Abstract
The c-fms gene encodes the receptor for macrophage colony-stimulating factor (CSF-1). The gene is expressed selectively in the macrophage and trophoblast cell lineages. Previous studies have indicated that sequences in intron 2 control transcript elongation in tissue-specific and regulated expression of c-fms. In humans, an alternative promoter was implicated in expression of the gene in trophoblasts. We show that in mice, c-fmstranscripts in trophoblasts initiate from multiple points within the 2-kilobase (kb) region flanking the first coding exon. A reporter gene construct containing 3.5 kb of 5′ flanking sequence and the downstream intron 2 directed expression of enhanced green fluorescent protein (EGFP) to both trophoblasts and macrophages. EGFP was detected in trophoblasts from the earliest stage of implantation examined at embryonic day 7.5. During embryonic development, EGFP highlighted the large numbers of c-fms–positive macrophages, including those that originate from the yolk sac. In adult mice, EGFP location was consistent with known F4/80-positive macrophage populations, including Langerhans cells of the skin, and permitted convenient sorting of isolated tissue macrophages from disaggregated tissue. Expression of EGFP in transgenic mice was dependent on intron 2 as no lines with detectable EGFP expression were obtained where either all of intron 2 or a conserved enhancer element FIRE (theFms intronic regulatory element) was removed. We have therefore defined the elements required to generate myeloid- and trophoblast-specific transgenes as well as a model system for the study of mononuclear phagocyte development and function.
Introduction
The differentiation of macrophages from bone marrow progenitors requires the coordinated expression of many genes needed for mature cell function. This process is controlled by the lineage-specific growth factor, macrophage colony-stimulating factor (CSF-1), which acts by binding to cell-surface receptors (CSF-1R) encoded by the c-fms proto-oncogene.1We2-4 and others5,6 have studied the transcriptional regulation of the c-fms gene as a route to understanding lineage commitment in the macrophage lineage. The c-fms mRNA is detectable in the earliest yolk sac phagocytes formed during mouse development, prior to many other markers, including the macrophage-restricted transcription factor PU.1, and the expression in the embryo and adult mouse is largely restricted to cells of the macrophage lineage.7,8 The only other major site where the c-fms gene is expressed is in placental trophoblasts.9,10 In humans, there is a trophoblast-specific promoter that lies at the 3′ end of the platelet-derived growth factor receptor–β (PDGFR-β) gene, some 20 kilobase (kb) upstream of the first coding exon.5 11
The murine exon 2 c-fms promoter was more active in transient transfections of a macrophage cell line, RAW264, than in untransformed fibroblasts,12 but it was also active in a wide range of tumor cell lines that do not express the full-length endogenous mRNA.3,13 Tumor cells in which the promoter was active were shown to produce c-fms transcripts that contained exon 2 and extended into the downstream intron 2, but did not have detectable full-length c-fms mRNA. Inclusion of intron 2 in reporter gene constructs abolished reporter gene expression in nonmacrophage tumor cells, but significant activity was retained in RAW264 macrophages.3 We showed elsewhere that intron 2 contains a DNase 1 hypersensitive site we refer to as theFms intronic regulatory element (FIRE). FIRE was required for maximal expression of a reporter gene in stably-transfected RAW264 macrophages.4 The intronic sequence without FIRE actually profoundly suppressed reporter gene expression driven by the exon 2 promoter.4
In this paper, we show that the proximal promoter of c-fmscombined with the first intron directed consistent expression of the enhanced green fluorescent protein (EGFP) reporter gene in the same locations as the endogenous gene, and appropriate expression required the intronic elements described previously. We demonstrate that thefms-EGFP reporter gene provides a definitive marker for cells of the mononuclear phagocyte lineage throughout embryonic development, in the bone marrow and peripheral blood and in all adult tissues. These findings provide a framework for the use of the c-fms promoter in applications where targeted manipulation of macrophage or trophoblast differentiation in transgenic animals is desired.
Materials and methods
Analysis of c-fms transcripts in placental RNA
Total RNA was extracted from embryonic day 14 (e14) placentae as described.9 Aliquots were subjected to 4 sets of 5′ rapid amplification of cDNA ends (RACE) experiments according to the manufacturer's instructions (Invitrogen, Carlsbad, CA) using reverse orientation (3′→5′) primers located in exon 2. Primer sequences for 5′RACE experiments are described in Table1. Products of the reactions were cloned into a TA vector (Invitrogen) and sequenced. The mouse c-fmssequence accession number is AF290879.
Primers used in the 5′RACE experiments
| Experiment set . | Primer name . | Sequence . | Purpose . |
|---|---|---|---|
| First set | JP170 | 5′-GCCAGCAAATTCCAAGAG-3′ | Reverse transcription |
| JP164 | 5′-ATCGCAGGGTCACCGTTTCACCCGGC-3′ | First PCR | |
| JP134 | 5′-GACCATGCCAAACTGTGGCCAGCA-3′ | Second PCR | |
| Second set | JP170 | 5′-GCCAGCAAATTCCAAGAG-3′ | Reverse transcription |
| JP169 | 5′-AAGCTCGGTACAACGGTA-3′ | First PCR | |
| JP164 | 5′-ATCGCAGGGTCACCGTTTCACCCGGC-3′ | Second PCR | |
| Third set | JP193 | 5′-CCAGGGGATAGCGTGAGC-3′ | Reverse transcription |
| JP192 | 5-GATGCAGGTTGGAGAGTCG-3′ | PCR | |
| Fourth set | JP194 | 5′-AGGCCCCAACTCCATAGC-3′ | Reverse transcription |
| JP193 | 5′-CCAGGGGATAGCGTGAGC-3′ | PCR |
| Experiment set . | Primer name . | Sequence . | Purpose . |
|---|---|---|---|
| First set | JP170 | 5′-GCCAGCAAATTCCAAGAG-3′ | Reverse transcription |
| JP164 | 5′-ATCGCAGGGTCACCGTTTCACCCGGC-3′ | First PCR | |
| JP134 | 5′-GACCATGCCAAACTGTGGCCAGCA-3′ | Second PCR | |
| Second set | JP170 | 5′-GCCAGCAAATTCCAAGAG-3′ | Reverse transcription |
| JP169 | 5′-AAGCTCGGTACAACGGTA-3′ | First PCR | |
| JP164 | 5′-ATCGCAGGGTCACCGTTTCACCCGGC-3′ | Second PCR | |
| Third set | JP193 | 5′-CCAGGGGATAGCGTGAGC-3′ | Reverse transcription |
| JP192 | 5-GATGCAGGTTGGAGAGTCG-3′ | PCR | |
| Fourth set | JP194 | 5′-AGGCCCCAACTCCATAGC-3′ | Reverse transcription |
| JP193 | 5′-CCAGGGGATAGCGTGAGC-3′ | PCR |
To construct a representative placental library, poly A+-selected e14 placental RNA was converted into cDNA using both random and oligo dT priming. This cDNA was cloned into a λ Zap vector (Clontech, Palo Alto, CA). Average library insert size was 1.5 kb and its complexity was 1.5 × 106 independent clones. This library was probed with a [32P]-dCTP–labeled full-length c-fms cDNA in the first round followed by a c-fms exon 2 probe in the second round. The 5′ ends of the selected inserts were sequenced.
For Northern blot analysis, 20 μg total RNA was separated by formaldehyde-agarose gel electrophoresis, transferred to nylon filters, and probed with a [32P]-dCTP–labeled cDNA probe specific for exon 1 or 2 of c-fms. Samples included RNA from trophoblastic stem cells (TS)14 (generously provided by Dr Janet Rossant, Toronto, ON, Canada), a macrophage cell line (BAC1.2F5),15 and e10 and e14 placentae, all of which are c-fms–positive9 10 as well as the c-fms–negative L-cell line and mouse embryo fibroblasts (MEF).
Plasmid constructs
Some of the reporter constructs that were used in this study have been described previously,3 except that the luciferase gene was replaced by an enhanced variant of the green fluorescent protein gene derived from the pEGFP-N1 vector (Clontech). Briefly, the constructs used were the 3.5 kb sequence flanking mouse c-fms exon 2 driving the EGFP gene (p3.5fms-EGFP), the 3.5 kb exon 2 promoter plus the whole intron 2 sequence driving the EGFP gene (p7.2fms-EGFP), and p7.2fms-EGFP with the deletion of conserved FIRE region on intron 2 driving the EGFP gene (p7.2fmsΔFIRE-EGFP). The 7.2fms promoter has been renamed from 6.7fms, to reflect accurate sizing based on the full-length sequence (accession number AF290879). All plasmids were prepared on QIAGEN affinity columns according to the manufacturer's instructions. For generation of transgenic mice, plasmid backbones were removed by restriction digestions, and the transgenes used for DNA injection were gel-purified by using the QIAquick gel extraction kit (QIAGEN, Clifton Hill, Victoria, Australia).
Generation of transgenic mice
Transgenic (TG) mice were generated at the Transgenic Animal Service of Queensland, Brisbane, Queensland, Australia (www.tasq.uq.edu.au) by injection of the transgenes into pronuclei of (C57BL/6 × CBA)F1 (BCBF1) fertilized eggs. TG mice generated were maintained under specific pathogen-free conditions. The integration of the transgenes was investigated by polymerase chain reaction (PCR) analysis of tail biopsy DNA, amplifying the EGFP gene by using primers 5′-CTGGTCGAGCTGGACGGCGACG-3′ (forward) and 5′-CACGAACTCCAGCAGGACCATG-3′ (reverse). The amplification temperatures were 1 minute at 95°C, 1 minute at 60°C, and 1 minute at 72°C for 25 cycles after an initial denaturing step of 5 minutes at 95°C. Southern blot analyses were also conducted on genomic DNAs of some mice for confirmation of genotype using standard protocols.
Cells and tissue culture
Thioglycollate-elicited peritoneal macrophages (TEPMs) were isolated from peritoneal cavities after intraperitoneal injections of 1 mL of 10% thioglycollate broth, followed by peritoneal lavage with phosphate-buffered saline (PBS) 3 to 4 days later. Pulmonary macrophages were obtained from broncho-alveolar lavage (BAL), whereas bone marrow–derived macrophages (BMMs) were obtained by isolation of bone marrow cells from the femurs of adult mice followed by differentiation of cells in complete RPMI medium (Invitrogen) supplemented with 10% fetal bovine serum (Serum Supreme, BioWhittaker, Walkersville, MD) and 2 mM l-glutamine (Glutamax, Invitrogen), 20 U/mL penicillin, 20 μg/mL streptomycin (Invitrogen), and 100 U/mL CSF-1 (Chiron) for 7 days.16 For dual-color fluorescence activated cell sorting (FACS) analysis of splenocytes (Figure 5A) using Mac-1 (CD11b) and F4/80 antibodies, spleen cells were mechanically disaggregated by mincing the tissues using sterile scalpel blades. Spleen adherent cells were enriched by overnight incubation on bacteriologic petri dishes, to which they adhere weakly, followed by removal of the tissue culture (TC) media and floating cells. The remaining adherent cells were harvested by squirting the petri dish surface with medium using a 20-mL syringe and an 18-gauge needle, and were subjected to FACS analysis after washing in PBS. For FACS analysis of tissue macrophages in spleen, liver, and lung, enzymatic digestions were performed with 0.1 U/mL Collagenase and 0.8 U/mL Dispase (Roche, Castle Hill, Australia) in PBS/20% fetal calf serum (FCS)7 for 1 hour at 37°C. Dispersed cells were washed in PBS and strained through 100-μm cell strainers (BD Falcon, Bedford, MA) and single cell suspensions were subjected to FACS. For isolation of cells from intestinal lamina propria, epithelial cells were washed in calcium- and magnesium-free Hanks balanced salt solution (Invitrogen) and minced tissues were enzymatically digested as described for other tissues above. Disaggregated cells were separated into low- and high-density fractions via centrifugation on Nycodenz (Nycomed Pharma, Norway) gradient prior to FACS analysis.17
Trophoblast cell examination in tissue culture was conducted by culturing the ectoplacental cone (EC)18 from e6.5 transgenic mice embryos. Briefly, the concepti were dissected free from the decidua under a stereomicroscope, the EC was then cultured on each well of 24-well TC plates, in modified Eagle medium (MEM) α-modification medium (Invitrogen) supplemented with 10% FCS, 20 U/mL penicillin, and 20 μg/mL streptomycin, in the presence or absence of 100 U/mL CSF-1 (Chiron).
Flow cytometry
Examination of EGFP+ cells by flow cytometry was done on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA) and the data were analyzed on CellQuest software (Becton Dickinson). Isolated macrophages were directly subjected to FACS analyses after being washed and resuspended on 1XPBS. For c-fms 2-color FACS analyses, isolated cells were washed once in ice-cold PBS containing 0.1% bovine serum albumin (BSA) and 0.1% NaN3. Nonspecific binding was blocked by incubation of cells in 0.1% goat serum for 15 minutes, followed by incubation with anti–c-fms antibody (1:100 dilution of hybridoma supernatant19) for 30 minutes. Cells were then washed with PBS containing 0.1% BSA and 0.1% NaN3, and incubated with goat anti–rat phycoerythrin (PE) F(ab′)2 (Serotec, Oxford, United Kingdom) for 30 minutes. Cells were subsequently washed with PBS containing 0.1% BSA and 0.1% NaN3, resuspended, and analyzed by FACS. For Mac-1 and F4/80 staining, the same procedure was conducted, except that PE-conjugated anti–Mac-1 or anti–F4/80 (Serotec) antibodies were used. For peripheral blood mononuclear cell (PBMC) analysis, mature red cells were lysed with lysis buffer (155 mM NH4CL, 10 mM KHCO3, and 0.1 mM EDTA [ethylenediaminetetraacetic acid]) prior to antibody staining.
Histology and EGFP examination
Various tissues were directly fixed for 2 hours in 4% paraformaldehyde in PBS after dissections, followed by an overnight incubation in 18% sucrose at 4°C. On the following day, tissues were embedded in Tissue-Tek OCT compound (Sakura Finetechnical, Tokyo, Japan) and frozen in liquid nitrogen. Frozen sections (8 μm to 12 μm thick) were cut at −16°C with a LEICA cryostat model CM3050 (Leica Instrument, Germany), and mounted in DAKO fluorescent mounting medium (DAKO, CA). The fluorescence of the EGFP was visualized under an Olympus AX70 microscope (Olympus Australia, Melbourne) and pictures were either taken using Kodak film or digitally acquired using Spot RT Colour digital camera model 2.2.0 (Diagnostic Instruments, Sterling Heights, MI). For Langerhans cell examination, mouse ears were separated into dorsal and ventral halves with forceps. Dorsal ear halves were incubated in 20 mM EDTA/PBS for 1 hour. Epidermal sheets were then mounted on glass slides. For whole-mount embryo observations, freshly dissected embryos were usually unfixed or fixed with 4% paraformaldehyde and visualized in PBS under an inverted Olympus AX70 microscope. Yolk sac examination was performed in embryos derived from nontransgenic mothers mated to transgenic males.
Results
Start site analysis
The region flanking the first coding exon (exon 2) in both mouse and human genes contains the transcription start site used in macrophages. Matrix alignment of the region flanking this exon in mouse (accession number AF290879) and human (U63963, X14 720) reveals no sequence homology outside of the 3′ end of the PDGFR gene and the 500-bp flanking exon 2 (data not shown). This finding suggested that the exon 1 promoter identified in the human c-fms gene, immediately distal to the PDGFR locus,5 11 might not be conserved in the mouse.
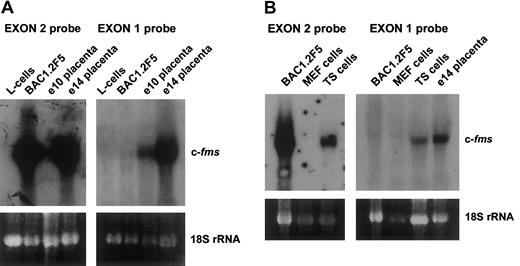
To determine whether mouse and human genes employ different transcriptional mechanisms to express c-fms mRNA in trophoblastic tissues, we performed systematic 5′RACE analyses on mouse e14 placental RNA utilizing primers based on the known mouse exon 2 sequence. The products were sequenced and revealed a heterogeneous set of spliced transcripts arising from the region from −280 to −400 relative to the fms ATG start codon in exon 2. To further explore the sequences of the trophoblastic transcriptional start site, an e14 placental library was probed with c-fms as described in “Materials and methods.” Sequences in the isolated clones upstream of exon 2 confirmed the heterogenous nature of the upstream sequences with no clones extending beyond −400. The start sites and splicing patterns of various transcripts are shown in Figure1A. These data indicate that mouse placenta utilizes a quite distinct promoter from the human choriocarcinoma lines previously studied. Figure 1B shows the splicing variants of exon 1. Of the splice donor sites used in the mouse transcripts, only the downstream site at −268 has a likely equivalent in the human gene. To confirm the 5′RACE data, separate probes were constructed to detect the putative exon 1 sequence and used in Northern analysis. Figure 2A-B shows that an exon 1 probe detected c-fms mRNA in the TS trophoblast cell line, and in e10 and e14 placentae, where c-fms mRNA in the macrophage line BAC1.2F5 was detected only with an exon 2 probe. No transcripts for either exon were detected in mouse L cells or MEFs, neither of which express c-fms. These data show the presence of immediate upstream sequences of exon 2 which are uniquely expressed in trophoblastic tissues.
DNA sequence of the upstream elements of the mouse c-fms gene encompassing exon 1.
Panel A shows 1 kb of the 5′ upstream sequence before the translational start site “ATG” (boxed) within exon 2. The arrow at “−72” indicates the beginning of exon 2. The arrowheads indicate the 3′ boundaries of exon 1. Boldfaced letters specify the splicing donor and acceptor sites. The sequence of exon 1 probe used for analysis in Figure 2 is underlined. (B) The splicing variants of exon 1. The number of arrowheads at each location represents the number of appearances of different exon 1 start sites from the sum of both the 5′RACE and e14 placental library screening experiments. GA and GT are splicing donor sites while AG and CAG are splice acceptors. The arrow shows the beginning of exon 2.
DNA sequence of the upstream elements of the mouse c-fms gene encompassing exon 1.
Panel A shows 1 kb of the 5′ upstream sequence before the translational start site “ATG” (boxed) within exon 2. The arrow at “−72” indicates the beginning of exon 2. The arrowheads indicate the 3′ boundaries of exon 1. Boldfaced letters specify the splicing donor and acceptor sites. The sequence of exon 1 probe used for analysis in Figure 2 is underlined. (B) The splicing variants of exon 1. The number of arrowheads at each location represents the number of appearances of different exon 1 start sites from the sum of both the 5′RACE and e14 placental library screening experiments. GA and GT are splicing donor sites while AG and CAG are splice acceptors. The arrow shows the beginning of exon 2.
Transgenic animal characterization
The 7.2fms-EGFP was microinjected into mouse embryos by standard methods, and progeny were screened for incorporation of the transgene by PCR and Southern blot (data not shown) analyses. There were 6 transgenic lines produced, but one did not transmit the transgene. For all of the 7.2fms-EGFP transgenic lines, EGFP was detected readily in peritoneal, bone marrow–derived, and broncho-alveolar lavage macrophages (Figure3B). The level of EGFP expression in macrophages from 3 lines tested by flow cytometry was remarkably consistent (Table 2).
Differential expression of murine exon 1 in trophoblast cells.
(A) Exon 1 is expressed only in mouse placenta. This panel shows 20 μg total RNA from L cells, a fibroblast cell line which does not express the fms gene, c-fms–positive macrophage cell line BAC1.2F5 cells, and e10 and e14 mouse placental RNA, probed with either an exon 2 or an exon 1 probe. (B) Exon 1 is expressed in trophoblastic cells. BAC1.2F5, mouse embryo fibroblasts (MEFs), trophoblastic stem cells (TSs), or e14 placentae were probed with exon 1 or exon 2 probes. In each panel the ethidium-bromide staining of 18S rRNA was used to monitor the loading of the RNAs.
Differential expression of murine exon 1 in trophoblast cells.
(A) Exon 1 is expressed only in mouse placenta. This panel shows 20 μg total RNA from L cells, a fibroblast cell line which does not express the fms gene, c-fms–positive macrophage cell line BAC1.2F5 cells, and e10 and e14 mouse placental RNA, probed with either an exon 2 or an exon 1 probe. (B) Exon 1 is expressed in trophoblastic cells. BAC1.2F5, mouse embryo fibroblasts (MEFs), trophoblastic stem cells (TSs), or e14 placentae were probed with exon 1 or exon 2 probes. In each panel the ethidium-bromide staining of 18S rRNA was used to monitor the loading of the RNAs.
Activity of the transgenes in isolated macrophages from 3 different transgenic mice.
This study generated 3 strains of transgenic mice. They were 3.5fms-EGFP (4 independent lines), 7.2fms-EGFP (6 independent lines), and 7.2fmsΔFIRE-EGFP (6 independent lines). Analyses were done on at least 3 independent lines for each strain. Macrophages isolated from peritoneum (thioglycollate–elicited peritoneal macrophages [TEPM]), derived from bone marrow macrophages (BMMs) and from bronchoalveolar lavage (BAL), were subjected to FACS analysis. Representative FACS profiles (filled histograms) of macrophages isolated from 3.5fms-EGFP TG mice are shown in the top row. Middle row profiles show the representative profiles of EGFP expression in macrophages isolated from 7.2fms-EGFP TG mice; histograms in the bottom row are the representative profiles of EGFP expression in macrophages isolated from 7.2fmsΔFIRE-EGFP TG mice. Dotted lines represent histograms of nontransgenic littermate controls.
Activity of the transgenes in isolated macrophages from 3 different transgenic mice.
This study generated 3 strains of transgenic mice. They were 3.5fms-EGFP (4 independent lines), 7.2fms-EGFP (6 independent lines), and 7.2fmsΔFIRE-EGFP (6 independent lines). Analyses were done on at least 3 independent lines for each strain. Macrophages isolated from peritoneum (thioglycollate–elicited peritoneal macrophages [TEPM]), derived from bone marrow macrophages (BMMs) and from bronchoalveolar lavage (BAL), were subjected to FACS analysis. Representative FACS profiles (filled histograms) of macrophages isolated from 3.5fms-EGFP TG mice are shown in the top row. Middle row profiles show the representative profiles of EGFP expression in macrophages isolated from 7.2fms-EGFP TG mice; histograms in the bottom row are the representative profiles of EGFP expression in macrophages isolated from 7.2fmsΔFIRE-EGFP TG mice. Dotted lines represent histograms of nontransgenic littermate controls.
Proportion and fluorescence intensities of EGFP+ macrophages in 3 different fms-EGFP transgenic mice in 3 different lines
| . | TEPM EGFP+cells, percentage . | TEPM mean fluorescence intensity . | BMM EGFP+cells, percentage . | BMM mean fluorescence intensity . |
|---|---|---|---|---|
| 3.5fms-EGFP transgenic mice | ||||
| Line no. 1 | 0.1 | 18.62 | 0.6 | 17.15 |
| Line no. 2 | 0.69 | 16.28 | 8.18 | 20.77 |
| Line no. 3 | 0.21 | 13.93 | 3.6 | 20.11 |
| Average ± SD | 0.3 ± 0.3 | 16.2 ± 2.3 | 3.8 ± 1.9 | 19.3 ± 1.9 |
| 7.2fms-EGFP transgenic mice | ||||
| Line no. 1 | 71.71 | 87.20 | 71.40 | 70.89 |
| Line no. 2 | 72.40 | 154.76 | 72.33 | 83.68 |
| Line no. 3 | 74.58 | 80.04 | 84.46 | 248.83 |
| Average ± SD | 72.89 ± 1.4 | 107.31 ± 41.1 | 76.06 ± 7.2 | 132.8 ± 96.3 |
| 7.2fmsΔFIRE-EGFP transgenic mice | ||||
| Line no. 1 | 2.92 | 20.43 | 2.54 | 17.59 |
| Line no. 2 | 0.9 | 13.72 | 0.69 | 13.38 |
| Line no. 3 | 0.8 | 15.01 | 1.36 | 40.87 |
| Average ± SD | 1.54 ± 1.1 | 16.3 ± 3.5 | 1.5 ± 0.9 | 23.9 ± 12.0 |
| . | TEPM EGFP+cells, percentage . | TEPM mean fluorescence intensity . | BMM EGFP+cells, percentage . | BMM mean fluorescence intensity . |
|---|---|---|---|---|
| 3.5fms-EGFP transgenic mice | ||||
| Line no. 1 | 0.1 | 18.62 | 0.6 | 17.15 |
| Line no. 2 | 0.69 | 16.28 | 8.18 | 20.77 |
| Line no. 3 | 0.21 | 13.93 | 3.6 | 20.11 |
| Average ± SD | 0.3 ± 0.3 | 16.2 ± 2.3 | 3.8 ± 1.9 | 19.3 ± 1.9 |
| 7.2fms-EGFP transgenic mice | ||||
| Line no. 1 | 71.71 | 87.20 | 71.40 | 70.89 |
| Line no. 2 | 72.40 | 154.76 | 72.33 | 83.68 |
| Line no. 3 | 74.58 | 80.04 | 84.46 | 248.83 |
| Average ± SD | 72.89 ± 1.4 | 107.31 ± 41.1 | 76.06 ± 7.2 | 132.8 ± 96.3 |
| 7.2fmsΔFIRE-EGFP transgenic mice | ||||
| Line no. 1 | 2.92 | 20.43 | 2.54 | 17.59 |
| Line no. 2 | 0.9 | 13.72 | 0.69 | 13.38 |
| Line no. 3 | 0.8 | 15.01 | 1.36 | 40.87 |
| Average ± SD | 1.54 ± 1.1 | 16.3 ± 3.5 | 1.5 ± 0.9 | 23.9 ± 12.0 |
Results are from individual animals of each of the transgenic lines. Mean fluorescence intensities of nontransgenic controls were 2.96 ± 0.9 (BMM) and 2.66 ± 1.1 (TEPM).
Intron 2 as well as the FIRE region required for the activity of the transgene in vivo
In stable transfections of the macrophage line RAW264, removal of the first intron greatly reduced EGFP expression, and removal of the FIRE sequence abolished it.4 To extend these observations, we produced 4 independent transgenic lines with constructs using the 3.5-kb promoter alone, and 6 independent lines with the 7.2-kb promoter without the FIRE sequence. In contrast to the thioglycollate-elicited peritoneal macrophages (TEPM) or BMMs from the 7.2fms-EGFP transgenic mice, those obtained from lines expressing EGFP from the 3.5-kb promoter exhibited fluorescence in a smaller number of cells and at a much lower level (Figure 3B, upper row and Table 2). On tissue sections of these transgenic mice, the level of EGFP was below the limits of detection (not shown). Deletion of the FIRE region completely abolished the activity of the transgene in these macrophage populations in each of the 6 transgenic lines examined (Figure 3B, lower row and Table 2). Again, there was no detectable expression of EGFP in tissue sections of any of the transgenic lines produced with the 7.2fmsΔFIRE-EGFP transgene. The data indicate that the intron, and particularly the FIRE sequence, is absolutely needed for the reproducible, position-independent expression, obtained with the 7.2fms-EGFP transgene.
c-fms/EGFP transgene is active in trophoblasts
The analysis of the start site above suggests that the 3.5-kb fms promoter could direct trophoblast expression. In order to assess the possible activities of the c-fmspromoter driving the EGFP reporter gene in these cells, EGFP expression in early embryonic development was determined. Fluorescence microscope observation of 7.2fms-EGFP TG embryos revealed the presence of green-fluorescent cells in the ectoplacental cone area of e7.5 embryos. Later in development, trophoblasts in the deciduum surrounding the embryo were also positive for EGFP, as were trophoblastic giant cells in e10.5 and e12.5 embryos (not shown). The locations of the EGFP+ cells in these embryos were similar to the expression of c-fms mRNA detected by in situ hybridization.8 20 The expression of EGFP in trophoblasts was also observed in giant cells grown in tissue-culture plates derived from the ectoplacental cones (Figure 4A). These observations support the data from the start site analysis, and demonstrated that the exon 2 c-fms upstream promoter region in the mouse is used by both macrophage lineage and trophoblast cells.
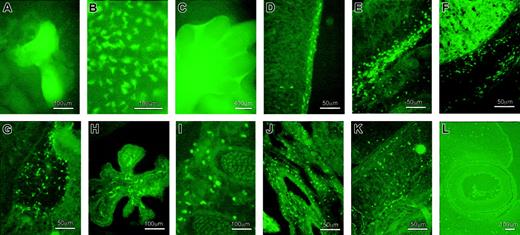
Expression of EGFP during embryonic development.
(A) In 7.2fms-EGFP TG mice, other than those of macrophage lineage, EGFP is also expressed in trophoblast cells. This panel shows the EGFP expression in cultured trophoblastic giant cells derived from the e6.5 ectoplacental cone. (B) Numerous EGFP+ stellate cells were observed in the yolk sac at e9.5. (C) EGFP+ cells were observed by whole-mount visualization in the interdigital region of the developing footplate of an e12 to e13 embryo. (D) Sagittal section of an e13.5 embryo showing numerous EGFP+ cells in the epidermal layer of the dorsal part of the embryo. (E) Numerous EGFP+ cells were observed in the mesenchymal area beneath the chest. (F) The liver and its surrounding area were occupied by a large number of EGFP+ cells. EGFP+ cells also present in the brain, especially in the pons-midbrain junction (G) and choroid plexus (H). (I) Section trough somites shows the presence of EGFP+ cells around the developing vertebrae. (J) Lung also contains EGFP+cells. (K) EGFP+ cells present in the intrinsic muscle of the tongue. (L) Few EGFP+ cells were observed in the developing eye.
Expression of EGFP during embryonic development.
(A) In 7.2fms-EGFP TG mice, other than those of macrophage lineage, EGFP is also expressed in trophoblast cells. This panel shows the EGFP expression in cultured trophoblastic giant cells derived from the e6.5 ectoplacental cone. (B) Numerous EGFP+ stellate cells were observed in the yolk sac at e9.5. (C) EGFP+ cells were observed by whole-mount visualization in the interdigital region of the developing footplate of an e12 to e13 embryo. (D) Sagittal section of an e13.5 embryo showing numerous EGFP+ cells in the epidermal layer of the dorsal part of the embryo. (E) Numerous EGFP+ cells were observed in the mesenchymal area beneath the chest. (F) The liver and its surrounding area were occupied by a large number of EGFP+ cells. EGFP+ cells also present in the brain, especially in the pons-midbrain junction (G) and choroid plexus (H). (I) Section trough somites shows the presence of EGFP+ cells around the developing vertebrae. (J) Lung also contains EGFP+cells. (K) EGFP+ cells present in the intrinsic muscle of the tongue. (L) Few EGFP+ cells were observed in the developing eye.
EGFP is widely expressed in phagocytic cells in tissues of the transgenic embryos
Our laboratory has previously described the analysis of c-fms gene expression by whole-mount in situ hybridization in mouse embryonic development.7,8 The ability of the 7.2fms-EGFP transgene to recapitulate c-fms mRNA expression was examined systematically. The first c-fms–expressing cells were detected in the yolk sac around e9.5 and the location is recapitulated by EGFP+ cells. Their abundance and distribution is striking. They are not focused in blood islands, but distributed throughout the yolk sac (Figure 4B). There is no possibility that the green-fluorescent cells observed in the yolk sac originated from infiltration of cells of maternal origin because all the embryos examined were derived from nontransgenic mothers mated with a transgenic male. The origin and function of these yolk sac–derived embryonic phagocytes has been reviewed recently.21 By e10 to e10.5, significant numbers of EGFP+ cells were observed infiltrating first into the head, then the liver of the embryo (not shown), again entirely consistent with the published pattern of c-fms mRNA expression7 and independent evidence of the infiltration of hematopoietic progenitors and stem cells into the liver which occurs at e10.5 to e11.22 The number of green-fluorescent cells escalated rapidly through development of the embryo to e12 to e13. In particular, c-fms–positive cells have been shown to be involved in removing apoptotic cells in the interdigital regions in developing footplates of the mouse embryo.7 8 Similarly, Figure 4C shows massive accumulation of green-fluorescent cells in the interdigital regions at e12.5.
Embryo sections at e13.5 revealed the presence of green-fluorescent cells throughout the body of the embryos, with extensive accumulation of the cells in regions with high tissue turnover and extensive cell death. Figure 4 shows some representative sites of high numbers of EGFP+ cell infiltration, including the epidermal layer of the dorsal part of the embryo (Figure 4D), as well as in the mesenchymal area beneath the chest (Figure 4E) and other surfaces of the body, the liver and its surrounding area (Figure 4F), and the brain. In the brain, EGFP+ cells were especially prevalent in the pons-midbrain junction area (Figure 4G), as well as in the choroid plexus (Figure 4H). Elsewhere in the embryo, numerous green-fluorescent cells were found in the mesenchymal area around the somites (Figure 4I), lung (Figure 4J), around the intrinsic muscles of the tongue (Figure 4K), and also in the developing eye (Figure 4L). Most importantly, the morphology, abundance, and location of EGFP+ cells are consistent with absolute restriction of the transgene to embryonic phagocytes, and to the known location of cells expressing c-fms mRNA and other macrophage-specific genes.7 8
EGFP is specifically expressed in myeloid cells in the bone marrow, blood, and tissues
Mononuclear phagocytes develop from bone marrow progenitors, enter the circulation as blood monocytes, and then leave to replenish tissue macrophage populations. Macrophages share progenitors with granulocytes, and the knockout of the PU.1 transcriptional regulator,23 which affects transcription of c-fms,24 has deficiencies in both lineages. Although c-fms mRNA and protein are absent from mature granulocytes, the more stable EGFP reporter could be retained in progeny from the common precursor. Dual-color fluorescence immunostaining was carried out on bone marrow and peripheral blood using the F4/80 and Mac-1 (CD11b) markers. Additionally, we examined CSF-1R (c-Fms) surface protein expression. Representative profiles are shown in Figure 5B. In bone marrow, most EGFP-expressing cells also expressed CD11b, indicating that the transgene expression is restricted to myeloid cells. By contrast, few CD11b− cells were EGFP+. About 50% of the EGFP+ cells also expressed the mature macrophage marker F4/80 and/or detectable surface CSF-1R. Among peripheral blood mononuclear cells, the EGFP+ cells all expressed both F4/80 and CD11b, and the proportion upon which surface CSF-1R was undetectable was smaller than in marrow (Figure 5B). Figure 5C shows the expression of CD11b and F4/80 on EGFP+ peritoneal and broncho-alveolar lavage cells. All EGFP+ cells in the peritoneum express both markers. By contrast, alveolar macrophages, as expected,25 lack CD11b but express F4/80 at low levels. The analysis of alveolar macophages is complicated by the relatively high background autofluorescence.
FACS analysis of spleen, bone marrow, peripheral blood mononuclear cells (PBMCs), peritoneal macrophages (TEPMs) and bronchoalveolar lavage (BAL) cells of the 7.2
fms-EGFP TG mice. (A) In the spleen, EGFP is expressed mainly on adherent cells. Mechanically disaggregated spleen cells were grown overnight in TC dishes, and adherent and nonadherent cells were subjected to FACS analysis (top panels). Splenocytes were also stained with anti–F4/80 and anti–Mac-1 antibodies as described in “Materials and methods” (bottom panels). (B) Coexpression of EGFP and cell-surface markers examined by dual-color FACS analysis in bone marrow and PBMCs. For CSF-1R staining, cells were incubated with rat anti–mouse c-fms antibody, and stained with PE-conjugated goat anti–rat F(ab′)2. Other markers used were PE-conjugated anti–Mac-1 and anti–F4/80 antibodies. Quadrants were set based on the profiles from non-TG littermate controls. (C). Mac-1 and F4/80 staining of peritoneal macrophages and bronchoalveolar lavage cells. Cells were stained with anti–Mac-1 and anti–F4/80 antibodies. EGFP+ cells were gated and presented as black histograms. Dotted histograms represent isotype control staining. All data are representative of 3 separate analyses.
FACS analysis of spleen, bone marrow, peripheral blood mononuclear cells (PBMCs), peritoneal macrophages (TEPMs) and bronchoalveolar lavage (BAL) cells of the 7.2
fms-EGFP TG mice. (A) In the spleen, EGFP is expressed mainly on adherent cells. Mechanically disaggregated spleen cells were grown overnight in TC dishes, and adherent and nonadherent cells were subjected to FACS analysis (top panels). Splenocytes were also stained with anti–F4/80 and anti–Mac-1 antibodies as described in “Materials and methods” (bottom panels). (B) Coexpression of EGFP and cell-surface markers examined by dual-color FACS analysis in bone marrow and PBMCs. For CSF-1R staining, cells were incubated with rat anti–mouse c-fms antibody, and stained with PE-conjugated goat anti–rat F(ab′)2. Other markers used were PE-conjugated anti–Mac-1 and anti–F4/80 antibodies. Quadrants were set based on the profiles from non-TG littermate controls. (C). Mac-1 and F4/80 staining of peritoneal macrophages and bronchoalveolar lavage cells. Cells were stained with anti–Mac-1 and anti–F4/80 antibodies. EGFP+ cells were gated and presented as black histograms. Dotted histograms represent isotype control staining. All data are representative of 3 separate analyses.
EGFP is expressed in tissue macrophage populations
Macrophages defined by the F4/80 surface marker in the mouse are a major component (10%-15%) of the cells in most tissues of the body.26-32 The images of F4/80 location have been loaded in a database at www.imb.uq.edu.au/groups/hume. F4/80 is not detected on all putative tissue macrophages, and, as evident from Figure 5C, is especially low in lung macrophages and macrophages of lymphoid organs. EGFP was detected in all of the major macrophage populations of the body. In the spleen, the red pulp macrophages were strongly EGFP+ (Figure 6A). Enzymatic digestion of the spleen followed by flow cytometry revealed approximately 18% of cells that were EGFP+. The majority of these cells adhered to bacteriologic plastic, supporting their identity as macrophages (Figure 5A). Similarly, in the liver EGFP was expressed specifically on sinusoidal stellate cells, resembling liver macrophages (Kupffer cells) with the characteristic concentration toward the periphery of liver lobules. In this organ, these cells constituted about 8% to 9% of the whole liver cells, as examined by FACS analysis following enzymatic disaggregation (Figure 6B). In the lung there is a large interstitial macrophage population in addition to those present in broncho-alveolar spaces.33 EGFP expression was detected on 8% of cells from enzymatically digested lung (Figure 6C). Finally, in the intestinal lamina propria there is a large population of macrophages that can be isolated following enzymatic digestion. Figure 6D shows the tissue section and the FACS profile of the enzymatically digested mouse intestinal wall, where 6% to 8% of cells were EGFP+. Nycodenz gradient centrifugation of this digested tissue could increase the proportion of EGFP+ cells up to 15%, consistent with previous studies on purification of these cells.17
Expression of EGFP in adult tissues examined by fluorescence microscopy.
Adult tissues were fixed on 4% paraformaldehyde, incubated overnight in 18% sucrose, embedded in optimal cutting temperature (OCT) media and frozen sectioned. (A) Left, splenic macrophages expressing EGFP. These cells are located mainly in the red pulp of the spleen. RP indicates red pulp; WP, white pulp. Right, FACS analysis of enzymatically digested spleen revealed about 18% of cells expressing EGFP. (B) In the liver, EGFP is expressed in the Kuppfer cells (left). These EGFP+ cells constituted about 8% of the cell population, as shown in the FACS profile of the liver (right). (C) Expression of EGFP in the alveolar and interstitial lung macrophages, which composed about 8% of the whole cell population. (D) Abundant EGFP+ cells were observed in the lamina propria of the small intestine and constituted about 7% of the whole cell population. EGFP+ cell gates (M2) in the FACS profiles were made based on the nontransgenic littermate control profiles. (E) Brain macrophages (microglia) expressed EGFP. (F) Examination of the cross section of the eye revealed the expression of EGFP in microglia (arrow) in the retina. Renal interstitial macrophages (G) are EGFP-positive, as are macrophages that reside in the interstitium of testis (H). (I) Bone-resorptive osteoclasts expressed EGFP. (J) A cross-section of the ear revealed the expression of EGFP in Langerhans cells (arrow) and epidermal macrophages. (K) EGFP expression in Langerhans cells in the epidermal sheet of the ear. (L) Thymic macrophages shown to express EGFP.
Expression of EGFP in adult tissues examined by fluorescence microscopy.
Adult tissues were fixed on 4% paraformaldehyde, incubated overnight in 18% sucrose, embedded in optimal cutting temperature (OCT) media and frozen sectioned. (A) Left, splenic macrophages expressing EGFP. These cells are located mainly in the red pulp of the spleen. RP indicates red pulp; WP, white pulp. Right, FACS analysis of enzymatically digested spleen revealed about 18% of cells expressing EGFP. (B) In the liver, EGFP is expressed in the Kuppfer cells (left). These EGFP+ cells constituted about 8% of the cell population, as shown in the FACS profile of the liver (right). (C) Expression of EGFP in the alveolar and interstitial lung macrophages, which composed about 8% of the whole cell population. (D) Abundant EGFP+ cells were observed in the lamina propria of the small intestine and constituted about 7% of the whole cell population. EGFP+ cell gates (M2) in the FACS profiles were made based on the nontransgenic littermate control profiles. (E) Brain macrophages (microglia) expressed EGFP. (F) Examination of the cross section of the eye revealed the expression of EGFP in microglia (arrow) in the retina. Renal interstitial macrophages (G) are EGFP-positive, as are macrophages that reside in the interstitium of testis (H). (I) Bone-resorptive osteoclasts expressed EGFP. (J) A cross-section of the ear revealed the expression of EGFP in Langerhans cells (arrow) and epidermal macrophages. (K) EGFP expression in Langerhans cells in the epidermal sheet of the ear. (L) Thymic macrophages shown to express EGFP.
Macrophages and c-fms expression in the brain are of particular interest because of the neurologic abnormalities in the CSF-1–deficientCsf1op/Csf1opmouse34 and the isolated claim that the human c-fms promoter drives transgene (lacZ) expression in astrocytes.35 The F4/80 antigen is readily detected on brain macrophages (microglial cells).36 In the brain, EGFP was detectable in macrophages associated with the microvasculature and meningeal surfaces (not shown) and in microglia (Figure 6E). In this organ, microglial processes were clearly evident and highlighted by the fluorescent protein expression. Expression of EGFP in microglia can also be clearly observed in the retina, where they spread in 2 dimensions in the plane of the inner and outer plexiform layers28 (Figure 6F). In neither brain nor retina was there any evidence of expression in glial cells other than microglia.
Macrophages defined by F4/80 antigen expression are abundantly associated with epithelia and in endocrine organs where they may perform specific physiologic functions.26,30,37 In an extensive survey, EGFP expression was generally consistent with previous data on F4/80. Examples shown include the lamina propria of the gastrointestinal tract (Figure 6D), the renal medulla (Figure 6G), and interstitium of the testis (Figure 6H). A number of additional examples and further images are displayed atwww.imb.uq.edu.au/groups/hume. Bone resorptive osteoclasts share a progenitor with macrophages and their production is CSF-1 dependent. Although they lose mature macrophage markers such as F4/80,29 mature osteoclasts express c-fms mRNA and CSF-1 can acutely regulate osteoclastic bone resorption.38-40 In the transgenic mice, osteoclasts expressed detectable EGFP both in culture and bone sections (Figure6I).
EGFP expression in lymphoid organs and dendritic cells
Cell populations such as the Langerhans cells (LCs) of the skin are considered to be immature dendritic cells (DCs). They are CSF-1 responsive,41,42 but in the absence of CSF-1 in theCsf1op/Csf1op mouse their numbers are unaffected.43,44 EGFP was detected in the Langerhans cells either in section (Figure 6J, arrow) or in en face view in epidermal sheets (Figure 6K). In the spleen, the EGFP marker was detectable at a low level in cells forming a reticular network within the white pulp, consistent with expression in interdigitating DCs.45 Similarly, in the mesenteric lymph node, EGFP was expressed at high levels on cells within the medullary cords, where F4/80 is also expressed,27 but also on presumed interdigitating cells within lymphoid follicles (not shown). The myeloid/DC component of thymus is complex.46 EGFP was very abundant in large stellate cells especially concentrated around the cortical medullary junction (Figure 6L). In the cortex, there was also a network of ramified EGFP expression that may correspond to thymic DCs.
Discussion
Transcriptional regulatory elements of the c-fms gene
The c-fms gene is expressed in trophoblasts and macrophages in both mice and humans. The first part of this study showed that the underlying mechanisms are different. Whereas the human gene reportedly utilizes a promoter in the 3′ end of the upstream PDGFR-β to direct expression in trophoblasts,5,11 the mouse gene clearly uses multiple transcription start sites within the 500-bp flanking exon 2. This region is actually conserved between mouse and human, and contains elements such as AP-1 sites that have been implicated in trophoblast-specific transcription. These proximal sequences might contribute to activation of the upstream promoter, or splicing, in the human gene expressed in trophoblasts. The 7.2-kb fms promoter could be utilized to direct expression of a trophoblast-specific transgene for functional/immunologic studies. Because the upstream promoter is not used in macrophages and is not required for promoter activity in transient transfections,3 we may be able to selectively delete the trophoblast control elements to eliminate trophoblast expression while retaining macrophage promoter activity.
Analysis of multiple independent transgenic lines confirmed that the activity of the 7.2-kb promoter construct was largely, or completely, abolished by either deletion of the first intron or elimination of the FIRE sequence. As observed in stably transfected cell lines,4 there was detectable residual activity in macrophages with the promoter alone, in keeping with reports of successful use of the corresponding human promoter region to drive low-level expression of a transgene.47 Elimination of FIRE in an intron-containing construct, on the other hand, abolished all activity.4 We have proposed that FIRE acts to overcome a block to transcription elongation in the first intron4; these findings extend the evidence favoring this model to a transgene in vivo. Based on the constitutive activity in a wide range of mouse tumor cell lines,3,13 we might have anticipated that the 3.5-kb c-fms promoter would generate ectopic expression in transgenic mice. In contrast to the data with tumor cells, the 3.5-kb promoter was very weakly active in untransformed 3T3 fibroblasts.12 The findings with the transgenic mice support the view that constitutive fms promoter activity in tumor lines reflects their malignancy.13 Interestingly, deletion of the trophoblast promoter region newly identified herein abolished c-fms promoter function in nonmacrophage tumor cells but not in RAW264 cells.13
Definition of the mononuclear phagocyte system
The mononuclear phagocyte system was defined as a family of cells arising from bone marrow progenitors, circulating as monocytes and entering the tissues where they form the resident macrophage population.48 In the mouse, F4/80, which detects a member of the EGF-TM7 family of surface receptors,49 has been most widely studied as a marker of tissue macrophages.26-32 The F4/80 immunoreactivity defined a population of cells in almost every organ of the body that shared characteristic locations and stellate morphology and included most known macrophagelike cells. However, F4/80 is not detectable on progenitor cells, and is difficult to detect on blood monocytes and several tissue macrophage populations, notably in lymphoid tissues, intestine, and lung.27,30 The pattern of the c-fms–EGFP transgene was indistinguishable from published pictures of F4/80 in many locations. In addition to the well-documented macrophage populations of the liver and the red pulp of spleen, EGFP, like F4/80, was expressed in the large peri-epithelial macrophagelike cell populations of the kidney and gastrointestinal tract26,30 as well as the sinusoidal and interstitial populations of endocrine organs.31 For this reason, we have dubbed fms-EGFP mouse lines the MacGreenmouse, recognizing that there will be many applications of such animals in the study of innate immunity and experimental pathology.
Tissue macrophages are thought to adapt to local environments to perform tissue-specific functions. For example, the interstitial macrophages of the testis shown clearly in Figure 6H are thought to regulate testosterone production37 and the CSF-1–deficient mouse has reproductive defects when they are reduced or absent.50 The transgene marker can be applied conveniently for purification of macrophages from each of these sites for phenotypic analysis. Figures 5 and 6 show the examples using bone marrow progenitor cells, lung interstitial macrophages, intestinal lamina propria, and splenic macrophages. Unlike F4/80, which appears only after the liver becomes the major hematopoietic site,21 the c-fms–EGFP transgene was expressed in phagocytelike cells throughout development. The expression in the embryo was indistinguishable from our earlier description of the location of c-fms mRNA.7 8 We have been able to sort fluorescent cells from enzymatically digested embryos (not shown), providing the opportunity to delineate further the ways that they differ from macrophages in adult tissues.
Given the central function of CSF-1 in macrophage differentiation, and the lack of macrophages in CSF-1–deficientCsf1op/Csf1op mice44 or in the recently characterized c-fms knock out,51 the CSF-1R is the most obvious definitive marker for the mononuclear phagocyte lineage. We infer that the expression of c-fms–EGFP correlates with expression of a functional CSF-1 receptor. Colocalization of EGFP and surface CSF-1R in bone marrow cells supported this view (Figure 5). There was a population of cells that were EGFP+ but lacked detectable surface CSF-1R, but these cells expressed myeloid markers, CD11b, and to a lesser extent, F4/80 (Figure 5). CSF-1R is acutely down-modulated from the cell surface by a range of stimuli including ligand,52 53whereas EGFP is stable. Hence, it is not surprising that the correlation is imperfect in progenitor cells and proliferating myeloid precursors. In peripheral blood, there was no detectable expression of EGFP in mature granulocytes. In the mononuclear cell fraction, and in peritoneal exudates, there was a very good correlation between EGFP and expression of F4/80 and CD11b, indicating that the transgene is restricted to monocyte macrophages and is absent from T or B lymphocytes.
CSF-1–independent populations of mononuclear phagocytes
The widespread expression of the transgene on all known F4/80-positive macrophage populations, including Langerhans cells, inMacGreen mice raises the issue of the existence of CSF-1–independent macrophage populations, and the relationship between macrophages and DCs. In the CSF-1–deficientCsf1op/Csf1op mouse, the majority of F4/80-positive macrophage populations are depleted, but not absent, whereas others, notably those of lymphoid organs and the skin, are unaffected.44 Even the CSF-1–dependent bone cells of theCsf1op/Csf1op mouse increase in number with age,54 a phenomenon that has been attributed to vascular endothelial growth factor A,55 which can signal macrophages through the fms-like receptor tyrosine kinase flt-1.56 Lymphoid organ T-cell area DC and skin LC numbers were also not substantially altered inCsf1op/Csf1opmice.43,44,57 This observation does not imply that subpopulations of F4/80-positive cells in vivo are CSF-1–unresponsive, or have differentiated from a separate progenitor cell from CSF-1–responsive monocyte-macrophages. There is, in fact, evidence that LCs and mature DCs respond to CSF-1 and that the factor can influence divergence of the macrophage/DC functional fate.42 We may infer that other factors in particular tissue locations can substitute for CSF-1. Ongoing studies in our laboratory will address the relationship between EGFP expression and other DC markers.
Conclusion
We have identified the elements within the c-fmsgene required for reproducible expression of a reporter gene in cells of the mononuclear phagocyte lineage. The c-fms–EGFP transgene will be a useful marker for further studies of the biology of the cells in this system.
Thanks to Ms Elizabeth Williams for help in all aspects of the production and analysis of the transgenic mice in this project.
Prepublished online as Blood First Edition Paper, September 12, 2002; DOI 10.1182/blood-2002-02-0569.
Supported by grants from the Australian National Health and Medical Research Council to D.A.H. R.T.S. and D.O. are recipients of the Australian International Postgraduate Research Scholarship and University of Queensland International Postgraduate Research Scholarship.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David A. Hume, Institute for Molecular Bioscience, University of Queensland, Brisbane, Q4072, Australia; e-mail: d.hume@imb.uq.edu.au.


![Fig. 3. Activity of the transgenes in isolated macrophages from 3 different transgenic mice. / This study generated 3 strains of transgenic mice. They were 3.5fms-EGFP (4 independent lines), 7.2fms-EGFP (6 independent lines), and 7.2fmsΔFIRE-EGFP (6 independent lines). Analyses were done on at least 3 independent lines for each strain. Macrophages isolated from peritoneum (thioglycollate–elicited peritoneal macrophages [TEPM]), derived from bone marrow macrophages (BMMs) and from bronchoalveolar lavage (BAL), were subjected to FACS analysis. Representative FACS profiles (filled histograms) of macrophages isolated from 3.5fms-EGFP TG mice are shown in the top row. Middle row profiles show the representative profiles of EGFP expression in macrophages isolated from 7.2fms-EGFP TG mice; histograms in the bottom row are the representative profiles of EGFP expression in macrophages isolated from 7.2fmsΔFIRE-EGFP TG mice. Dotted lines represent histograms of nontransgenic littermate controls.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/3/10.1182_blood-2002-02-0569/4/m_h80333727003.jpeg?Expires=1764990459&Signature=aNJnsRaSVj0MrV6qiq8sll2smqejhmjMEyfAxRudVwGhssLgLfnY5oal83reH6~OJvxowPRr0445d2ZS7sSZmU94tbwrxYIMzEU6Rj5Xk8tFsqf9PCKrcb1XnQlypj9~wDfysTULHBUUmdSy3AacBVcOC2ANk9asdDxEnnPuioPb4-A8ghJsyHlkzX2u23~INORqTDs4nEhjnhf8y9chf1HDQPDB~QObLp~JJGN3O63RxX4YroPrYEZdJnt33JNW4ipizoqep4AJePmYNjQ5~FYNw2q9WdCIurmjkBP91vMf8LZUfjr2aDcndIgv3xsvaEEhqcdGBaH63jVFqUF30Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal