Abstract
Accumulating evidence in various species has suggested that the origin of definitive hematopoiesis is associated with a special subset of endothelial cells (ECs) that maintain the potential to give rise to hematopoietic cells (HPCs). In this study, we demonstrated that a combination of 5′-flanking region and 3′ portion of the first intron of the Flk-1 gene (Flk-1 p/e) that has been implicated in endothelium-specific gene expression distinguishes prospectively the EC that has lost hemogenic activity. We assessed the activity of this Flk-1 p/e by embryonic stem (ES) cell differentiation culture and transgenic mice by using theGFP gene conjugated to this unit. The expression ofGFP differed from that of the endogenous Flk-1gene in that it is active in undifferentiated ES cells and inactive in Flk-1+ lateral mesoderm. Flk-1 p/e becomes active after generation of vascular endothelial (VE)–cadherin+ ECs. Emergence of GFP− ECs preceded that of GFP+ ECs, and, finally, most ECs expressed GFP both in vitro and in vivo. Cell sorting experiments demonstrated that only GFP− ECs could give rise to HPCs and preferentially expressed Runx1 and c-Myb genes that are required for the definitive hematopoiesis. Integration of both GFP+ and GFP− ECs was observed in the dorsal aorta, but cell clusters appeared associated only to GFP−ECs. These results indicate that activation of Flk-1 p/e is associated with a process that excludes HPC potential from the EC differentiation pathway and will be useful for investigating molecular mechanisms underlying the divergence of endothelial and hematopoietic lineages.
Introduction
During early embryogenesis, hematopoietic cells (HPCs) are generated in close association with the development of the vascular system. In the blood islands of the yolk sac where the earliest hematopoietic cells appear, both hematopoietic and endothelial cell (EC) lineages arise almost simultaneously from extraembryonic mesoderm, thereby forming structures in which primitive erythrocytes are surrounded by a layer of angioblasts. These histologic observations have led to the hypothesis that the 2 lineages arise from a common precursor, the hemangioblasts.1 This concept is supported by the shared expressions of a number of different genes by both lineages.2-5 Following the process known as primitive hematopoiesis, the major hematopoietic site shifts to the fetal liver at midgestation and finally to the bone marrow. Before colonizing fetal liver, the definitive type hematopoietic progenitors were thought to be generated in a restricted region within the embryo.6-8About the cellular origin of definitive hematopoiesis, several possibilities have been proposed.9 An intraembryonic origin for definitive hematopoiesis is supported by histologic observations that clusters of hematopoietic cells are attached to the luminal wall of the dorsal aorta, as if budding from the endothelial cells.10 As the presence of such intra-aortic clusters has been observed over many vertebrate species and correlates well with development of the definitive hematopoietic cells,11-14 it was proposed that at least a certain portion of definitive hematopoiesis derives from “the hemogenic endothelium.”15 The concept of hemogenic endothelium is also supported by functional studies investigating the potential of ECs to give rise to HPCs. In avian systems, clonogenic analyses have demonstrated that multipotent hematopoietic progenitors are generated only from the aortic region.16 Jaffredo et al17 showed that hematopoietic cells are derived from endothelial cells that had been labeled by low-density lipoproteins injected into the circulation of chick embryos, indicating that endothelial cells of the dorsal aorta can function as hematopoietic progenitors. Moreover, we have demonstrated that vascular endothelial (VE)–cadherin+ cells that were purified from murine embryos can give rise to hematopoietic cells.18
It is of importance to know biologic significance and mechanisms of the emergence of HPCs from a subset of ECs during embryogenesis. We have established a culture system in which embryonic stem (ES) cells differentiate into HPCs and ECs through the proximal lateral mesoderm.19-21 Flk-1+ VE cadherin− cells that represent lateral mesodermal cells were induced first from ES cells. The divergence of primitive type HPCs and Flk-1+ VE-cadherin+ ECs was shown to occur during the mesodermal stage.22 The definitive type HPCs eventually diverged from the Flk-1+ VE-cadherin+ECs.22 This in vitro culture system enables us to dissect the differentiation process of HPCs and ECs from mesoderm. For further investigation of molecular mechanisms underlying the divergence of EC and HPC lineages, it is useful to drive the genes of interest in a specific lineage at specific stages.
Recently, cis-acting regulatory elements of the murine fetal liver kinase-1 (Flk-1) were studied by Kappel et al23 who showed that a combination of a 5′-flanking region and 3′ portion of the first intron (Flk-1 promoter/enhancer) was sufficient to direct EC-specific gene expression in vivo, although activity of thecis element in Flk-1+ mesodermal cell was not known. We analyzed the activity of this promoter/enhancer in ES cell differentiation in vitro and unexpectedly found that these Flk-1 regulatory elements are active only after commitment to ECs but not in lateral mesodermal stage. A more interesting finding is that only ECs negative for green fluorescence protein (GFP) driven by the Flk-1 promoter/enhancer (p/e) could give rise to HPCs. In this study, we will show both in vitro and in vivo that Flk-1 p/e is a useful marker for monitoring the EC maturation, which is inversely correlated with the hemogenic potential.
Materials and methods
Monoclonal antibodies (MoAbs), cell staining, and sorting
The MoAbs AVAS12 (anti–Flk-1)24 and VECD1 (anti–VE-cadherin)25 were purified from hybridoma culture supernatants by protein G-Sepharose columns (Pharmacia, Uppsala, Sweden) and labeled with allophycocyanin (APC) by standard methods. The phycoerythrin (PE)–conjugated MoAbs, anti-CD45, Ter119 (erythroid lineage marker), and anti-CD31 (platelet endothelial cell adhesion molecule-1 [PECAM-1]) were purchased from PharMingen (San Diego, CA).
Cells were blocked with normal mouse serum and labeled with combinations of the above MoAbs. Stained cells were resuspended in Hank balanced salt solution (GIBCO BRL) containing 1% bovine serum albumin (Sigma, St Louis, MO) and 5 μg/mL propidium iodide (PI; Sigma) to exclude dead cells. Cells were analyzed and sorted by fluorescence-activated cell sorter (FACS) Vantage (Becton Dickinson Immunocytometry Systems, San Jose, CA). Data were analyzed using the software CellQuest (Becton Dickinson Immunocytometry Systems).
Plasmids
The pGLacZ-Flk-1p/e contains the β-galactosidase (LacZ) gene sequence under the control of the −640 bp/+299 bp promoter fragment ofFlk-1 and the 2.3-kb XhoI/BamHI fragment of the first intron as an enhancer.23 pFlkp/eGFP was generated by substitution of the LacZ sequence betweenHindIII and BamHI restriction sites of pGLacZ-Flk-1 p/e with GFP sequence (HindIII andAflII fragment) of pEGFP.N3 (Clontech, Palo Alto, CA) (Figure 1A). The pFlkp/eGFP was linearized by PvuI digestion before transfection into ES cells. The pPGKpuro.bpA, which carries a puromycin-resistant gene, was cotransfected or, alternatively, subcloned into the SalI site of the pFlk-1p/eGFP in a direction reverse to GFP before linearization. For transgenic mice, the splice donor and acceptor sequences of pSVβ (Clontech, Palo Alto, CA) were ligated to the GFP cDNA in pFlkp/eGFP.
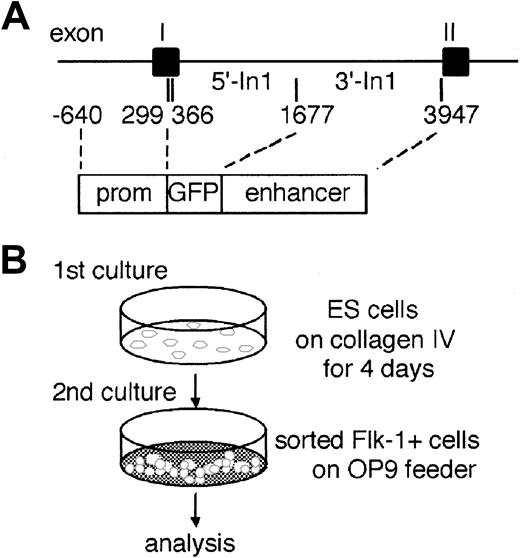
Outlines of experimental procedure.
(A) Partial structure of the murine Flk-1 locus and a reporter gene construct. The GFP gene is flanked by aFlk-1 promoter fragment (prom) spanning bp −640 to bp +299 and an enhancer sequence between bp +1677 and bp +3947 in the first intron. (B) For induction of differentiation in vitro, undifferentiated ES cells were transferred to a type IV collagen–coated plate and incubated for 4 days in the absence of LIF. Cultured cells were harvested and analyzed for expression of GFP and Flk-1 by flow cytometry. The Flk-1+ cells were sorted for secondary culture on OP9 stromal cells. These cells were recovered after 1 to 5 days and analyzed for expression of GFP, VE-cadherin, CD45, and TER119 or for hemangiogenic ability.
Outlines of experimental procedure.
(A) Partial structure of the murine Flk-1 locus and a reporter gene construct. The GFP gene is flanked by aFlk-1 promoter fragment (prom) spanning bp −640 to bp +299 and an enhancer sequence between bp +1677 and bp +3947 in the first intron. (B) For induction of differentiation in vitro, undifferentiated ES cells were transferred to a type IV collagen–coated plate and incubated for 4 days in the absence of LIF. Cultured cells were harvested and analyzed for expression of GFP and Flk-1 by flow cytometry. The Flk-1+ cells were sorted for secondary culture on OP9 stromal cells. These cells were recovered after 1 to 5 days and analyzed for expression of GFP, VE-cadherin, CD45, and TER119 or for hemangiogenic ability.
Cell lines
EB5 (a kind gift from Dr Hitoshi Niwa, RIKEN, Kobe, Japan) is a subline derived from E14tg2a ES cells.26 This line was generated by targeted integration of an Oct3/4-IRES-BSD-pA vector into the Oct-3/4 allele and carries the blasticidin S resistant selection marker gene driven by the Oct-3/4 promoter, which is active under the undifferentiated status.27Undifferentiated ES cells were maintained on gelatin-coated dishes in Glasgow minimum essential medium (G-MEM; GIBCO-BRL) supplemented with 1% fetal calf serum (FCS), 10% knockout serum replacement (KSR; GIBCO-BRL), 0.1 mM nonessential amino acids, 1 mM sodium pyruvate (GIBCO-BRL), 0.1 mM 2-mercaptoethanol (2ME), 1000 U/mL leukemia inhibitory factor (LIF) (GIBCO-BRL), and 20 μg/mL blasticidin S to eliminate differentiated cells. OP9 stromal cell line was maintained in alpha minimum essential medium (MEM) (GIBCO BRL) supplemented with 20% FCS (HyClone Laboratories, Logan, UT).28
The ES cells were electroporated with linearized plasmids and then selected for resistance to puromycin (2 μg/mL). In this study, 5 independent transfectants were analyzed. No substantial differences in differentiation ability among the transfected clones and the parental cell line were observed.
In vitro differentiation of ES cells
Induction of ES cell differentiation was carried out as described previously.19 21 Briefly, 3 × 104undifferentiated ES cells were transferred to each well of a type IV collagen-coated 6-well plate (BIOCOAT; Becton Dickinson Labware, Bedford, MA) and incubated for 4 days in alpha MEM (GIBCO BRL) supplemented with 10% FCS and 50 μM 2ME (induction medium) in the absence of LIF (Figure 1B). Cultured cells were harvested with cell dissociation buffer (GIBCO BRL) and analyzed for expression of GFP and Flk-1 by flow cytometry. The Flk-1+ cells were sorted from the harvested cells for a second round of induction. Sorted Flk-1+ cells (3-10 × 104) were transferred to each well of a 6-well plate (Becton Dickinson) that was preseeded with OP9 stromal cells. These cells were recovered after 1 to 5 days and analyzed for expression of GFP, VE-cadherin, CD45, and Ter119 by flow cytometry. The VE-cadherin+CD45−Ter119− population in the harvested cells was sorted into GFP+ and GFP− fractions and cultured for further induction of hematopoietic or endothelial cells.
For the measurement of frequency of hematopoietic precursors, sorted cells were transferred into a 6-well plate that was preseeded with OP9 stromal cells and incubated in the induction medium supplemented with a mixture of recombinant growth factors containing 200 U/mL murine interleukin-3 (IL-3), 2 U/mL human erythropoietin (Epo), 100 ng/mL murine granulocyte colony-stimulating factor (G-CSF), and 100 ng/mL murine stem cell factor (SCF). Recombinant Epo and G-CSF were purchased from R&D Systems. Recombinant IL-3 and SCF were prepared as previously described.29 After 24 hours, medium was replaced with a fresh semisolid medium consisting of the induction medium, a mixture of growth factors, and 1.2% methylcellulose (Muromachi Kagaku, Tokyo, Japan). Cells were cultured for 6 days, and hematopoietic colonies were scored under a microscope.
For induction of endothelial cell growth, sorted cells were put into a 6-well plate that was preseeded with OP9 cells and incubated in the induction medium. After 10 days, the cultures were fixed in situ with 4% paraformaldehyde and stained with either rat anti–Flk-1 or rat anti–PECAM-1 MoAbs. Flk-1+/PECAM-1+endothelial cell colonies were detected by alkaline phosphatase–conjugated antirat immunoglobulin G (IgG) antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) and nitroblue tetrazolium (NBT)/BCIP (5-bromo-4-chloro-3-indolylphosphate) substrate solution (Boehringer Mannheim, Mannheim, Germany).
Single cell deposition assay for hemangiogenic potential
Single cell deposition of sorted cells into separate wells of 96-well plates (Becton Dickinson) was carried out by the Clon-Cyt system of FACS Vantage (Becton Dickinson). Sorted single cells were cocultured with OP9 stromal cells with 100 ng/mL SCF, 200 U/mL IL-3, 2 U/mL Epo, and 100 ng/mL G-CSF for 7 days. The presence of hematopoietic colonies was judged morphologically and by May-Giemsa staining. The endothelial colonies were immunostained with anti–PECAM-1 MoAb.
Staining of DiI-labeled acetylated low-density lipoprotein (DiI-Ac-LDL)
Staining of DiI-Ac-LDL was performed as previously described.20 Cultured cells were incubated in alpha MEM supplemented with 10 mg/mL DiI-Ac-LDL (Biomedical Technologies, Stoughton, MA) in chamber slides for 4 hours. Cells were then washed with alpha MEM and observed by fluorescence microscopy (Axiovert 135M; Zeiss, Jena, Germany).
Transgenic mice
Linearized plasmids were purified by NACS PREPAC (GIBCO BRL), adjusted to 5 ng/mL, and injected into mouse oocytes as described.30 Transgenic integration was confirmed by polymerase chain reaction (PCR) of genomic DNA obtained from mouse ears. The primers used were Flk-1 p/e, 5′-AGTCTGTGCCTGAGAACTGG-3′, and GFP, 5′-GTAGTTGTACTCCAGCTTGTGC-3′. Three independent transgenic lines were established and analyzed for the expression of GFP.
Embryos were harvested at 10.5 to 12.5 days after coitus (dpc) as described21 and subjected to immunofluorescence, FACS, or cell sorting analyses. For immunofluorescence, embryos were fixed in 4% paraformaldehyde, embedded in optimum cutting temperature (OCT) compound, and cryosectioned. Sections (9-12 μm) were stained with rabbit anti-GFP and rat anti–PECAM-1 for primary antibodies and Alexa 488–conjugated antirabbit IgG (H+L) and Alexa 564-conjugated antirat IgG (H+L) (Molecular Probe) for secondary antibodies.
Reverse transcribed-polymerase chain reaction (RT-PCR)
Total RNA was prepared from sorted cells or cultured cells using ISOGEN (Nippon Gene, Toyama, Japan). RNA was reverse-transcribed with Superscript II reverse transcriptase (GIBCO BRL) and oligo (dT)12-18 primer (GIBCO BRL) according to the manufacturer's instructions. PCR assays were performed in the reaction mixture containing 1× ExTaq Buffer (Takara Shuzo, Osaka, Japan), 200 μM dNTPs (Pharmacia), 25 U/mL ExTaq DNA polymerase (Takara Shuzo), several dilutions of cDNA, and 2 μmol/L of specific primers. Sequences of primers and conditions for PCR were described elsewhere: GATA2,31 Runx1,32 SCL,33 and c-Myb.34 PCR products were electrophoresed through 1% agarose gel and analyzed by staining with ethidium bromide.
In situ hybridization
Digoxigenin-11–uridine triphosphate (UTP)–labeled single-stranded RNA probes were prepared by using the DIG RNA labeling kit (Roche Diagnostics, Mannheim, Germany). To generate the Runx1 probe, a nucleotide 1-1038 fragment of Runx1 cDNA was cloned into theEcoRI and BamHI sites of pBluescript KS+. This plasmid was either linearized with XhoI and transcribed by T7 RNA polymerase to generate an antisense probe or linearized withNotI and transcribed by T3 RNA polymerase to generate a sense probe. Embryos (10.5 dpc) were fixed in 4% paraformaldehyde, embedded in OCT compound, and cryosectioned. These specimens were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 minutes and then washed with PBS for 2 minutes. The sections were treated with 7.5 μg/mL proteinase K in PBS at 37°C for 1 hour, washed with PBS for 2 minutes, refixed with 4% paraformaldehyde in PBS, again washed with PBS for 2 minutes, and placed in 0.2 M HCl for 10 minutes. After washing with PBS for 2 minutes, the specimens were acetylated by incubation in 0.1 M triethanolamine-HCl, pH 8.0, for 1 minute and further in 0.1 M triethanolamine-HCl, 0.25% acetic anhydride for 10 minutes. After washing with PBS for 2 minutes, the samples were incubated with 3% hydrogen peroxide for 1 hour, washed in PBS for 2 minutes, and dehydrated through a series of ethanols. Hybridization was performed with probes at concentrations of 500 ng/mL in a hybridization solution (50% formamide, 5× saline sodium citrate [SSC], 1% sodium dodecyl sulfate [SDS], 50 μg/mL tRNA, and 50 μg/mL heparin) at 55°C for 16 hours. After hybridization, the specimens were washed in 5× SSC at 55°C for 15 minutes and then in 50% formamide, 2× SSC at 55°C for 15 minutes, followed by RNase treatment in 50 μg/mL RNase A in 10 mM Tris (tris(hydroxymethyl)aminomethane)–HCl, pH 8.0, 1 M NaCl, and 1 mM EDTA (ethylenediaminetetraacetic acid). Then the sections were washed twice with 2× SSC at 50°C for 15 minutes, twice with 0.2× SSC at 50°C for 15 minutes, and once with TBST (0.1% Tween 20 in Tris-buffered saline [TBS]) for 5 minutes. After treatment with 1% blocking reagent (Roche Diagnostics) in TBST for 1 hour, the samples were incubated with antidigoxigenin peroxidase (POD), Fab fragment (Roche Diagnostics) diluted 1:100 with the blocking reagent for 1 hour. The sections were washed 3 times with 0.1% Tween 10 in PBS (PBST), incubated with tyramide signal amplification (TSA) plus Cy3 system for coloring reaction, and washed 3 times with PBST.
Results
Activity of Flk-1 promoter/enhancer in undifferentiated ES cells
Kappel et al23 demonstrated in a previous study thatFlk-1 p/e is active in most embryonic ECs throughout embryogenesis. Endogenous Flk-1, however, was shown to be expressed not only in ECs but also in lateral mesoderm and some fractions of hematopoietic cells.24 35-37 Thus, whetherFlk-1 p/e accurately represents the endogenousFlk-1 cis-regulating region or only part of its activity remained to be investigated. To assess the activity ofFlk-1 p/e in more detail, we took advantage of the ES cell culture system that allows detailed dissection of the differentiation process of ECs from mesoderm (Figure 1B). For this purpose, we established ES cell lines that were stably transduced with aGFP gene conjugated to the Flk-1 p/e (Figure1A).
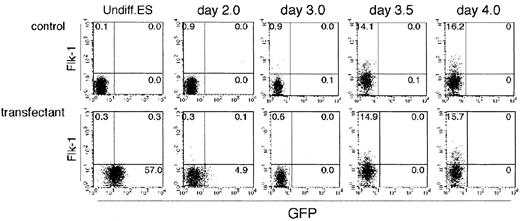
To our surprise, all stably transformed ES cell lines expressed GFP before induction of differentiation (Figure2). As all independent cell lines expressed GFP, it is likely that this expression pattern is specific to the Flk-1 p/e rather than variations among ES cell lines or sites to which the transgene was integrated. As Flk-1 is not expressed in ES cells,19 24 it appears that Flk-1 p/e activity differs from that of the endogenous regulatory regions. This ectopic activity disappeared rapidly on induction of ES cell differentiation by removing LIF from cultures on collagen IV-coated dishes. Of note, however, is that GFP expression could not be detected even at 4 days of incubation under our culture conditions, although more than 15% of cells expressed endogenous Flk-1, representing differentiation of lateral mesodermal cells. This result indicates again the difference between Flk-1 p/e and the endogenouscis-regulatory region of the Flk-1 gene.
Flow cytometric analysis of the differentiating ES cells.
Undifferentiated parental ES cells (top row) and ES cells transfected with GFP under the control of Flk-1 promoter/enhancer (bottom row) were cultured on type IV collagen–coated dishes in the absence of LIF and analyzed for the expression of Flk-1 and GFP for 4 days. The results shown are representative of 3 independent experiments.
Flow cytometric analysis of the differentiating ES cells.
Undifferentiated parental ES cells (top row) and ES cells transfected with GFP under the control of Flk-1 promoter/enhancer (bottom row) were cultured on type IV collagen–coated dishes in the absence of LIF and analyzed for the expression of Flk-1 and GFP for 4 days. The results shown are representative of 3 independent experiments.
Activity of Flk-1promoter/enhancer in endothelial cells
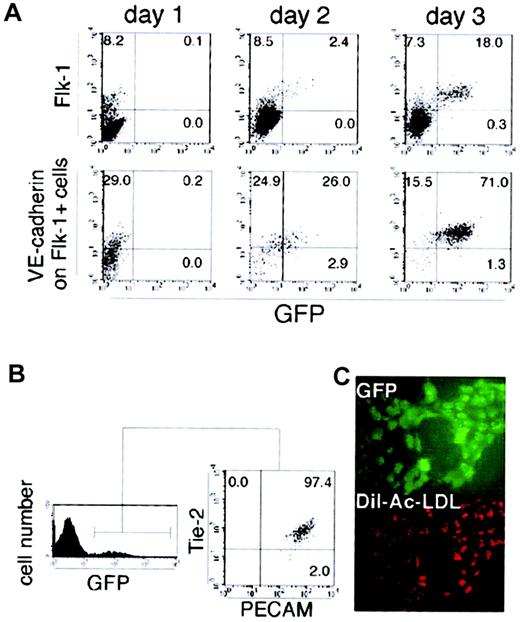
Purified Flk-1+ VE-cadherin−GFP− mesoderm cells could rapidly give rise to GFP+ cells on an OP9 stromal cell layer. To specify the cells in which Flk-1 p/e is active, we analyzed the expression of other surface markers in GFP+ cells. As shown in Figure 3A, VE cadherin+GFP− cells appeared within 24 hours, followed by the emergence of VE-cadherin+ GFP+ cells. The GFP signals were restricted to the VE-cadherin+ fraction, and most GFP+ cells harvested from day 3 cultures of Flk-1+ GFP− cells coexpressed a series of EC markers such as Flk-1, PECAM-1, and Tie-2 (Figure 3B). In addition, GFP+ cells formed sheetlike colonies on the OP9 feeder layer, which could be labeled by acetylated-LDL (Figure 3C). All of these observations are consistent with an idea that Flk-1p/e is active in mature EC cells.
Endothelial nature of cells expressing GFP driven by theFlk-1 promoter/enhancer.
(A) Sorted Flk-1+ cells derived from ES cells were cultured on OP9 stromal cells and analyzed for the expression of GFP and Flk-1 on wild-type cells (top row) or analyzed for the expressions of GFP and VE cadherin on Flk-1+ cells (bottom row) from day 1 to day 3. (B) After a 3-day culturing of the sorted Flk-1+ cells, Tie-2 and PECAM-1 expression in GFP+ cells were analyzed by flow cytometry. All the GFP+ cells express Tie-2 and PECAM-1. (C) Incorporation of DiI-labeled acetylated LDL (red) by GFP+ cells (green) was examined by fluorescent microscopy on day 3 of Flk-1+ cell culture on OP9 cells. Original magnification ×200. The results shown are representative of 3 independent experiments.
Endothelial nature of cells expressing GFP driven by theFlk-1 promoter/enhancer.
(A) Sorted Flk-1+ cells derived from ES cells were cultured on OP9 stromal cells and analyzed for the expression of GFP and Flk-1 on wild-type cells (top row) or analyzed for the expressions of GFP and VE cadherin on Flk-1+ cells (bottom row) from day 1 to day 3. (B) After a 3-day culturing of the sorted Flk-1+ cells, Tie-2 and PECAM-1 expression in GFP+ cells were analyzed by flow cytometry. All the GFP+ cells express Tie-2 and PECAM-1. (C) Incorporation of DiI-labeled acetylated LDL (red) by GFP+ cells (green) was examined by fluorescent microscopy on day 3 of Flk-1+ cell culture on OP9 cells. Original magnification ×200. The results shown are representative of 3 independent experiments.
Absence of hemogenic potential in GFP+ endothelial cells
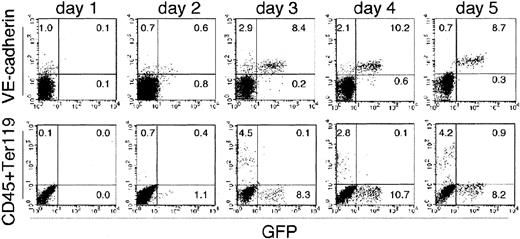
Flk-1+ VE-cadherin− GFP−mesoderm cells could also give rise to CD45+ or Ter119+ HPCs in the culture (Figure4). Although HPCs and GFP+ECs were already present in cultures of the same stage (from day 2 to day 5), none of the GFP+ cells expressed CD45 or Ter119. During the course of this study, we noticed that sorted GFP+ cells could not give rise to HPCs. It is thus likely that the GFP+ population represents a subset of ECs that is excluded from hemogenic potential. This possibility may also account for the absence of CD45+ GFP+ cells that may represent a transitory stage from EC to HPC, as no HPCs are generated from GFP+ cells. To examine this possibility, we sorted the VE-cadherin+CD45−Ter119−GFP− cells (Flk-GFP− ECs) and VE-cadherin+CD45−Ter119−GFP+ cells (Flk-GFP+ ECs) from the culture of Flk-1+ cells and assessed the frequency of precursors that give rise to HPCs or ECs (Figure 5A-F). Except for GFP expression, the 2 populations were indistinguishable in terms of the expression of Flk-1, VE-cadherin, PECAM, CD34, and Tie-2 (data not shown), although it is difficult to formally rule out the possibility of contamination of other cell lineages. Flk-GFP− ECs were observed in the culture of Flk-1+ VE cadherin− cells from day 1 to day 4 (Figure 4) and harbor potential to give rise to definitive type HPCs (Figure 5A,B). In contrast, neither CD45 nor Ter119 were expressed in the culture of GFP+ population (Figure5A).
Expression of hematopoietic markers and that of GFP were reciprocally exclusive.
Sorted Flk-1+ cells derived from ES cells were cultured on an OP9 stromal cell layer and analyzed for the expression of hematopoietic markers (CD45 and Ter119; bottom row), an endothelial marker (VE cadherin; top row), and GFP for 5 days. The results shown are representative of 3 independent experiments.
Expression of hematopoietic markers and that of GFP were reciprocally exclusive.
Sorted Flk-1+ cells derived from ES cells were cultured on an OP9 stromal cell layer and analyzed for the expression of hematopoietic markers (CD45 and Ter119; bottom row), an endothelial marker (VE cadherin; top row), and GFP for 5 days. The results shown are representative of 3 independent experiments.
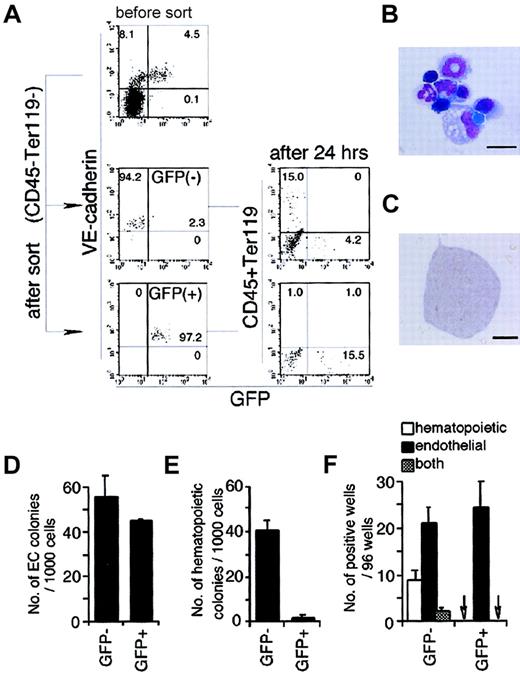
Hemogenic ability of GFP− or GFP+ endothelial cells.
(A) GFP− or GFP+ endothelial cells (VE cadherin+, CD45−, and Ter119−) were harvested from 2.5-day culture of sorted Flk-1+ cells and cultured on OP9 cells. Then the expression of hematopoietic markers and GFP were analyzed by flow cytometry 24 hours later. The result shown is a representative of 3 independent experiments. (B) May-Grünwald-Giemsa staining of hematopoietic cells formed in the culture of GFP− endothelial cells. Erythroblasts, monocytes, macrophages, and polymorphonuclear cells were observed. The scale bar represents 10 μm. (C) Morphology of a PECAM-1+colony generated from either GFP− or GFP+endothelial cells. The scale bar represents 200 μm. (D) Frequency of cells capable of formation of endothelial colony in the indicated fractions. Error bars indicate standard deviations for 3 independent determinations. (E) Frequency of hematopoietic colony-forming cells in the indicated fractions. Error bars indicate standard deviations for 3 independent determinations. (F) Incidence of hematopoietic and endothelial cell differentiation from single GFP− or GFP+ endothelial cells (VE cadherin+, CD45−, and Ter119−) on OP9 stromal cell layer in the presence of SCF, IL-3, Epo, and G-CSF. Arrows indicate that no colony-forming cell was detected. Error bars indicate standard deviations for 3 independent determinations.
Hemogenic ability of GFP− or GFP+ endothelial cells.
(A) GFP− or GFP+ endothelial cells (VE cadherin+, CD45−, and Ter119−) were harvested from 2.5-day culture of sorted Flk-1+ cells and cultured on OP9 cells. Then the expression of hematopoietic markers and GFP were analyzed by flow cytometry 24 hours later. The result shown is a representative of 3 independent experiments. (B) May-Grünwald-Giemsa staining of hematopoietic cells formed in the culture of GFP− endothelial cells. Erythroblasts, monocytes, macrophages, and polymorphonuclear cells were observed. The scale bar represents 10 μm. (C) Morphology of a PECAM-1+colony generated from either GFP− or GFP+endothelial cells. The scale bar represents 200 μm. (D) Frequency of cells capable of formation of endothelial colony in the indicated fractions. Error bars indicate standard deviations for 3 independent determinations. (E) Frequency of hematopoietic colony-forming cells in the indicated fractions. Error bars indicate standard deviations for 3 independent determinations. (F) Incidence of hematopoietic and endothelial cell differentiation from single GFP− or GFP+ endothelial cells (VE cadherin+, CD45−, and Ter119−) on OP9 stromal cell layer in the presence of SCF, IL-3, Epo, and G-CSF. Arrows indicate that no colony-forming cell was detected. Error bars indicate standard deviations for 3 independent determinations.
We also measured the frequency of clonogenic progenitors for HPCs and ECs in each population. The frequency of cells that can give rise to EC colonies on OP9 are nearly the same between the 2 populations (Figure5C,D), whereas that of hematopoietic progenitors in Flk-GFP− ECs is 20-fold higher than that in Flk-GFP+ ECs (Figure 5E). This finding was further confirmed by single cell deposition assay of each population into 96-well dishes that support generation of ECs and HPCs. As shown in Figure 5F, no hemogenic activity was observed in Flk-GFP+ECs, whereas 10% of the GFP− population contained hematopoietic progenitors, among which 3% can also give rise to ECs.
We have previously shown that α4-integrin is a marker for the earliest precursor of HPC lineage derived from ECs.21 When we analyzed the VE-cadherin+ cells derived from Flk-1+ mesodermal cells, expression of α4-integrin and that of GFP were exclusively reciprocal (data not shown). These results indicate that Flk-1 p/e activity is specific to ECs that have already been excluded from a HPC fate.
Differential expression of transcriptional regulators in the EC populations
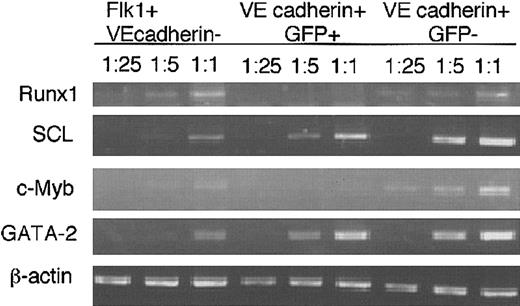
All of the above-mentioned results indicate that nonhemogenic ECs can be distinguished prospectively by the Flk-1 p/e activity. Thus, it is interesting to assess the expressions of molecules such as Runx1, c-Myb, SCL, and GATA2 that have been shown to play a requisite role in HPC differentiation.32 38-40
VE-cadherin+ cells were divided into GFP+ and GFP− populations, and the expression of various genes was assessed by RT-PCR (Figure 6). Expressions of Runx1 and c-Myb that are essential for the differentiation of the definitive HPCs were detected preferentially in GFP− population, whereas only low-level expression was observed in GFP+ population. In contrast, both populations expressed GATA2 and SCL at equal levels.
mRNA expression of transcription factors.
Flk-1+ VE cadherin− cells, GFP−VE cadherin+ CD45−Ter119− cells, and GFP+ VE cadherin+CD45−Ter119− cells were induced in vitro from ES cell-derived Flk-1+ cells for 2.5 days, and different dilutions of cDNA prepared from sorted cells were subjected to PCR amplification using primers specific for Runx1, SCL, c-Myb, GATA-2, and β-actin. PCR products were separated on 1% agarose gel and stained with ethidium bromide. The results shown are representative of 3 independent experiments.
mRNA expression of transcription factors.
Flk-1+ VE cadherin− cells, GFP−VE cadherin+ CD45−Ter119− cells, and GFP+ VE cadherin+CD45−Ter119− cells were induced in vitro from ES cell-derived Flk-1+ cells for 2.5 days, and different dilutions of cDNA prepared from sorted cells were subjected to PCR amplification using primers specific for Runx1, SCL, c-Myb, GATA-2, and β-actin. PCR products were separated on 1% agarose gel and stained with ethidium bromide. The results shown are representative of 3 independent experiments.
Flk-1 promoter/enhancer activity in the embryo
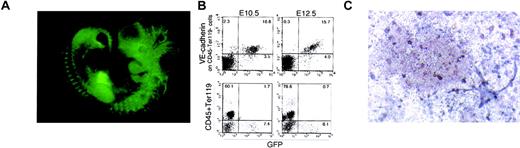
Given that GFP expression can specify the ECs that are excluded from HPC fate, it is of great interest to determine the regions in the embryo where GFP+ and GFP− ECs are present. For this purpose, we generated transgenic mouse strains harboring the same construct as used in the ES experiment. As previously reported, nearly all the vessels expressed GFP in developing embryos (Figure7A). To correlate the data of the transgenic mice with that of ES cell-derived cells, we dissociated 10.5-dpc embryos and analyzed the expression of GFP by flow cytometry (Figure 7B). Consistent with the data in ES cultures, VE-cadherin+ cells were observed in both GFP+(16.8%) and GFP− fractions (2.3%), and the number of cells in the GFP− fraction decreased in 12.5-dpc embryo (0.3%). The expression of hematopoietic markers (CD45 or Ter119) and GFP were exclusively reciprocal. We next sorted VE-cadherin+ GFP+ and VE-cadherin+GFP− populations (CD45− and Ter119−) from 10.5- or 11.5-dpc embryos and analyzed the ability to give rise to HPCs or ECs (Figure 7C and Table1). PECAM-1+ endothelial sheet colonies were found in wells seeded with either GFP−or GFP+ ECs. In complete agreement with the results obtained from ES cell experiments, HPCs were generated only in the cultures of GFP− endothelial cells (Figure 7C).
Activity of the Flk-1 promoter/enhancer in vivo.
(A) Direct fluorescent image of a 10-dpc mouse embryo transgenic for a reporter gene construct in which the GFPgene is under the control of the Flk-1 promoter/enhancer. (B) Expression of VE cadherin (top row) and GFP in CD45−, Ter119− (bottom row), and GFP and hematopoietic markers in cells dissociated from 10.5- and 12.5-dpc transgenic mouse embryos. The result shown is a representative of 3 independent experiments for 3 independent clones. (C) Morphology of hematopoietic and PECAM+ endothelial colonies generated from either GFP− endothelial cells obtained from 10.5-dpc transgenic embryos. Original magnification ×100.
Activity of the Flk-1 promoter/enhancer in vivo.
(A) Direct fluorescent image of a 10-dpc mouse embryo transgenic for a reporter gene construct in which the GFPgene is under the control of the Flk-1 promoter/enhancer. (B) Expression of VE cadherin (top row) and GFP in CD45−, Ter119− (bottom row), and GFP and hematopoietic markers in cells dissociated from 10.5- and 12.5-dpc transgenic mouse embryos. The result shown is a representative of 3 independent experiments for 3 independent clones. (C) Morphology of hematopoietic and PECAM+ endothelial colonies generated from either GFP− endothelial cells obtained from 10.5-dpc transgenic embryos. Original magnification ×100.
Hemogenic ability of VE cadherin+(CD45− Ter119−) cells obtained from transgenic embryos
| Embryo . | No. of cells/well . | No. of positive wells/no. of wells analyzed . | |||||
|---|---|---|---|---|---|---|---|
| VE cadherin+GFP− . | VE cadherin+GFP+ . | ||||||
| HPC colonies . | EC colonies . | Both . | HPC colonies . | EC colonies . | Both . | ||
| 10.5-dpc | |||||||
| Exp 1 | 30 | 2/6 | 4/6 | 1/6 | 0/6 | 4/6 | 0/6 |
| Exp 2 | 50 | 4/6 | 4/6 | 3/6 | 0/6 | 6/6 | 0/6 |
| 11.5-dpc | |||||||
| Exp 1 | 15 | 2/4 | 2/4 | 1/4 | 0/4 | 3/4 | 0/4 |
| Exp 2 | 30 | 9/12 | 10/12 | 8/12 | 0/9 | 8/9 | 0/9 |
| Embryo . | No. of cells/well . | No. of positive wells/no. of wells analyzed . | |||||
|---|---|---|---|---|---|---|---|
| VE cadherin+GFP− . | VE cadherin+GFP+ . | ||||||
| HPC colonies . | EC colonies . | Both . | HPC colonies . | EC colonies . | Both . | ||
| 10.5-dpc | |||||||
| Exp 1 | 30 | 2/6 | 4/6 | 1/6 | 0/6 | 4/6 | 0/6 |
| Exp 2 | 50 | 4/6 | 4/6 | 3/6 | 0/6 | 6/6 | 0/6 |
| 11.5-dpc | |||||||
| Exp 1 | 15 | 2/4 | 2/4 | 1/4 | 0/4 | 3/4 | 0/4 |
| Exp 2 | 30 | 9/12 | 10/12 | 8/12 | 0/9 | 8/9 | 0/9 |
Indicated numbers of cells were sorted from embryos transgenic for GFP gene under the control of Flk-1 promoter/ enhancer and cocultured with OP9 stromal cells with SCF, IL-3, Epo, and G-CSF for 10 days. Existences of hematopoietic colonies were judged by their morphology, and those of endothelial colonies were by immunostaining with anti-PECAM-I MoAb. Exp, experiment.
Presence of GFP− endothelial cells in the dorsal aorta
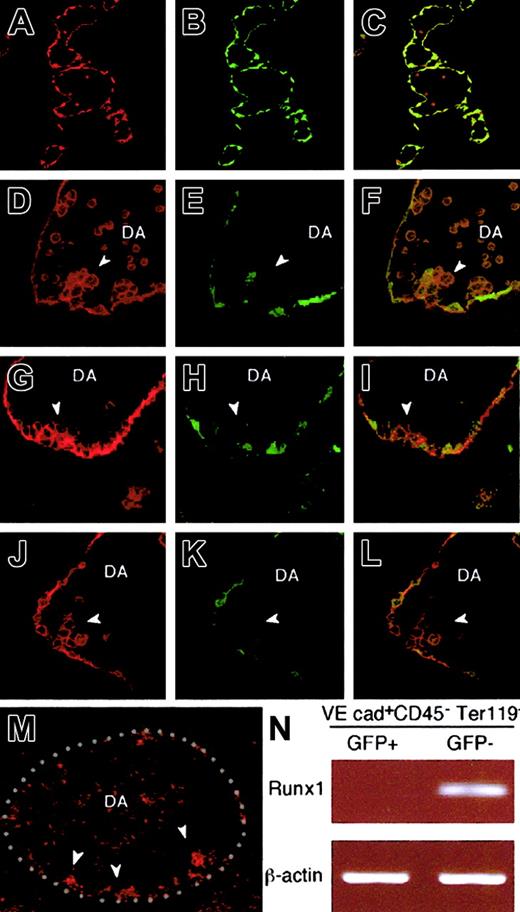
To determine the regions in the embryos where GFP− ECs exist, we sectioned 10.5-dpc transgenic embryos and stained them with anti-GFP (green) and anti–PECAM-1 antibodies (red). As previously reported,23 GFP was expressed by nearly all the endothelial cells (Figure8A-C). PECAM-1+ endothelial cells expressed GFP, and, in addition, there were PECAM-1+round cells circulating within the vessels but were negative for GFP. These findings corroborate with the observation from ES cell experiments. Outside the endothelium, GFP was expressed only in a limited portion of the neural tube (data not shown). Taking into consideration the fact that GFP− ECs contain hemogenic ECs, we analyzed GFP expression in dorsal aorta that has been implicated as the site of HPC generation from ECs. As shown in Figure 8D-L, integration of GFP− endothelial cells was observed in the ventral wall of the dorsal aorta, where hematopoietic clusters were formed, and the rest of the ECs were GFP+ (Figure 8D-L). It was recently shown that a subset of endothelium that expresses Runx1 includes a potent progenitor for definitive hematopoiesis.41 By in situ hybridization, we also confirmed that expression of Runx1 was observed at the cell clusters on the ventral wall of the dorsal aorta in 10.5-dpc embryo, whereas other cells of inner linings of blood vessels were negative for the transcripts (Figure 8M). To show that Runx1 expression is exclusive to Flk-GFP, we sorted GFP+ and GFP− endothelial cells from 10.5-dpc transgenic mouse and analyzed the expression of Runx1 (Figure 8N). Consistent with the results in ES cell experiments, Runx1 was preferentially expressed by Flk-GFP–negative endothelial cells. These data show that Runx1+ endothelial cells are integrated within the ventral wall of dorsal aorta and they are negative in GFP.
Localization of GFP− and GFP+endothelial cells in mouse embryos.
E10.5 transgenic embryos were stained with anti-GFP (green) and anti–PECAM-1 (red) antibodies. Nearly all the PECAM-1+endothelial cells express GFP, and, in addition, there were PECAM-1+ round cells circulating within the vessels, but they were negative for GFP (A-C). (D-L) GFP−endothelial cells (arrowheads) were found integrated in the endothelium at the ventral wall of the dorsal aorta, where hematopoietic clusters were formed, whereas the rest of the ECs were GFP+. (M-N) expressions of the transcripts for Runx1 were shown by using in situ hybridization (M) or RT-PCR (N). In situ hybridization was performed as described in “Materials and methods,” and red dots indicate the expression of Runx1. The sense probe did not give any signal (data not shown). White-dotted line indicates the outlines of dorsal aorta (M). (N) GFP− VE cadherin+CD45−Ter119− cells and GFP+ VE cadherin+ CD45−Ter119− cells were sorted from 10.5-dpc transgenic embryos and subjected to RT-PCR using primers specific for Runx1. PCR products were separated on 1% agarose gel and stained with ethidium bromide. The result shown is representative of 3 independent experiments. DA, dorsal aorta.
Localization of GFP− and GFP+endothelial cells in mouse embryos.
E10.5 transgenic embryos were stained with anti-GFP (green) and anti–PECAM-1 (red) antibodies. Nearly all the PECAM-1+endothelial cells express GFP, and, in addition, there were PECAM-1+ round cells circulating within the vessels, but they were negative for GFP (A-C). (D-L) GFP−endothelial cells (arrowheads) were found integrated in the endothelium at the ventral wall of the dorsal aorta, where hematopoietic clusters were formed, whereas the rest of the ECs were GFP+. (M-N) expressions of the transcripts for Runx1 were shown by using in situ hybridization (M) or RT-PCR (N). In situ hybridization was performed as described in “Materials and methods,” and red dots indicate the expression of Runx1. The sense probe did not give any signal (data not shown). White-dotted line indicates the outlines of dorsal aorta (M). (N) GFP− VE cadherin+CD45−Ter119− cells and GFP+ VE cadherin+ CD45−Ter119− cells were sorted from 10.5-dpc transgenic embryos and subjected to RT-PCR using primers specific for Runx1. PCR products were separated on 1% agarose gel and stained with ethidium bromide. The result shown is representative of 3 independent experiments. DA, dorsal aorta.
Discussion
Expression of Flk-1 has been evaluated by using MoAb and mice in which marker genes such as LacZ were knocked into theFlk-1 locus.24,42,43 Previous results indicated that Flk-1 expression is detected during successive stages from early lateral mesoderm to endothelial stages.19 Moreover, it was also reported that Flk-1 expression is maintained in nascent HPCs for a short interval after differentiation from Flk-1+progenitors.37 In contrast to commonly used endothelial markers such as Tie-2 or CD34 that are expressed also in adult hematopoietic stem cells,44,45 Flk-1 expression is unique in that it is restricted to mesodermal cells and ECs and is absent in adult HPCs. Kappel et al23 investigated thecis-regulatory region of the Flk-1 gene and showed that a combination of a 5′-flanking promoter region together with the 3′ portion of the first intron, Flk-1 p/e, can direct gene expression specifically in the EC population in the embryo after 7.8-dpc. Although the activity of Flk-1 p/e decreases in the ECs of adult mice, it appears again in the ECs during neoangiogenesis.46 Although these studies suggested thatFlk-1 p/e can largely dictate the expression pattern of the endogenous Flk-1 gene, it remained unclear whether it is also active in the Flk-1+ nonendothelial populations such as lateral mesoderm and also nascent HPCs.
In this study, using both the ES cell differentiation system and transgenic mice, we showed that the Flk-1 p/e is not identical to the endogenous regulatory unit of the Flk-1gene. First, it is active in undifferentiated ES cells, although endogenous Flk-1 expression is not detectable in ES cells either by flow cytometry with anti–Flk-1 MoAb or RT-PCR analyses.19 37 The expression level of GFP driven by this promoter is fairly strong and found in all 5 independent ES cell lines, indicating that the observed activity represents an autonomous activity of Flk-1 p/e rather than a reflection of integrated sites. Of note is that this activity is specific to LIF-maintained ES cells, as its activity disappears rapidly on removal of LIF. This ectopic activity of Flk-1 p/e indicates the presence of additionalcis-regulatory elements outside this region, which repressFlk-1 gene activation in ES cells. The second difference between Flk-1 p/e and the endogenouscis-regulatory region is that the former is not active in Flk-1+ lateral mesoderm cells and Flk-1+hematopoietic cells, although the endogenous Flk-1 gene is continuously expressed throughout these successive stages. Activation of Flk-1 p/e occurs only after the differentiation of Flk-1+ VE-cadherin− lateral mesoderm to Flk-1+ VE-cadherin+ cells. This result is interesting in that it demonstrates that Flk-1 gene expression is regulated differently even in the successive stages during which Flk-1 expression is maintained at the same level. The shift of gene regulatory units from mesodermal type to EC type appears to be associated with the process of EC commitment from lateral mesoderm. However, it should be noted that GFP− ECs exist and are maintained for several days after formation of the vascular network in the embryos, although their proportion decreases as embryogenesis progresses. Thus, we want to view this process as generating a new EC population in which Flk-1 p/e is active during the process of EC maturation.
Accumulating data indicated that ECs are a highly diverse population, which is defined by morphology, anatomical location, differential expression of molecules, and also functional properties. We and others have proposed that the ability to give rise to HPCs is a way of defining EC diversity.17,18 This study demonstrated that HPCs are generated only from a Flk-1 p/e inactive population. Interestingly, Flk-1+ mesoderm contains a direct precursor of HPCs, although its differentiation is restricted to the primitive HPCs.22 Thus, both before and after expression of VE-cadherin, Flk-1 p/e activity is correlated inversely with the ability to generate HPCs. The same results were obtained by sorting GFP+ and GFP− ECs from transgenic mice harboring Flk-1 p/e–GFP gene, indicating that this is unlikely because of an in vitro artifact.
The inverse relationship between hemogenic potential andFlk-1 p/e activity was also suggested by the differential expression of transcriptional regulators that are expressed in both HPCs and ECs and are essential for HPC differentiation. Our present study showed that Runx1 and c-Myb are expressed preferentially in the GFP− population, whereasSCL and GATA2 are expressed in both populations. Previous gene targeting studies demonstrated that the former 2 are involved in the process of generating the definitive HPCs. Moreover, we recently demonstrated that Runx1 is required for the differentiation process of VE-cadherin+ cells to HPCs.47 Accepting the hypothesis that ECs are the precursors of the definitive HPCs, the restriction of Runx1 and c-Myb to GFP− ECs suggests that Flk-1 p/e activation may reflect the process to generate fully committed ECs through exclusion of hemogenic potential. On the other hand, SCL and GATA2 were shown to have roles in vascular development. By mutational analysis, binding sites for SCL, GATAs, and Ets transcription factors were identified as critical elements for the EC specificity of theFlk-1 p/e.48 However, as GATA2 and SCL are expressed equally in both GFP+ and GFP−populations, there should be involvement of additional molecules and sequences for regulation of the restricted expression ofFlk-1p/e in nonhemogenic ECs.
Nonetheless, Flk-1 p/e allows us to distinguish prospectively the nonhemogenic ECs in the embryonic tissues. Previous histologic studies demonstrated that transition of ECs to HPCs is observed most consistently in the dorsal aorta. Moreover, expression of Runx1 that is essential for this process is observed in this region (Figure 8M and North et al41 49). Our study showed that GFP− ECs are integrated into the EC sheet that lines the luminal wall of the dorsal aorta, indicating that both hemogenic and nonhemogenic VE-cadherin+ cells are indeed an integral part of the EC lining rather than existing separately in different tissues. Consistent with our expectation, many HPC clusters were found associated with ECs that were GFP− but rarely GFP+ ECs. As only GFP− ECs had potential to give rise to HPCs, it is likely that the HPC clusters attaching to the luminal wall of dorsal aorta are indeed the cells generated de novo from the GFP− fraction of ECs.
In conclusion, Flk-1 gene regulation appears to be under a complex control mechanism and different sets of molecules. Additionalcis-regulatory regions that dictate the endogenous Flk-1 expression remain to be identified. As revealed in this study, however, the Flk-1 p/e identified by Kappel et al,23although representing only a part of the Flk-1 regulatory region, is useful both for dissecting the process of EC commitment and as a tool for gene transduction to fully committed ECs.
We thank Mariko Moriyama (Riken, Center for Developmental Biology, Kobe, Japan) for her critical help in the in situ hybridization procedure and Dr Ruth Yu for a critical reading of the manuscript.
Prepublished online as Blood First Edition Paper, September 19, 2002; DOI 10.1182/blood-2002-02-0655.
Supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (nos. 12770506, 12670301, and 07CE2005), the Cell Science Research Foundation, and Japanese Society for the Promotion of Science “Research of Future” program.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hideyo Hirai, Department of Microbiology, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan; e-mail: hhirai@basic.kpu-m.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal