Abstract
The localization of adult hemopoiesis to the marrow involves developmentally regulated interactions between hemopoietic stem cells and the stromal cell–mediated hemopoietic microenvironment. Although primitive hemopoietic cells exhibit a broad repertoire of adhesion molecules, little is known about the molecules influencing the site of cell lodgment within the marrow following transplantation. However, our recent studies indicate that hierarchically dependent patterns of migration of transplanted hemopoietic cells result in the retention of primitive cells within the endosteal and lineage-committed cells in the central marrow regions. Herein, we now demonstrate that these 2 subpopulations exhibit a striking difference in the expression of a cell surface adhesion molecule, with populations enriched for murine and human hemopoietic stem cells expressing the carbohydrate hyaluronic acid (HA). Furthermore, the presence of this glycosaminoglycan appears critical for the spatial distribution of transplanted stem cells in vivo. In addition, we also demonstrate that the binding of HA by a surrogate ligand results in marked inhibition of primitive hemopoietic cell proliferation and granulocyte differentiation. Collectively, these data describe an important yet previously unrecognized role for HA in the biology of primitive hemopoietic progenitor cells.
Introduction
Considerable evidence supports the proposal that the localization of hemopoiesis to the bone marrow (BM) in adult mammals involves developmentally regulated interactions between primitive hemopoietic stem cells (HSCs) and the stromal cell–mediated hemopoietic microenvironment (HM) of the marrow (for a review, see Simmons et al1).
Although the anatomic location of maturing hemopoietic cells within the BM is well documented,2,3 the spatial distribution of more primitive cells is less well studied. Previous studies in the mouse have established that lineage-restricted clonogenic hemopoietic progenitor cells (HPCs) also conform to a well-defined spatial distribution across the axis of the femur with greatest numbers near the central longitudinal vein.4 In contrast, hierarchically more primitive progenitors, such as spleen colony-forming units, exhibit the converse distribution with low numbers in the central region of the marrow and greatest enrichment in a region adjacent to bone, the endosteum.5 Until recently, it has not been possible to define the spatial distribution of HSCs within the BM. This is due to the rarity of HSCs and the lack of a single, unique antigenic marker allowing their unambiguous identification in situ.
To circumvent these problems, we recently developed a novel approach to the study of hemopoietic cell homing and lodgment within the BM that involves transplantation of phenotypically defined populations of fluorescently labeled cells and histologic analysis of BM sections to track individual cells lodging in recipient mice.6 This approach was used for studies performed in mice, which were not myeloablated prior to transplantation. Although transplantation into myeloablated recipients represents the standard means by which patients are given an HSC graft, the most appropriate method for analyzing the spatial distribution of cells within the BM and the factors that regulate this process is one in which the HM has not been altered by preparative ablation.
Transplants using different BM subpopulations demonstrated that although the majority of cells entered the BM from the central vessels, their subsequent localization varied according to their phenotype.6 Populations enriched in HSCs such as lineage-negative (Lin−) cells showing low retention of rhodamine 123 (Lin−Rh123dull)7exhibited selective migration to and lodgment within the endosteal region, whereas hemopoietic cells expressing surface markers associated with lineage commitment (designated Lin+) demonstrated high selectivity for the central BM region. Thus, the distribution of transplanted hemopoietic cells within the BM is not random and closely reflects that previously defined for related cell populations in steady-state adult mouse BM.4 These data therefore demonstrate for the first time that the discrete spatial localization of transplanted hemopoietic cells within the BM appears to be the result of specific, hierarchically dependent patterns of migration that culminate in the retention of these populations at anatomically distinct sites. It is therefore proposed that the endosteal region of the BM represents the site of HSC “niches.”8
The re-establishment of hemopoiesis by intravenously infused cells requires several coordinated events including homing, migration, and lodgment of HPCs within the BM HM. The initial event, homing, is the specific recruitment of circulating HSCs to the BM and involves the selective recognition of HSCs by the microvascular endothelium of the marrow and transendothelial cell migration into the extravascular hemopoietic space. In contrast, lodgment encompasses events following extravasation and is defined as the selective migration of cells to suitable HM niches in the extravascular compartment. Current data suggest that homing involves a similar cascade of cell adhesion molecules (CAMs) to those participating in the extravasation of mature leukocytes into tissues.9 Primitive hemopoietic cells exhibit a broad repertoire of CAMs including various members of the integrin, sialomucin, immunoglobulin superfamily (IgGSF), and CD44 families. Current data suggest key roles for the sialomucin receptor for P-selectin, P-selectin glycoprotein ligand (PSGL-1),10the β1 integrin very late antigen 4 (VLA-4),11 and the receptor for stromal derived factor 1 (SDF-1), CXCR4,12 in HSCs homing to the BM. In contrast, very little is known about the molecules that influence the site of HSC lodgment following homing to the BM. To identify cell surface molecules with potential roles in this process, we conducted an extensive analysis of CAM expression on Lin+ and Lin−Rh123dull populations. This encompassed 21 distinct CAMs including a range of β1 and β2 integrins, various members of the IgSF, L-selectin, CD44, and a range of sialomucins including CD34 and PSGL-1. One very striking difference between these 2 populations was the restricted cell surface expression by the Lin−Rh123dull cells of the single-chain carbohydrate hyaluronic acid (HA), a molecule with roles in regulating cell adhesion, motility, proliferation, and differentiation of a wide variety of cell types.
HA is a single-chain high-molecular-mass polysaccharide of repeating disaccharide units (glucuronic acid and N-acetylglucosamine) that is synthesized by 1 of 3 hyaluronic acid synthases(HASs), encoded by 1 of 3 HAS genes (Has-1, Has-2, Has-3).13-15 The mode of HA synthesis is unique among macromolecules. The synthases are integral membrane glycosyltransferases, located in the plasma membrane and HA is translocated as a free linear polymer to the pericellular space,16 not covalently linked to proteins on the cell surface.17 HA is present in many different organs and is a component of the extracellular matrix (ECM) within the BM microenvironment.18 Cell surface HA significantly affects the adhesion, motility, and growth of a wide variety of cell types, both normal and neoplastic. Due to its multivalency (which allows cross-bridging of multiple receptors on adjacent cells), the interaction of endogenous cell surface HA with its primary receptor, CD44, mediates aggregation of several cell types.19 There are many examples of increased cell movement or invasion following either the exposure of cells to HA or the ectopic expression of HA.20 Moreover, inhibition of cell movement also occurs as a consequence of either HA degradation or the blocking of HA receptors.20 HA also influences cell proliferation, differentiation, and tissue repair,16 and a substantial body of data implicates HA in the pathogenesis and dissemination of tumors.21
In the present study we have shown for the first time that hemopoietic cells synthesize and express HA. HA synthesis and expression was shown in both murine and human hemopoiesis and was restricted to primitive hemopoietic cells in particular. HA was shown to be critical in the lodgment of transplanted HSCs within the BM as demonstrated by the significant alteration in spatial distribution resulting from the enzymatic removal of HA from HSCs. In addition, the binding of HA on the surface of HSCs by a surrogate ligand in vitro resulted in a profound suppression of HSC proliferation and differentiation. Overall, our data are suggestive of a key role for this polysaccharide in HSC biology.
Materials and methods
Umbilical cord blood
Samples of umbilical cord blood (CB) were taken after informed consent was obtained following normal and cesarean section deliveries at the Mercy Hospital for Women (East Melbourne, Australia); samples were anticoagulated with sodium citrate.
Mice
BALB/c and congenic C57Bl/6J (Ly5.2) and PTRPA (Ly5.1) mice were purchased from Animal Resources Center (Perth, Australia) and housed clean conventionally for at least a week prior to experimental use. CD44−/− mice22 were kindly donated by Dr Tak Mak (Amgen Institute, Ontario Cancer Institute, University of Toronto, ON, Canada). All mice received mouse chow (Barastok, St Arnaud, Victoria, Australia) and acidified water ad libitum.
Irradiation
The ability of cells to reconstitute hemopoiesis was analyzed in mice receiving a near-lethal dose of irradiation (9.5 Gy) in 2 equal fractions separated by a 4-hour interval, delivered from 2 opposing137Cs sources (Gammacell 40; Atomic Energy of Canada, Ottawa, QC, Canada) at a dose-rate of 1.4 Gy/min.
Isolation of hemopoietic cells
Mice were killed by cervical dislocation. BM was routinely collected from femurs, tibiae, and iliac crests. These bones were thoroughly ground in phosphate-buffered saline (PBS) supplemented with 2% heat-inactivated (HI) fetal calf serum (FCS; Hyclone, Logan, UT). Bone fragments were washed, and the supernatant cell suspension was filtered through a 40-μm filter (Becton Dickinson, Franklin Lakes, NJ). The marrow was centrifuged (400g, 5 minutes) and resuspended in fresh buffer. The cell supernatant was refiltered through a 40-μm filter and diluted to 107 cells/mL PBS supplemented with 2% HI FCS (buffer).
Strategies for enriching hemopoietic cells
Murine.
BM mononuclear cells of low density (< 1.077 g/cm3) were isolated by discontinuous density centrifugation using Nycoprep for animals (Accurate Chemical and Scientific, Westbury, NY). Isolated cells were washed prior to Lin− cells being separated in a similar manner to that previously described.6 Briefly, low-density cells were labeled with a cocktail of biotinylated or unconjugated primary rat antimouse antibodies: B220 (CD45R; B cells); Mac-1 (CD11b; macrophages); Gr-1 (Ly-6G; neutrophils); Lyt-2 (CD8), L3T4 (CD4), CD3, and CD5 (T cells); and Ter119 (erythroid cells). Lin+ cells were then removed by immunomagnetic selection using the magnetic-activated cell sorting (MACS) system (Miltenyi Biotec, Bergisch, Gladbach, Germany) as previously described.6
123Rh labeling.
The remaining Lin− cells were washed, resuspended at 1 × 106/mL, and incubated in a final Rh (Molecular Probes, Eugene, OR) concentration of 0.1 μg/mL for 20 minutes at 37°C in the dark, then allowed to efflux for 15 minutes at 37°C in the dark in fresh buffer. Cells were incubated with a final concentration of 6.8 μg/mL goat antirat phycoerythrin (PE)–conjugated secondary antibody (Biosource International; Camarillo, CA) on ice, in the dark for a further 20 minutes. Finally, the cells were washed and resuspended at 5 × 106cells/mL prior to fluorescence-activated cell sorting (FACS).
Stem cell antigen 1 and c-kit labeling.
Lin− cells were washed and resuspended in a cocktail of stem cell antigen 1 (Sca-1)–fluorescein isothiocyanate (FITC; 1 μg/5 × 106 cells; Pharmingen, San Diego, CA), c-kit-PE (Pharmingen; 1 μg/ 5 × 106 cells), and streptavidin-Red 670 (Gibco, Grand Island, NY; 1:160 final concentration) on ice in the dark for 20 minutes. Cells were washed and resuspended at 5 × 106 cells/mL prior to fluorescence-activated cell-sorter (FACS) analysis.
Human.
Mononuclear cells of low density were isolated from CB by discontinuous density centrifugation using Ficoll-Hypaque density gradient (d = 1.077 g/mL, Pharmacia Biotech, Uppsala, Sweden). Isolated cells were washed in PBS prior to Lin− cells being separated using the following cocktail of lineage antibodies: anti-CD19, -CD20, and -CD24 (B cells and erythroid cells); anti-CD3, -CD2, and -CD56 (T cells and natural killer [NK] cells); anti-CD16, -CD66b, and -CD11b (granulocytes and monocytes); and anti-CD14, -CD36 (monocytes, platelets, and erythroid cells). Lin+ cells were removed by immunomagnetic selection using goat antimouse IgG microbeads and the magnetic-activated cell-separation (MACS) system.
CD34, CD38, and CD15 labeling.
Lin− cells were washed and resuspended in a cocktail of CD34-FITC and CD38-PE, or CD15-FITC (Becton Dickinson, San Jose, CA; at the concentration recommended by the manufacturer) on ice in the dark for 20 minutes. Finally, the cells were washed and resuspended at 5 × 106 cells/mL prior to FACS analysis or separation.
HA labeling
The presence of HA on the cell surface was detected using a final concentration of 20 μg/mL biotinylated hyaluronic acid–binding protein (HABP; Seikagaku, Tokyo, Japan) on ice for 15 minutes. Cells were washed, and labeled with streptavidin-PE (Biosource; 1:400 final concentration) or Red 670 as described in “Stem cell antigen 1 and c-kit labeling.”
Hyaluronidase treatment
To ensure that HABP labeling was specific, as well as to assess the importance of cell surface HA on the spatial distribution of engrafting cells, BM subpopulations were treated with 0.1 UStreptomyces hyaluronidase (HY; Sigma Aldrich, Castle Hill, New South Wales, Australia) for 15 minutes at 21°C to remove HA. Cells were washed in PBS 0.5% HI FCS.
Cell culture
Cells were sorted directly into 96-well plates containing 100 μL serum-deprived media containing multiple cytokines consisting of either stem cell factor (SCF; 75 ng/mL), interleukin 11 (IL-11), FLT3 ligand, and IL-6 (all 100 ng/mL) or granulocyte colony-stimulating factor (G-CSF), SCF, FLT3 ligand, megakaryocyte growth and development factor (MGDF) (all 100 ng/mL), and IL-6 and IL-3 (both 10 ng/mL) for murine and human HSCs, respectively, and 1, 5, 10, and 20 μg/mL HABP. Murine cells were cultured in Iscove modified Dulbecco medium (Gibco BRL) containing 1% bovine serum albumin (BSA), 10 μg/mL human insulin, 200 μg/mL human transferrin, 0.05 mM β2-mercaptoethanol, and 21 μg/mL low-density lipoprotein. Human cells were cultured in X Vivo 10 media (Biowhittaker, Walkersville, MD) containing 0.5% human serum albumin (Buminate; Baxter, Glendale, CA). Cells were cultured at 37°C in the presence of 5% O2, 10% CO2, and 85% N2 for 6 days.
HA synthesis
The ability of CB HPCs to synthesize HA was determined by culturing the cells as described, with the addition of 5 μCi/mL (0.185 MBq) d-[6-3H]–glucosamine hydrochloride (Perkin Elmer, Boston, MA) for 60 hours at 37°C in the presence of 5% O2, 10% CO2, and 85% N2. The presence of [3H]-HA was determined by digestion of cell-free supernatant with Streptomyces hyalurolyticus hyaluronidase (Calbiochem, Darmstadt, Germany). Cell culture supernatant was adjusted to pH 6 using Sorenson buffer to give a final concentration of 9 mM KH2PO4 and 7 mM Na2HPO4. Hyaluronidase was reconstituted in 9 mM KH2PO4 and 7 mM Na2HPO4 · 2H20, pH 6, and 5 turbidity-reducing units (TRU) were added to the sample at 0 and 3 hours. The digestion was incubated at 37°C for 24 hours. Conditioned media was subjected to size exclusion chromatography in a Superose 12 column (1 × 30 cm) equilibrated in 0.15 M NaCl/PO4 buffer containing 0.2% Teric X-10 and eluted at 20 mL/h in fractions of 0.5 mL. The radioactive content was then determined for each fraction using a Wallac 1410 scintillation counter (LKB, Melbourne, Victoria, Australia) by adding 3 mL HiSafe 2 scintillant. Counts due to unincorporated glucosamine (known to be eluted in fractions 35-40) and the nonspecific binding of glucosamine to human serum albumin (as shown by pronase pretreatment, and known to be eluted in fractions 24-26) were removed, and the amount of degradation products under the glucosamine peak presented as [3H] eluted per 0.5-mL fraction versus fraction number. The molecular weight of synthesized HA was determined by comparison with eluted purified HA of known molecular size.
Flow cytometry
Labeled cells were sorted on a FACStarPLUS cell sorter equipped with a 5-W argon ion laser (Coherent Innova 90, Palo Alto, CA) emitting 488 nm light at 200 mW. Light-scatter signals were collected through a 488 ± 10-nm band pass filter and a 1-decade logarithmic neutral density filter in the forward light scatter path. Rh-emitted green fluorescence pulses were collected through a 530 ± 15-nm band pass filter. Orange fluorescence pulses emitted following excitation of PE were reflected through a 440 dichroic short pass mirror, and collected through a 575 ± 26-nm band pass dichroic filter. Pulses emitted following the excitation of Red 670 were collected through a long-pass RG655 filter.
CFSE labeling
Cells to be transplanted for spatial distribution analysis were labeled with the fluorescent dye 5– (and 6)–carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes) as previously described.6 Briefly, cell populations were resuspended in PBS 0.5% HI FCS at a density of 106 cells/mL and preincubated at 37°C for 3 minutes. CFSE was diluted to 5 mM in dimethyl sulfoxide (DMSO) and then to 5 μM in PBS. CFSE was added to the cells to give a final concentration of 0.5 μM, and the dye solution/cell mixture was incubated at 37°C for a further 10 minutes. Staining was stopped by adding 10 times the dye solution/cell volume of ice-cold PBS containing 20% FCS.
Transplants
Cells were transplanted by injection in 0.2 mL PBS into the lateral tail vein. The actual numbers were 500 Lin−Sca+HA+, 500 Lin−Sca+HA−, 5000 to 6000 Lin−HA+, 5 to 6 × 105Lin−HA−, 1.1 × 105Lin−Rhdull, and 5.5 × 105Lin−Rhbright untreated and hyaluronidase-treated cells.
Analysis of transplanted cell spatial distribution
The spatial distribution of CFSE+ cells was analyzed 15 hours after transplantation, as previously described.6The location of CFSE-labeled cells (positive cells) from at least 6 longitudinal sections per transplant recipient was recorded. Central longitudinal sections were analyzed as opposed to transverse sections, because each individual section encompasses more of the entire femur. To ensure that individual cells were only analyzed once, every alternate 3.5-μm section was analyzed. The location of positive cells was designated as either endosteal (previously arbitrarily defined as within 12 cells of the endosteum23) or central (> 12 cells from either endosteum).6
RNA extraction
RNA was extracted using an RNAzol B (Geneworks, Adelaide, Australia) extraction method. Briefly, cells isolated by FACS were centrifuged and RNA prepared by lysing with 0.8 mL RNAzol B/106 cells. The homogenate was extracted with chloroform and precipitated with isopropanol. RNA was washed, dried, and resuspended in sterile water.
RT-PCR
Template cDNA was prepared using random hexamers (Pharmacia Biotech) and Superscript II reverse transcriptase (RT; Gibco BRL, Gaithersburg, MD). Polymerase chain reaction (PCR) was performed using the following oligonucletide primers: murine Has-1 sense, 5′ CGTGGACTACGTGCAGGTCTGTG 3′ and antisense, 5′ GAGCGCGAGGTATACTTGGTAGC 3′; murine Has-2 sense, 5′ GACCACACAGACAGGCGGA 3′ and antisense, 5′ TCCCAGGGTAGGTCAGCCTT 3′; murine Has-3 sense, 3′ 5′ 5′ GAGCGTGTGC GAGCTGTGGT GTG 3′ and antisense, 5′ GAAGCATCTCAATGGTGCAGGCT 3′; human HAS-1, sense 5′ GCTACCAAGTACACCTCCAGGTC 3′ and antisense, 5′ CGCGTAGAACAGACGCAGCACAG 3′; human HAS-2 sense, 5′ GCTCGCAACACGTAACGCAA 3′ and antisense 5′ GGCACTTAGATCGAGCTGTG 3′; and human HAS-3 sense, 5′ AGCCTGCAGGAGGGCATGGA 3′ and antisense 5′ GGAGCGCGCGGTATACTTAGTTCGG 3′ synthesized by Geneworks (Adelaide, Australia). All PCRs were performed in a 25-μL volume under 30 μL paraffin oil in a gene machine (Innovonics, Melbourne, Australia). Each PCR consisted of 1 ×Taq gold buffer, 200 μM dNTPs (Boehringer Mannheim, Branchburg, NJ), 4 μg/mL of each primer, 1.5 mM MgCl2, 10% DMSO, and 0.5 U Taq gold (Boehringer Mannheim). PCRs were performed with a profile of 10 minutes ofTaq gold activation at 94°C for the first cycle and 30 seconds of denaturing for subsequent cycles, 30 seconds of annealing at 60°C for 10 cycles followed by 30 seconds of annealing at 55°C for a further 25 cycles, and 30 seconds of extension at 72°C for the 35 cycles followed by a final 5-minute extension at 72°C.
Statistical analysis
Differences between means were evaluated by one-way analysis of variance (ANOVA) or Student t test where appropriate.
Results
HA expression on murine hemopoietic cells
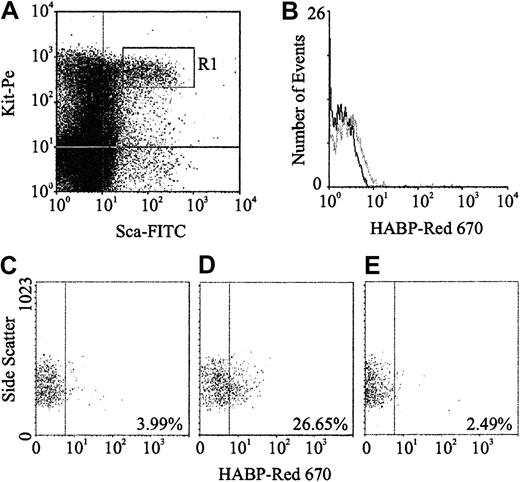
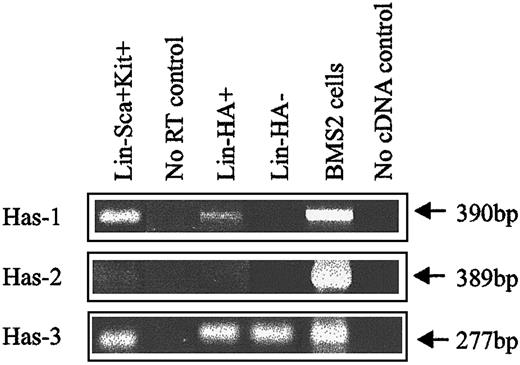
Expression of HA was demonstrated by flow cytometric analysis through the binding of a biotinylated form of HABP, which demonstrates absolute specificity for HA.24 Using this approach, HABP binding was detected on 2 subpopulations enriched for murine HSCs: Lin−Sca+Kit+ (Figure1A-B) and Lin−Rhdull (Figure 1D). Although the level of HA expression on these populations was of low intensity, it was clearly above that observed for the control (Figure 1C) and was completely eliminated by prior treatment of the cells with the specific HA-cleaving enzyme hyaluronidase (HY) (Figure 1E). In addition, a similar proportion of Lin−Rh123dull cells isolated from CD44−/− mice22 exhibited binding of HABP (25.0% compared with 26.5%), demonstrating that HA detected on the cell surface is not due to the binding of exogenous HA to its major receptor, CD44.19 RT-PCR analysis demonstrated that Lin−Sca+Kit+, Lin−HA+ but not HA− cells express transcripts for Has-1, Has-2, and Has-3 (Figure2). Interestingly, HABP−cells expressed Has-3, but synthesize undetectable levels of HA. This phenomenon has previously been reported in adenocarcinoma cells.25
HA expression on murine BM HSCs.
(A) Sca-1 and Kit expression characteristics of Lin−murine BM cells; R1 sort region for HSCs. (B) HA expression on all R1 HSCs (gray line) as detected using HABP. (C) Control (Red 670 alone) for HA expression on Lin−Rhdull cells. (D) HABP binding to Lin−Rhdull cells. (E) HABP binding to Lin−Rhdull cells following hyaluronidase pretreatment. The percentages represent the proportion of positive cells.
HA expression on murine BM HSCs.
(A) Sca-1 and Kit expression characteristics of Lin−murine BM cells; R1 sort region for HSCs. (B) HA expression on all R1 HSCs (gray line) as detected using HABP. (C) Control (Red 670 alone) for HA expression on Lin−Rhdull cells. (D) HABP binding to Lin−Rhdull cells. (E) HABP binding to Lin−Rhdull cells following hyaluronidase pretreatment. The percentages represent the proportion of positive cells.
HA synthase expression in murine BM subpopulations.
Has-1, Has-2, and Has-3 PCR products were specifically amplified under RT-PCR conditions described in “Materials and methods.” BMS2 stromal cell line: positive control known to express HA synthases.
HA synthase expression in murine BM subpopulations.
Has-1, Has-2, and Has-3 PCR products were specifically amplified under RT-PCR conditions described in “Materials and methods.” BMS2 stromal cell line: positive control known to express HA synthases.
HA expression on human HPCs
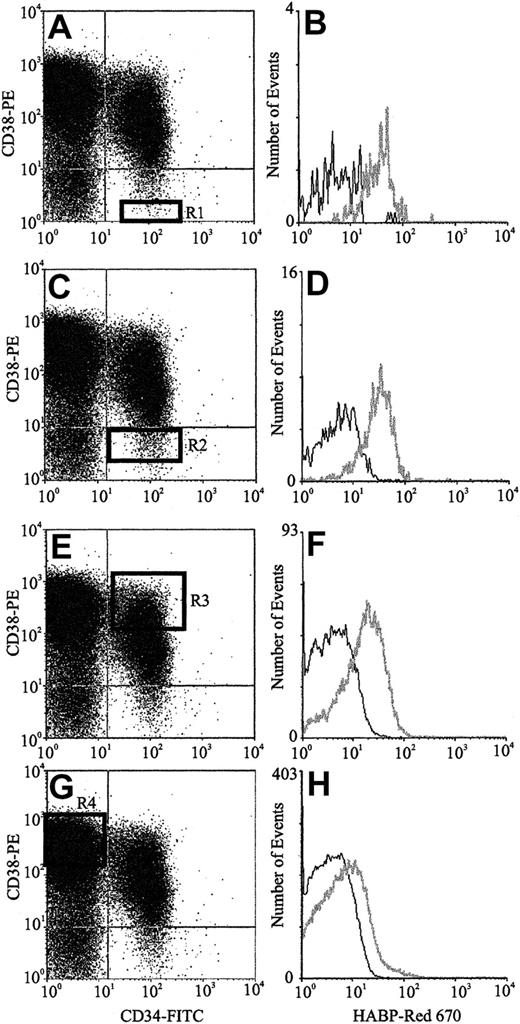
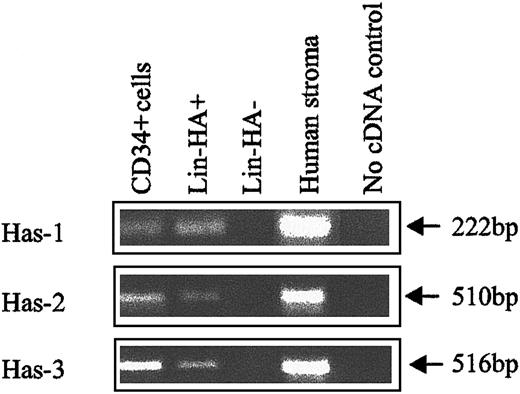
The expression of HA by primitive BM cells is not a unique feature of the mouse but is also a characteristic of human HPCs. FACS analysis of human CB (Figure 3) using CD34, CD38, and HABP showed that putative human HSCs (CD34+CD38− cells) exhibited the highest levels of HA expression. Furthermore, as demonstrated by the histograms of Figure 3B,D,F,H, HA expression decreased as HPC matured, as evidenced by acquisition of CD38 and decreased expression of CD34. In accordance with data in the mouse, isolated human CD34+and Lin−HA+ cells but not Lin−HA− also express HAS-1, HAS-2, and HAS-3 (Figure 4).
HA expression on human CB progenitors.
Scatter properties of Lin− CB cells. (Panels A,C,E,G) CD34 and CD38 expression of maturing progenitors: R1 to R4, respectively (Panels B,D,F,H). HA expression on each progenitor subpopulation (gray line) as detected using HABP.
HA expression on human CB progenitors.
Scatter properties of Lin− CB cells. (Panels A,C,E,G) CD34 and CD38 expression of maturing progenitors: R1 to R4, respectively (Panels B,D,F,H). HA expression on each progenitor subpopulation (gray line) as detected using HABP.
HA synthase expression in human CB subpopulations.
Has-1, Has-2, and Has-3 PCR products were specifically amplified under RT-PCR conditions described in “Materials and methods.” Human stroma: positive control known to express HA synthases.
HA synthase expression in human CB subpopulations.
Has-1, Has-2, and Has-3 PCR products were specifically amplified under RT-PCR conditions described in “Materials and methods.” Human stroma: positive control known to express HA synthases.
HA synthesis by human HPCs
To exclude the possibility that the HA detected on primitive hematopoietic cells was bound to the cells by a CD44-independent mechanism, but was actually produced by these cells, we examined whether primitive human HPCs synthesize and secrete HA. Putative human HSCs (CD34+CD38−) synthesized HA of 3 different molecular weights (high, > 100 000 Da; intermediate, 48 000 Da; and low, 20 000 Da) when cultured for 3 to 4 days in serum-free conditions in the presence of multiple cytokines and D-[6-3H]-glucosamine hydrochloride (Figure5). Isolated HSCs were CD45+, and in none of the synthesis assays did any of the cells adhere to the plastic culture wells, demonstrating an absence of contaminating stromal cells. These results are consistent with the expression of the 3-hyaluronan synthases Has-1, Has-2, and Has-3 by this population of HPCs (Figure 4). Collectively these data demonstrate that HA is synthesized by and expressed on human hemopoietic populations enriched for HSCs.
HA synthesis by human HPCs.
HA synthesis by CB CD34+CD38− cells following 60 hours in culture demonstrated using Superose 12 size exclusion chromatography by the difference in [3H] counts eluted at fractions 15 to 25 before digestion withStreptomyces hyaluronidase (○) versus total digestion of high-molecular-weight material to the enzymatic degradation products tetrasaccharides and hexasaccharides (●). The 3 peaks represent HA within the cell supernatant. The molecular weights of synthesized HA were determined by comparison with eluted purified HA of known molecular size. For calculation of recovery, counts were taken as significant when they were more than 3 SDs above the mean background (n = 15).
HA synthesis by human HPCs.
HA synthesis by CB CD34+CD38− cells following 60 hours in culture demonstrated using Superose 12 size exclusion chromatography by the difference in [3H] counts eluted at fractions 15 to 25 before digestion withStreptomyces hyaluronidase (○) versus total digestion of high-molecular-weight material to the enzymatic degradation products tetrasaccharides and hexasaccharides (●). The 3 peaks represent HA within the cell supernatant. The molecular weights of synthesized HA were determined by comparison with eluted purified HA of known molecular size. For calculation of recovery, counts were taken as significant when they were more than 3 SDs above the mean background (n = 15).
HA is expressed on cells capable of reconstituting the hemopoietic compartment
To determine whether the HA+ cells were capable of reconstituting the hemopoietic compartment of a lethally ablated recipient, FACS was used to isolate Lin−HA+and Lin−HA− subpopulations and the cells were assayed in vivo using the congenic Ly5.1/Ly5.2 mouse transplant model.26 A total of 5000 to 6000 Lin−HA+ cells reconstituted all hemopoietic lineages in the peripheral blood of lethally irradiated recipients 8 weeks after transplantation (Table 1), whereas an equivalent number of Lin−HA− cells failed to do so, with the recipients dying 14 to 16 days after transplantation. Isolating the more enriched HSC Lin−Sca+ population, and subfractionating on the basis of HA, resulted in reconstitution of multiple hemopoietic lineages of lethally irradiated recipients 4 weeks after transplantation of 500 Lin−Sca+HA+cells (Table 1). However, recipients given transplants of 500 Lin−Sca+HA+ cells exhibited an average of only 9.4% donor engraftment, which is significantly lower than predicted from infused unfractionated Lin−Sca+ cells. Studies by Spangrude and Scollay demonstrate that transplanting as few as 100 Lin−Sca+ cells results in the survival of greater than a third of recipients with more than 75% donor cells in the peripheral blood 12 weeks after transplantation.27 In only 1 of 6 recipients were HA− cells capable of rescuing lethally irradiated recipients (following a transplant of 500 Lin−Sca+HA− cells), suggesting that fewer long-term repopulating cells (LTRCs) are contained within this population compared with the HA+ fraction. In the one surviving recipient, there was an equivalent proportion of donor cells to that seen following a transplant of HA+cells (about 9%). The reason for this lower than expected level of engraftment remains unclear, but as discussed, is suggestive of HA playing an active role in homing or lodgment of HSCs to and within the BM.
Analysis of the ability of Lin− cells expressing HA to reconstitute hemopoiesis in vivo
Mice were injected with either 5000 to 6000 Lin−HA+ cells (4 mice) or 500 Lin−Sca+HA+ cells (2 mice) and analyzed 4 to 8 weeks after transplantation. Peripheral blood was collected and the red blood cells lysed using ammonium chloride. The percentage of donor cells was determined using Ly5.1 antibody labeling and was 9% ± 1.7% and 9.4% ± 0.02% following transplantation of 5000 to 6000 Lin−HA+ cells or 500 Lin−Sca+HA+ cells, respectively. White blood cells were labeled for macrophages and granulocytes (Mac-1/Gr-1), B cells (B220), and T cells (CD4/CD8) and the proportion of donor analyzed using an Ly5.1 antibody.
UN indicates unknown.
Percent of each subpopulation within the donor cells detected following a transplant of each subpopulation.
When 10-fold more Lin−Sca+HA−cells were transplanted, all recipients were reconstituted, demonstrating that not all LTRCs are in the HA fraction. Due to the presence of HABP on the surface of the cell appearing to affect the ability of transplanted cells to home and lodge, it is presently not possible to do competitive reconstituting experiments to accurately fractionate the LTRCs between HA+ and HA−subpopulations.
The potential role of HA in the lodgment of transplanted murine HSCs
Transplantations were performed using HSCs and HPCs incubated with and without HY to investigate the role of HA in cell lodgment. When Lin−Rhdull cells were treated with HY prior to transplantation, there was an approximate 40% reduction in the proportion of cells located at the endosteum 15 hours after transplantation compared with untreated cells (39% ± 3% and 65% ± 8%, respectively; n = 4). In contrast, HY treatment of Lin−Rhbright cells, shown not to synthesize HA or transcribe the HAS genes (data not shown), did not alter their spatial distribution at the same time point (41% ± 3% and 37% ± 4% respectively; n = 3). This suggests a specific functional role for HA in determining the spatial localization of HSCs but not HPCs.
The potential role of HA as a negative regulator of HSC proliferation
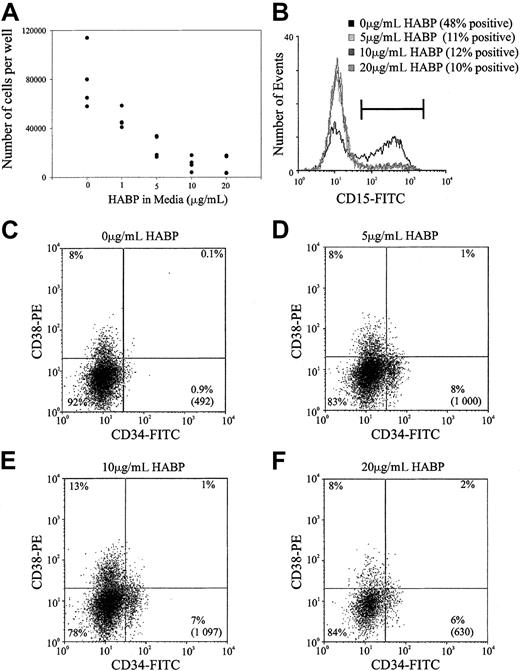
Previous studies within this and other laboratories have demonstrated that ligation of certain cell surface adhesion molecules on primitive hemopoietic cells such as PSGL-1, CD164, and CD43 can result in the in vitro inhibition of HSC proliferation.28Accordingly, the possibility that the cross-linking of HA by its surrogate ligand HABP could result in similar growth inhibition was investigated in stroma-free, cytokine-dependent cultures. As demonstrated in Figure 6, the ligation of HA by HABP results in a profound suppression of human HSC proliferation stimulated by a potent combination of early acting hemopoietic growth factors. This resulted in a dose-dependent inhibition of cell proliferation that was maximal at 10 μg/mL HABP in both the human (Figure 6A) and murine (data not shown) cultures. In addition to growth inhibition, incubating human HSCs in increasing amounts of HABP inhibited myeloid differentiation as evidenced by the decreased number of CD15+ cells generated in culture (Figure 6B). In accordance with the inhibition of cell differentiation, incubating cells with increasing amounts of HABP also maintained a population of cells characterized by a CD34+CD38− phenotype (Figure 6C). It should be noted that the presence of this phenotype in culture does not necessarily indicate immature progenitors and future transplant experiments after culture in the presence of HABP are required to confirm this.
Representative experiment of HA-HABP binding negatively regulating HSC proliferation and differentiation.
(A) Number of progenitors produced per 320 CD34+CD38− CB HSCs seeded in serum-free culture for 6 days with various concentrations of HABP. (B) CD15 expression of cells cultured in various concentrations of HABP in Figure 6A. (C-F) CD34 and CD38 expression of cells cultured in various HABP concentrations in Figure 6A. The percentage in each quadrant represents the proportion of cultured cells expressing the particular phenotype. The number in parentheses represents the total number of CD34+CD38− cells in the culture compared with the original 320 cells seeded.
Representative experiment of HA-HABP binding negatively regulating HSC proliferation and differentiation.
(A) Number of progenitors produced per 320 CD34+CD38− CB HSCs seeded in serum-free culture for 6 days with various concentrations of HABP. (B) CD15 expression of cells cultured in various concentrations of HABP in Figure 6A. (C-F) CD34 and CD38 expression of cells cultured in various HABP concentrations in Figure 6A. The percentage in each quadrant represents the proportion of cultured cells expressing the particular phenotype. The number in parentheses represents the total number of CD34+CD38− cells in the culture compared with the original 320 cells seeded.
Discussion
HA is a major constituent of the ECM in a wide variety of tissues. For example, HA is a major ECM constituent within the vitreous of the human eye, and although representing less of the physical mass of the tissue HA serves as an essential structural element within the ECM of cartilage. In the BM, HA is the third main glycosaminoglycan produced by stromal cells, with HA synthesis hypothesized to be an initial step in ECM organization.29 HA has been shown to account for 40% of the glycosaminoglycans in BM stromal cultures17,30and interacts through its principal ligand CD4419 with T and B cells and neutrophils.31 However, to date there are no reports of hemopoietic cells synthesizing HA.
This study is the first report of hemopoietic cells themselves synthesizing and expressing HA. Of great interest but as yet unknown significance, this synthesis and expression was largely restricted to very primitive cells in both murine and human systems. It would also be of great interest to determine if stem cells in other tissues and organs similarly are distinguished by their synthesis of HA.
Our data demonstrate that HSCs synthesize and express HA from all 3 of the HAS synthases. Previous analysis of the size distribution of HA generated in vitro by the recombinant proteins demonstrated that HAS-1 and HAS-2 synthesize HA with molecular masses of 2 × 105to about 2 × 106 Da. In contrast HAS-3 synthesized shorter HA of 1 × 105 to 1 × 106Da.32 Has-1 is known to produce HA that is extruded from the cell. In contrast Has-2 produces HA that forms a pericellular coat, has been correlated with increased cell migration, and been associated with increased HA production in solid tumors.21 The physiologic significance of why cells transcribe the 3 differentHAS genes and produce 3 molecular weight species of HA remains unknown, but may provide a mechanism for the flexible control of HA synthesis and resulting function. In the current study, putative HSCs isolated from human CB were shown to extrude HA of 3 molecular weights into the culture media. In contrast, no HA was detected in the cell lysates (data not shown), suggesting that the higher molecular weight HA was predominately synthesized by HAS-1 and not HAS-2, and the lower molecular weight HA was synthesized by HAS-3. This is further supported by the abundance of HAS-1 mRNA detected from these cells compared to HAS-2.
Our recent development of a methodology allowing us to track transplanted cells at the individual cell level as they engraft and lodge within the BM has provided a unique and very powerful tool for identifying and characterizing key molecules involved in these processes.6 HA is the first molecule identified in this manner that appears to have a significant impact on the lodgment of engrafting HSCs. In a previous study we showed that more than 60% of an enriched population of HSCs lodged within the endosteal region 15 hours after transplantation.6 However, following HY pretreatment, the proportion of cells residing at the endosteum at the same time point was significantly reduced to about 40% of HSCs. Further studies will help to resolve whether HA is playing a dual role in both the migration and anchoring of HSCs in this lodgment process.
In addition, the present study also showed the binding of HA on HSCs by HABP, acting as a surrogate ligand, inhibits the proliferation and differentiation of these cells. This role is in accordance with a recent report showing that the addition of HA to in vitro long-term bone marrow cultures (LTBMCs) results in perturbation of hemopoiesis.33 In this study, exogenous HA was added to murine LTBMCs, resulting in enhanced production of both progenitors and mature BM cells, whereas the addition of HY resulted in the inhibition of cell production. In these cultures, HA was thought to be regulating hemopoiesis through a progenitor ancestral to the common myeloid and lymphoid progenitor. In addition, our observed growth regulatory role of HA is very similar to that recently described for the sialomucin PSGL-1/P-selectin interaction.28 In addition to its well-documented role as an adhesion molecule, PSGL-1 has now been shown to be a potent negative regulator of human hemopoietic progenitors in vitro.
In vivo there are a number of potential receptors of HA in the marrow, of which CD44H from the Link superfamily is the most widely recognized. CD44 is present on many cell types in a nonfunctional form that requires some activation process to acquire HA-binding capabilities. The regulation of CD44 activity is highly complex and is influenced by glycosylation status, differential splicing to generate distinct isoforms, cytoskeletal attachment, receptor density on the cell surface, and phosphorylation of the intracytoplasmic domain.34,35 Receptor for hyalurenan acid–mediated motility (RHAMM) is also an HA receptor expressed in the BM, which has recently been identified as the major HA-binding receptor expressed by mobilized blood HPCs.36 Furthermore, the study by Pilarski et al37 suggests that RHAMM and CD44 played key roles in cell motility and trafficking after mobilization versus adhesion to the ECM by marrow HPCs, respectively. Further studies will help to define whether the interactions of HA with these 2 receptors may also be responsible for the functions of HA on HSC described in this study.
Overall, our data comprise the first demonstration of the synthesis and expression of HA by HSCs in vitro and in vivo, representing a very specific but as yet unrecognized characteristic of primitive hemopoietic cells in at least 2 mammalian species. The presence of HA on HSCs is functionally significant in stem cell biology, specifically in the regulation of HSC lodgment after transplantation. In addition, demonstrating inhibition of cell proliferation and differentiation through the interaction of HA and the surrogate ligand HABP suggests an important role for this glycosaminoglycan in vivo in regulating HSCs within the hemopoietic “niche.” Potentially, these processes could be occurring through interactions with the 2 HA receptors RHAMM and CD44H.
The authors would like to acknowledge Kate Sells for her help with the animal work, Brenda Williams for the isolation of different BM subpopulations, and Ralph Rossi and Andrew Fryga for their help with flow cytometry. The authors would also like to acknowledge Angie Dinning at the Mercy Hospital for Women for her help in coordinating cord blood collections and Dr Tak Mak for providing the CD44 knockout mice.
Prepublished online as Blood First Edition Paper, September 12, 2002; DOI 10.1182/blood-2002-05-1344.
S.K.N. is an RD Wright Fellow, granted from the National Health and Medical Research Council (Canberra, ACT, Australia).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Susan K. Nilsson, Stem Cell Laboratory, Peter MacCallum Cancer Institute, Locked Bag No. 1, A'Beckett St, Melbourne, 3000, Australia; e-mail:s.nilsson@pmci.unimelb.edu.au.

![Fig. 5. HA synthesis by human HPCs. / HA synthesis by CB CD34+CD38− cells following 60 hours in culture demonstrated using Superose 12 size exclusion chromatography by the difference in [3H] counts eluted at fractions 15 to 25 before digestion withStreptomyces hyaluronidase (○) versus total digestion of high-molecular-weight material to the enzymatic degradation products tetrasaccharides and hexasaccharides (●). The 3 peaks represent HA within the cell supernatant. The molecular weights of synthesized HA were determined by comparison with eluted purified HA of known molecular size. For calculation of recovery, counts were taken as significant when they were more than 3 SDs above the mean background (n = 15).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/3/10.1182_blood-2002-05-1344/4/m_h80333733005.jpeg?Expires=1769475497&Signature=sR4JiKMT09bTPQe1tDpRIWexBqYGamJHxRazlr3JWrFqJ8tSZjeIUo32UA4kuW2Dkv7dzqAyedQmB~e8axi-R~lDyXk5jvdgBCXARJko079zERbVT9AsWsLvGVU6UnnRvzfh9lsay90y~symzvkP4HJcPxZoSvkqIhuO-QQmXz-5~OsZ6bJIu53vRDuayobSQXDzZSjT2rodsHCO8ifdRiXGhjfS~J3Mwq~aGlZkxS4ZJhNwOVL7MRxTto7TgbRrB9-qWNp0eOFuo0J~w2RVP~lRAi7EWNOiZmYOFuBBc4RHQ49rjg6iinWeUnFJRCkzNFffP0BtvQSNqXmkyvCnrg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal