a promoter appears to be the underlying molecular mechanism in most of these tumors.182
Frequency of p16INK4a alterations in B-cell lymphomas
| Lymphoma . | Deletion, % . | Mutation, % . | Hypermethylation, % . | Reference . |
|---|---|---|---|---|
| All NHL | — | — | 32 (21 indolent, 50 aggressive) | Baur et al172 |
| 12 (4 indolent, 23 aggressive) | 3 | — | Pinyol et al173 | |
| 19 (0 indolent, 30 aggressive) | 4 | 26 (0 indolent, 35 aggressive) | Villuendas et al171 | |
| FL | 0 nontransformed, 73 transformed | — | — | Elenitoba-Johnson et al44 |
| MCL | 0 typical, 50 aggressive variants | 0 | — | Pinyol et al174 |
| 41 | — | — | Dreyling et al175 | |
| B-CLL | 0 | — | — | Delmer et al176 |
| 10 | — | — | Haidar et al177 | |
| LBCL | 16 | 3 | 37 | Sanchez-Beato et al178 |
| 15 | — | Koduru et al179 | ||
| 19 | 0 | 8 | Moller et al180 | |
| BL | — | — | 42 | Klangby et al181 |
| HL | — | — | 61 | Garcia et al182 |
| Lymphoma . | Deletion, % . | Mutation, % . | Hypermethylation, % . | Reference . |
|---|---|---|---|---|
| All NHL | — | — | 32 (21 indolent, 50 aggressive) | Baur et al172 |
| 12 (4 indolent, 23 aggressive) | 3 | — | Pinyol et al173 | |
| 19 (0 indolent, 30 aggressive) | 4 | 26 (0 indolent, 35 aggressive) | Villuendas et al171 | |
| FL | 0 nontransformed, 73 transformed | — | — | Elenitoba-Johnson et al44 |
| MCL | 0 typical, 50 aggressive variants | 0 | — | Pinyol et al174 |
| 41 | — | — | Dreyling et al175 | |
| B-CLL | 0 | — | — | Delmer et al176 |
| 10 | — | — | Haidar et al177 | |
| LBCL | 16 | 3 | 37 | Sanchez-Beato et al178 |
| 15 | — | Koduru et al179 | ||
| 19 | 0 | 8 | Moller et al180 | |
| BL | — | — | 42 | Klangby et al181 |
| HL | — | — | 61 | Garcia et al182 |
— indicates not analyzed.
There is ample evidence supporting a role forp16INK4a inactivation in the transformation of low-growth fraction lymphomas into their aggressive variants. The histologic progression of FL to aggressive large cell lymphoma is frequently accompanied by 9p21 deletions (absent in the low-grade tumor) that often result in reduction or loss of p16INK4aexpression.44 In MCL, p16INK4adeletions are associated with aggressive variants,174 a finding that interestingly suggests the possibility that cyclin D1 overexpression and p16INK4a inactivation might not be completely redundant alterations. In composite tumors (lymphomas consisting of a low-grade—FL or MALT—and a high-grade component—LBCL), the low-growth fraction component (small cells) expresses p16INK4a, but the high-growth fraction component (large cells) usually exhibits loss of p16INK4a expression associated with promoter hypermethylation or LOH.171
The relatively high frequency of p16INK4a/Rbalterations—mainly restricted to aggressive lymphomas—and their association with the transformation from low- to high-grade tumors indicates that inactivation of the Rb pathway (predominantly byp16INK4a silencing and more rarely byRb alteration or CDK4/6–cyclin Dhyperactivation) is one of the key events in the progression of lymphomas (Figure 4B).
Alterations in the p53 pathway in lymphomas
p53 is a transcription factor that induces the expression of genes involved in cell cycle arrest or apoptosis in response to a variety of toxic or oncogenic stimuli, the final cellular response depending on the biologic context. Negative regulation of cell cycle progression by p53 is partially mediated by induction ofp21CIP1, a CKI of the CIP/KIP family that neutralizes the kinase activity of CDK2–cyclin E complexes. One of the target genes of p53 is MDM2, which inhibits p53 and targets it for degradation by virtue of its E3-ubiquitin ligase activity. In response to DNA damage or hyperproliferative stimulation, the p53-MDM2 interaction is disrupted by phosphorylation or antagonized by p14ARF induction, respectively, resulting in the activation of p53.150 This “p53 pathway” is also frequently altered in most human cancers, either by inactivation of the tumor suppressor genes p14ARF or p53or by hyperactivation of the MDM2 oncogene. In fact,p53 is the most frequently mutated gene in human cancer (> 50% of all cases).
Mutations of p53 are detected in 17% to 25% of cases of LBCL and BL.178,180,186-190 This frequency is relatively low compared with other types of cancer but is still significantly higher than that observed in most low-growth fraction BCLs, which supports the hypothesis that p53 inactivation is one of the events leading to high-grade progression (Table3). Mutations of p53 in lymphomas tend to be clustered in conserved regions that are part of the sequence encoding the DNA binding domain. In aggressive BCL, the inability of mutant p53 to transactivate its target genes is reflected by the p53+p21− immunophenotype in contrast with the p53−p21+ phenotype observed in most cases with wild-type p53.192
Frequency of p53 mutations in B-cell lymphomas
| Lymphoma . | Frequency of mutation, % . | References . |
|---|---|---|
| All NHL | 11 | Koduru et al188 |
| 25 | Wilson et al191 | |
| 8 indolent, 33 aggressive | Pinyol et al186 | |
| 26 | Villuendas et al192 | |
| FL | 0 nontransformed, 80 transformed | Lo Coco et al45 |
| 12 nontransformed, 26 transformed | Sander et al46 | |
| MALT | 19 nontransformed, 33 transformed | Du et al193 |
| MCL | 6 typical, 29 aggressive variants | Greiner et al194 |
| 0 typical, 38 aggressive variants | Hernandez et al195 | |
| B-CLL | 15 | el Rouby et al98 |
| HCL | 28 | Konig et al196 |
| LBCL | 17 | Sanchez-Beato et al178 |
| 18 | Ichikawa et al187 | |
| 19 | Koduru et al188 | |
| 22 | Moller et al180 | |
| BL | 19 | Preudhomme et al197 |
| HL | 0 | Montesinos-Rongen et al198, Kupper et al199 |
| 26 | Chen et al200 |
| Lymphoma . | Frequency of mutation, % . | References . |
|---|---|---|
| All NHL | 11 | Koduru et al188 |
| 25 | Wilson et al191 | |
| 8 indolent, 33 aggressive | Pinyol et al186 | |
| 26 | Villuendas et al192 | |
| FL | 0 nontransformed, 80 transformed | Lo Coco et al45 |
| 12 nontransformed, 26 transformed | Sander et al46 | |
| MALT | 19 nontransformed, 33 transformed | Du et al193 |
| MCL | 6 typical, 29 aggressive variants | Greiner et al194 |
| 0 typical, 38 aggressive variants | Hernandez et al195 | |
| B-CLL | 15 | el Rouby et al98 |
| HCL | 28 | Konig et al196 |
| LBCL | 17 | Sanchez-Beato et al178 |
| 18 | Ichikawa et al187 | |
| 19 | Koduru et al188 | |
| 22 | Moller et al180 | |
| BL | 19 | Preudhomme et al197 |
| HL | 0 | Montesinos-Rongen et al198, Kupper et al199 |
| 26 | Chen et al200 |
Missense mutations of p53 usually result in the stabilization of the protein; therefore, increased levels of p53 are typically detected in association with p53mutations.201 Although this is essentially true in low-growth fraction lymphomas such as B-CLL, FL, and MCL, the relationship between p53 mutation and expression in LBCL and BL is not a direct one,188,190 suggesting the existence of alternative mechanisms for the stabilization of p53. Overexpression of the p53 protein is relatively common in high-growth fraction NHL (de novo as well as those that progress from low grade)202 but not in low-growth fraction tumors. However, only a fraction of the NHL cases overexpressing p53 have an underlying p53mutation.188,190 Moreover, some NHLs with a wild-typep53 gene have the p53+MDM2−p21− phenotype, characteristically found as a consequence of p53 mutation and indicative of the malfunction of p53 as a transcription factor.192 Still more intriguingly, p53 typically accumulates in RS cells in HL, but, according to most reports, these cells very rarely harbor p53mutations198-200,203,204 and regularly express MDM2 and p21CIP1,205 raising the question of whether or not this p53 protein is active. However, the absence of p53 expression cannot be regarded as an unequivocal sign of a wild-type gene, because rare nonsense or frameshift mutations produce truncated, rapidly degraded p53 proteins that fail to accumulate.201
There is firm evidence that inactivation of p53 plays a key role in the progression of low-grade lymphomas. Mutations of p53are strongly associated with large cell transformation of MALT lymphomas193 and FL45,46 and with blastoid variants of MCL.195 They are frequently detected in progressed tumors but appear at lower frequencies in their low-grade counterparts. In any case, and independently of the presence of morphologic signs of progression, p53 mutations and stabilization of the p53 protein are associated with more aggressive clinical behavior and treatment resistance in all histologic types of low-growth fraction lymphomas, such as B-CLL,98 FL, MZL, and MCL.194,195 The contribution of p53inactivation to the aggressive phenotype has also been observed at the clinical level in LBCL, in which p53 overexpression or mutation has been reported as being associated with lower survival probability.187,191 206
The relatively low incidence of p53 mutations in lymphomas has prompted the examination of possible alternative mechanisms of inactivation of the p53 pathway. Mutation, deletion, and hypermethylation of p21CIP1 have only very rarely been described in BCL, although recent data show an increased incidence of p21CIP1 promoter hypermethylation associated with poor prognosis in ALL.207 The p53-activating kinase ATM is mutated in B-CLL and MCL (see “Other alterations observed in low-growth fraction lymphomas”).
Overexpression of MDM2 may represent a mechanism of inactivation of p53 that is not dependent on p53 mutations. This might be particularly true in HL, a tumor that consistently displays accumulation of the MDM2 protein in RS cells205,208 and at the same time exhibits an especially low frequency of p53mutations. Overexpression of MDM2 also occurs at significant frequencies in leukemias209 and NHL,180,210,211 suggesting a contribution to tumorigenesis that is further supported by the association between high levels of MDM2 and shorter overall survival in LBCL,158 low-growth fraction NHL,210 and treatment resistance in ALL.212 It is not completely clear whether MDM2 overexpression has a genetic basis: several studies coincide in finding no amplifications of the MDM2 gene in lymphomas.213 However, amplifications of the 12q13 region, which resulted in an effective increase in the MDM2 gene copy number, were detected in 14% of LBCLs,56 and gains in 12q with increased MDM2 copy number have been described in HL.199
Alterations of the p14ARF gene that result in the loss of its expression are relatively rare in LBCL and BL.186,214 Unlike p16INK4a,p14ARF promoter hypermethylation has rarely been found; point mutations are very infrequent (0%-4%); and monoallelic or biallelic deletions at 9p21 potentially affectingp16INK4a and p14ARF have been observed in 10% to 20% of cases. Consistent with these findings, only a small group of lymphomas lack p14ARF expression in tumoral cells, whereas the rest show variable levels of the p14ARF protein.214
p27KIP1 alterations in lymphomas
At the G1/S transition, mitogen-dependent phosphorylation of Rb by CDK4/6–cyclin D is shifted to mitogen-independent action of CDK2–cyclin E complexes, which, in addition to triggering DNA replication, are able to phosphorylate Rb further, thus contributing to the irreversibility of the transition to S phase. Negative regulation of CDK2–cyclin E activity is mediated by the CIP/KIP family of CKI (p21CIP1, p27KIP1, p57KIP2). These proteins also bind CDK4–cyclin D, but these complexes may preserve kinase activity; CIP/KIP proteins may even be required for the assembly of active CDK4 holoenzymes. Therefore, it has been postulated that the sequestration of CIP/KIP may be another function of CDK4–cyclin D, which would facilitate the activation of CDK2–cyclin E.150
Multiple roles have been attributed to p27KIP1, not only as a mediator of G1 arrest in response to TGF-β, cell-cell contact, cyclic adenosine monophosphate (cAMP), or rapamycin, but also as a factor involved in differentiation, apoptosis, and chemotherapeutic response (for a review see Lloyd et al215). p27KIP1 is phosphorylated by CDK2–cyclin E, and this modification signals the proteolysis of p27KIP1 via ubiquitination-proteasomal degradation.
Biallelic genetic alterations of p27KIP1 in lymphomas and other tumors are very rare.216-219 However, there is evidence that p27KIP1 behaves as a haploinsufficient gene for tumor suppression,220 in such a way that a reduction in its levels (resulting from monoallelic silencing,221 increased degradation, or other mechanisms) may be sufficient to contribute to tumorigenesis.
In reactive lymphoid tissue, the expression of p27KIP1 is inversely related to proliferation219 222-224 (Figure 3). Strong p27KIP1 expression is detected in quiescent cells (mantle lymphocytes, medullary thymocytes), whereas p27KIP1is down-regulated in proliferating cells (germinal center B cells, large interfollicular lymphocytes, and cortical thymocytes).
As a general rule, the expression of p27KIP1 in lymphomas follows the same inverse relationship with proliferation.219,222-224 In low-growth fraction BCL, p27KIP1 is strongly detected in small cells, medium-sized cells stain more weakly, and occasional large cells are negative. Possible exceptions are typical MCL219 and HCL,225 in which p27KIP1 appears to be undetectable by immunohistochemistry despite the low proliferative index of these tumors. In contrast, most high-growth fraction BCLs do not show p27KIP1 expression by tumoral cells (Figure 3). In HL, p27KIP1 protein is only rarely detected in the highly proliferative RS cells, whereas small surrounding lymphocytes preserve p27KIP1 expression.222 Similar observations have been made in different tumoral types, implying a role of p27KIP1 down-regulation in tumor progression. Loss of p27KIP1 is indicative of poor outcome (shorter survival) in NHL, considered as a whole,224 and also in the discrete group of MCL.226 Mechanisms underlying this down-regulation (Figure 5) may include transcriptional repression (p27KIP1 is a target of c-Myc–mediated repression136) and increased degradation, as suggested by the observation that increased expression of SKP2 (ubiquitin ligase for p27KIP1) is positively correlated with cell proliferation and grade of malignancy in human lymphomas and inversely correlated with p27KIP1expression.227 MCL and a group of LBCLs are exceptions to this pattern, implying that there are alternative pathways regulating p27KIP1.
Mechanisms of p27KIP1 inactivation in aggressive B-cell lymphomas.
BCL6 and c-Myc could both contribute to p27KIP1inactivation through diverse mechanisms, leading to the increased activity of cyclin E–CDK2 complexes. Transcriptional repression ofp27KIP1, or induction of its proteasomal degradation, causes a decrease in p27KIP1 levels. Alternatively, in a subset of aggressive lymphomas featuring inactivation of p16INK4a and/or p21CIP1, p27KIP1 is sequestered in an inactive form by cyclin D–CDK4 complexes. K2, CDK2; K4, CDK4; E, cyclin E; D, cyclin D; Ub, ubiquitin.
Mechanisms of p27KIP1 inactivation in aggressive B-cell lymphomas.
BCL6 and c-Myc could both contribute to p27KIP1inactivation through diverse mechanisms, leading to the increased activity of cyclin E–CDK2 complexes. Transcriptional repression ofp27KIP1, or induction of its proteasomal degradation, causes a decrease in p27KIP1 levels. Alternatively, in a subset of aggressive lymphomas featuring inactivation of p16INK4a and/or p21CIP1, p27KIP1 is sequestered in an inactive form by cyclin D–CDK4 complexes. K2, CDK2; K4, CDK4; E, cyclin E; D, cyclin D; Ub, ubiquitin.
An unexpected finding was the observation that a group of LBCL and BL had an unusually high level of p27KIP1 expression in tumoral cells219,222-224 (Figure 3). A more detailed analysis of a large series of LBCLs228 led to the following conclusions: (1) a broad gradient of p27KIP1expression exists in LBCL, ranging from absence of protein to overexpression; (2) there is no positive or negative correlation between p27KIP1 and proliferation in LBCL; and (3) a high level of p27KIP1 expression is an adverse prognostic factor for LBCL, strongly associated with shorter overall survival and disease-free survival. Anomalous high expression of p27KIP1associated with a more aggressive course has also been described in B-CLL; high p27KIP1 expression levels are associated with lower spontaneous cell death rate of B-CLL cells in culture, suggesting that p27KIP1 may contribute to apoptosis impairment.229 These findings suggest that overexpressed p27KIP1 might be inactive in cell cycle inhibition and that mechanisms of p27KIP1 inactivation other than down-regulation should exist, which allowed tumoral cells to overcome the antiproliferative activity of this CKI.
The previous findings and the observation that high levels of p27KIP1 in LBCL and BL were significantly related to cyclin D3 overexpression162,223 (but not to that of cyclins D1, D2, or E), suggested that p27KIP1 might be sequestered in CDK4–cyclin D3 complexes. In this context, p27KIP1 might be protected from CDK2-mediated degradation and at the same time be inactive as a CKI, because CDK4–cyclin D–KIP complexes appear to preserve kinase activity. Consistent with this hypothesis, p27KIP1 was immunoprecipitated in cyclin D3–containing complexes in BL cell lines with high levels of both proteins (Raji, BL40); part of p27KIP1 was also bound to cyclin D3 in tumor samples with intermediate p27KIP1levels.162,223 In lymphomas overexpressing p27KIP1 and cyclin D3, both proteins were colocalized in the nucleus of most tumoral cells, in sharp contrast with the observation in reactive lymphoid tissue and all other lymphomas that expression patterns of cyclin D3 and p27KIP1 were mutually exclusive.162
These data allow us to conclude that several mechanisms operate in high-growth fraction BCL to inactivate p27KIP1. In some tumors, transcriptional down-regulation or increased degradation may predominate, causing a decrease in p27KIP1 levels. However, in a subset of aggressive lymphomas, p27KIP1 is redistributed from CDK2–cyclin E complexes, in which p27KIP1 is active as a CKI and is unstable (susceptible to CDK2-signaled degradation) to CDK4/6–cyclin D complexes, in which p27KIP1 is stabilized but inactive (Figure 5).
Accumulated alterations in tumor suppressor pathways
The results discussed above indicate that the inactivation of individual CKI is biologically and clinically significant and plays a major role in the progression of lymphomas. Do these individual events operate in a concerted way in the pathogenesis of aggressive lymphomas? The combined analysis of the major tumor suppressor pathways p16INK4a-Rb, p14ARF-p53, and p27KIP1-CDK2 has revealed that simultaneous inactivation of several cell cycle regulators does occur in a subset of lymphomas, bestowing an advantage on the tumor.
Highly aggressive lymphomas frequently show alteration of at least one of these pathways with a significantly higher frequency than in low-grade tumors.178,186,230 Moreover, the simultaneous inactivation of the p16INK4a-Rb and p14ARF-p53 pathways occurs in a significant proportion of aggressive BCLs, mainly through 2 alternative mechanisms: (1) the combination ofp16INK4a hypermethylation and p53mutation predominates in LBCL; (2) deletions at 9p21 occur more frequently in BL, and their presence and p53 mutation are mutually exclusive. This is consistent with the prediction that 9p21 deletions normally affect both p16INK4a andp14ARF, and, thus, a single event accounts for the impairment of both pathways (in this context, the additional advantage provided by a p53 mutation would be markedly reduced). The difference between LBCL and BL suggests that mechanisms of inactivation of tumor-suppressor pathways are not “chosen” at random but depend on underlying genetic alterations. Simultaneous inactivation of both pathways predicts poor outcome in aggressive BCL230 and in MCL.186
Overexpression of p27KIP1 occurs, in most LBCL cases, in tumors with inactivation of p16INK4a and/orp53.178 This relationship is not as obvious in BL, implying a greater complexity of the mechanisms regulating p27KIP1 expression. Moreover, in LBCL, p27KIP1accumulation is more frequent in tumors with thep16INK4a and p53 double alteration than in those with a single inactivating event. This implies that p27KIP1 sequestration is a secondary event, derived from the absence of p16INK4a and/or p21CIP1 (Figure5). Under these conditions, p16INK4a or p21CIP1do not compete with p27KIP1 to bind CDK4–cyclin D, there is no displacement of p27KIP1 to CDK2–cyclin E complexes and p27KIP1 remains inactive, sequestered by cyclin D–dependent complexes. Therefore, although abnormal accumulation of p27KIP1 behaves as a marker of the inactivation ofp16INK4a/p53, it is not a mere “reflection” of the malfunction of these pathways. Instead, there is evidence that this mechanism of inactivation of p27KIP1 contributes by itself to the pathogenesis of the lymphoma. Thus, the accumulation of alterations in p53,p16INK4a, and p27KIP1 confers higher aggressivity on tumors, which manifests itself by increased proliferative activity and shorter overall survival. The group of tumors with the triple alteration has the highest proliferation index and a significantly worse outcome. From this we may conclude that alterations in different CKIs cooperate to give a proliferative advantage to lymphomas.
In the light of the previous data, it may be worth reconsidering the role of p14ARF. In a normal cell, p14ARFantagonizes MDM2 and activates p53-mediated responses as a protective mechanism against hyperproliferative stimuli.150,187 To this end, p14ARF expression is induced by oncogene products such as c-Myc, E1A, Ras, and v-Abl. Thep14ARF promoter is responsive to the E2F-1 transcription factor, a property that links Rb to the p14ARF-p53 pathway and allows p53 to sense and respond to oncogenic stimulation transduced through the Rb pathway. As a negative-feedback mechanism, wild-type p53 represses the transcription of the p14ARF gene. A group of aggressive BCLs (20%) abnormally overexpresses the p14ARF protein (this is not confined to the nucleolus, as in normal cells).214This group of cases was characterized by higher aggressivity: p14ARF overexpression was significantly related to a higher growth fraction and shorter overall survival. Overall, these tumors had alterations in at least one of the major tumor suppressor pathways (p16INK4a, p53, or p27KIP1), and p14ARF overexpression was more frequently detected in lymphomas with simultaneous inactivation of several of these tumor suppressor genes. This could be explained as a “normal” response of the p14ARF gene to cell cycle malfunction in a tumoral cell: inactivation of p16INK4a or p27KIP1 results in increased phosphorylation of Rb by CDK4/6 or CDK2, respectively, and active E2F-1 is generated, thereby inducing p14ARF expression. Activated oncogenes (Ras, Myc) also up-regulate p14ARF. Finally,p53 mutations disrupt the p53-p14ARFnegative-feedback loop. As a consequence, p14ARF expression reflects the degree of malfunction of the cell cycle, which, as shown before, is related to tumoral aggressivity (Figure6).
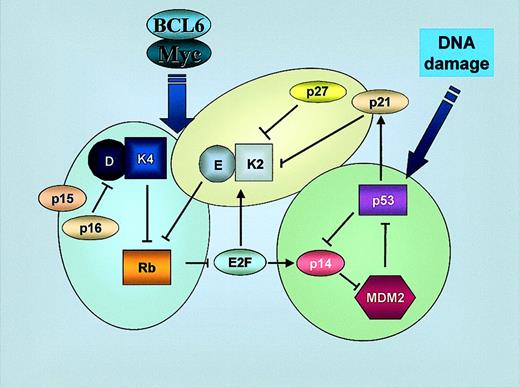
Interconnections between the 3 main pathways that regulate G1/S transition.
The 3 main pathways that regulate the G1/S checkpoint (p16INK4a-cyclin D- CDK4-Rb, p14ARF-MDM2-p53-p21CIP1, and p27KIP1-cyclin E-CDK2) are closely interconnected: cyclin D/CDK4 phosphorylates Rb, releasing E2F which itself regulates p14ARF and, consequently, p53 activity. Activation of p53 induces p21CIP1 that inhibits cyclin E/CDK2, closing the circle. Alterations in one or more of them alter cell cycle control and give an increasing proliferative advantage to tumoral B cells. K2, CDK2; K4, CDK4; E, cyclin E; D, cyclin D.
Interconnections between the 3 main pathways that regulate G1/S transition.
The 3 main pathways that regulate the G1/S checkpoint (p16INK4a-cyclin D- CDK4-Rb, p14ARF-MDM2-p53-p21CIP1, and p27KIP1-cyclin E-CDK2) are closely interconnected: cyclin D/CDK4 phosphorylates Rb, releasing E2F which itself regulates p14ARF and, consequently, p53 activity. Activation of p53 induces p21CIP1 that inhibits cyclin E/CDK2, closing the circle. Alterations in one or more of them alter cell cycle control and give an increasing proliferative advantage to tumoral B cells. K2, CDK2; K4, CDK4; E, cyclin E; D, cyclin D.
Conclusion
The mechanisms and the extent to which the physiologic balance between cell division and cell death are disrupted in lymphoma cells determine the characteristics and evolution of the tumor. In low-growth fraction lymphomas, cell accumulation can be achieved through the inhibition of apoptosis without a very severe deregulation of cell proliferation. In contrast, in high-growth fraction BCL, proliferation is stimulated through the coordinated action of cell cycle regulators such as BCL6 or c-Myc. A fraction of both low- and high-growth fraction lymphomas acquire additional changes that allow tumoral cells to bypass the multiple mechanisms by which cell cycle progression is inhibited in normal cells, including the tumor suppressor pathways represented by p16INK4a-cyclin D-CDK4-Rb, p14ARF-MDM2-p53-p21CIP1, and p27KIP1-cyclin E-CDK2. These lymphomas have a very high clinical aggressivity that is related to the degree of malfunction of these pathways. Tumoral cells often undergo accumulated disruption of more than one of these cascades, taking advantage of their simultaneous and concerted inactivation (Figure7).
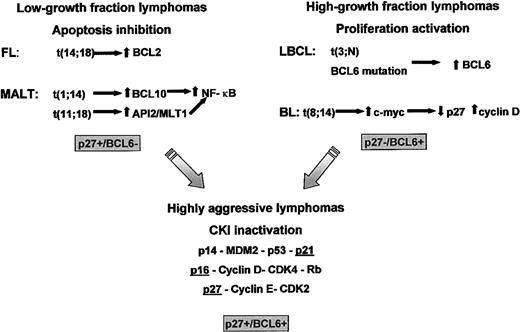
Lymphoma genesis and progression.
In most low-growth fraction lymphomas, cell accumulation is achieved through the inhibition of apoptosis. In contrast, in high-growth fraction BCL, proliferation is stimulated through the deregulated action of cell cycle regulators such as BCL6 or c-Myc. A fraction of both low- and high-growth fraction lymphomas acquire additional alterations affecting the tumor suppressor pathways represented by p16INK4a-cyclin D-CDK4-Rb, p14ARF-MDM2-p53-p21CIP1, and p27KIP1-cyclin E-CDK2. As a consequence, tumoral cells can bypass the multiple mechanisms by which cell cycle progression is negatively regulated in normal cells.
Lymphoma genesis and progression.
In most low-growth fraction lymphomas, cell accumulation is achieved through the inhibition of apoptosis. In contrast, in high-growth fraction BCL, proliferation is stimulated through the deregulated action of cell cycle regulators such as BCL6 or c-Myc. A fraction of both low- and high-growth fraction lymphomas acquire additional alterations affecting the tumor suppressor pathways represented by p16INK4a-cyclin D-CDK4-Rb, p14ARF-MDM2-p53-p21CIP1, and p27KIP1-cyclin E-CDK2. As a consequence, tumoral cells can bypass the multiple mechanisms by which cell cycle progression is negatively regulated in normal cells.
However, despite the important progress that has been made toward the elucidation of the mechanisms of lymphomagenesis, many points still need to be clarified, such as the interrelationships between BCL6, c-Myc, and p27KIP1 and the plethora of molecules and pathways that may be affected by their alterations. Gene expression profiling experiments are providing us with unprecedented information regarding new molecules and pathways involved in tumorigenesis. Whole genome analysis will surely contribute to our better understanding of the mechanisms governing the initiation and progression of lymphomas and allow the establishment of correlations between patterns of gene expression and precise biologic and clinical features.
We apologize to those many authors whose work could not be appropriately cited in this review because of space limitations. We thank Marcos Malumbres, Ana I. Sáez, Raquel Villuendas, Juan F. Garcı́a, Marı́a J. Artiga, and Francisca I. Camacho for critical reading and advice and Phil Mason for his collaboration in the preparation of the manuscript.
Prepublished online as Blood First Edition Paper, September 12, 2002; DOI 10.1182/blood-2002-07-2009.
Supported by grants from the Fondo de Investigaciones Sanitarias (FIS 98/993), Ministerio de Sanidad y Consumo, Comisión Interministerial de Ciencia y Tecnologı́a (1FD97-0431), Comunidad Autónoma de Madrid (08.1/0028/2000 1), and the Ministerio de Ciencia y Tecnologı́a (SAF2001-0060), Spain. A.S.-A. is supported by a predoctoral fellowship from the Consejerı́a de Educacion de la Comunidad de Madrid, Spain, and the European Social Fund.
References
Author notes
Miguel A. Piris, Molecular Pathology Program, Centro Nacional de Investigaciones Oncológicas (CNIO), Melchor Fernández Almagro 3, E-28029 Madrid, Spain; e-mail:mapiris@cnio.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal