Tumoral cells in Hodgkin lymphoma (HL) display an increased growth fraction and diminished apoptosis, implying a profound disturbance of the cell cycle and apoptosis regulation. However, limitations of molecular techniques have prevented the analysis of the tumor suppressor pathways and cell-cycle checkpoints. Tissue microarray (TMA) is a powerful tool for analyzing a large number of molecular variables in a large series of tumors, although the feasibility of this technique has not yet been demonstrated in heterogeneous tumors. The expression of 29 genes regulating the cell cycle and apoptosis were analyzed by immunohistochemistry and in situ hybridization in 288 HL biopsies using TMA. The sensitivity of the technique was validated by comparing the results with those obtained in standard tissue sections. The results revealed multiple alterations in different pathways and checkpoints, including G1/S and G2/M transition and apoptosis. Striking findings were the overexpression of cyclin E, CDK2, CDK6, STAT3, Hdm2, Bcl2, Bcl-XL, survivin, and NF-κB proteins. A multiparametric analysis identified proteins associated with increased growth fraction (Hdm2, p53, p21, Rb, cyclins A, B1, D3, and E, CDK2, CDK6, SKP2, Bcl-XL, survivin, STAT1, and STAT3), and proteins associated with apoptosis (NF-κB, STAT1, and RB). The analysis also demonstrated that Epstein-Barr virus (EBV)–positive cases displayed a characteristic profile, confirming the pathogenic role of EBV in HL. Survival probability depends on multiple biologic factors, including overexpression of Bcl2, p53, Bax, Bcl-XL, MIB1, and apoptotic index. In conclusion, Hodgkin and Reed-Sternberg cells harbor concurrent and overlapping alterations in the major tumor suppressor pathways and cell-cycle checkpoints. This appears to determine the viability of the tumoral cells and the clinical outcome.
Introduction
Hodgkin lymphoma (HL) is a distinct primary solid malignancy of the immune system in which the exuberant production of cytokines and chemokines is associated with an abundance of a component of inflammatory cells that outnumber the recognized tumoral subpopulation of Hodgkin and Reed-Sternberg (H/RS) cells. These H/RS cells harbor clonally rearranged and somatically mutated immunoglobulin genes, indicating that, in most cases, they are derived from germinal center B cells.1
Despite the progress made in the understanding of the biology of HL, the transforming events in these cells remain to be elucidated. In normal lymphoid tissues, germinal center B cells lacking expression of functional high-affinity antibody (Ab) inevitably suffer apoptosis under normal physiologic conditions,2 whereas H/RS cells escape programmed cell death and instead proliferate and disseminate clonally by unknown means.
The analysis of cell-cycle regulation in different types of lymphoid and epithelial neoplasms reveals a relationship whereby increased clinical aggressiveness is associated with the accumulation of genetic and epigenetic alterations,3 which lead to the dismantling and inactivation of the main tumor suppressor pathways: p14ARF-p53-p21WAF,1p16INK4a-Rb, and p27KIP1. Thus, although some low-grade lymphomas exhibit alterations that are limited to apoptosis control, large-cell and Burkitt lymphomas tend to display concurrent and concerted inactivation of these tumor suppressor pathways,3 4 which finally turns them into neoplasms with an extremely high growth fraction and resistance to standard chemotherapy.
The alterations in the main pathways responsible for controlling the cell cycle in HL have only rarely been studied and are poorly understood, mainly because the techniques used for molecular analyses in this disease have been limited by the scarcity of tumoral cells. Previous studies have shown some relationships between the expression of cell-cycle proteins and patient outcome.5 The incidence of p53 mutations has been investigated in H/RS cells and is known to be significantly lower than in other neoplasias.6Nonetheless, proteins regulating the function of p53, such as p14ARF and Hdm2, are expressed aberrantly and may have an indirect role in regulating the function of p53.7 On the other hand, the Rb pathway is disrupted by p16INK4amethylation in some cases.8 The NF-κB (p65/RelA) pathway is constitutively activated in different HL-derived cell lines and tumors,9,10 partially explaining the resistance of H/RS cells to apoptosis. The NF-κB activation can be related to IκBα gene mutations in Epstein-Barr virus (EBV)–negative HL cases,11 but not in EBV-positive cases, indicating a characteristic regulation of these cases. Other studies have shown disorder in the S-phase,12,13 frequent aneuploidy14,15 of the tumoral cells, and suggested the presence of structural anomalies during mitosis that lead to the formation of the characteristic multinucleated cells.16
The high proliferative index17,18 and resistance to apoptosis in germinal center–derived cells,19 20 such as H/RS cells, that produce no functional immunoglobulin, suggest that the control of the cell cycle in these cells is seriously disrupted, and highlights the interest in additional studies that might clarify this issue.
The recently developed tissue microarray (TMA) technology allows simultaneous analyses of hundreds of tissue specimens for numerous molecular targets21,22 by using the techniques of immunohistochemistry (IHC), in situ hybridization (ISH), and fluorescence in situ hybridization (FISH). This technology also has the potential to significantly accelerate progress in the search for associations between molecular changes and clinical traits.23 This tool has proved to be remarkably useful in evaluating different cancer types, particularly those with homogeneous histology. However, the feasibility of this technique for analyzing highly heterogeneous tumors such as HL remains to be elucidated. Recent studies using TMA24 have shown the potential of this technology for restricted analyses in HL.
IHC and ISH techniques can contribute significantly to the identification of an expression profile of cell-cycle markers in H/RS cells. However, due to the paucity of such cells, we decided to investigate whether TMA technology can also be used in this heterogeneous tumor and in large-scale analyses. With the aim of evaluating the alterations in the expression of essential cell-cycle regulatory proteins, we performed a series of IHC and ISH analyses of 29 different markers in 288 HL cases. This revealed different alterations affecting the main checkpoints of the cell cycle, in G1/S and G2/M transition, and involving all the major pathways simultaneously.
Materials and methods
HL samples and cell lines
The collaborating members of the Spanish Hodgkin Lymphoma Study Group collected 288 retrospective cases of HL. Cases were randomly selected, and were diagnosed in the period between 1994 and 1998, with stages being evaluated according to standard protocols and treated with standard polychemotherapeutic regimes (including adriamycin in the majority of cases) with or without adjuvant radiotherapy. Paraffin blocks were selected only on the basis of the availability of suitable formalin-fixed, paraffin-embedded tissue (at least 1-mm thick). All the included samples represent at-diagnosis biopsies; any relapse biopsies were discarded beforehand. The histologic confirmation of HL and subtype was achieved in 277 cases by central review using standard tissue sections, and diagnoses were made according to the criteria of the World Health Organization (WHO) classification,25 with the help of CD30, CD15, CD20, ALK, and HE staining. Cases included 149 cases of nodular sclerosis HL, 99 cases of mixed cellularity HL, 12 cases of lymphocyte-rich classical HL, 7 cases of lymphocyte-depletion HL, 9 cases of nodular lymphocyte-predominant HL (NLPHL), and 1 case of unclassifiable HL.
Paraffin-embedded blocks from reactive lymphoid tissue and different B- and T-cell lymphoma samples were obtained from the tissue archives of the Centro Nacional de Investigaciones Oncológicas (CNIO) Tumor Bank.
We obtained 5 HL-derived cell lines (L428, HDLM2, L540, KMH2, and HD-MY-Z) from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweigh, Germany).
Tissue microarray design
We used a Tissue Arrayer device (Beecher Instrument, Silver Spring, MD) to construct the TMAs. All HL cases were histologically reviewed and the richest areas of H/RS cells were marked in the paraffin blocks. In each case, 2 selected 1-mm-diameter cylinders from 2 different areas were included, along with 43 different controls to ensure the quality, reproducibility, and homogenous staining of the slides. Thus, 4 different TMA blocks were constructed, each containing 187 cylinders.
Internal controls were provided by cell lines, normal lymphoid tissue, and different B- and T-cell lymphomas. We used 5 EBV-negative HL-derived cell lines, with known cell-cycle alterations: L428, HDLM2, L540, KMH2, and HD-MY-Z. These cell lines were grown following standard protocols, fixed in formalin and included in paraffin as previously described.26 Included in each TMA were a representation of reactive lymphoid tissue (10 samples, including tonsil, spleen, and lymphadenitis) and the most frequent non-Hodgkin lymphomas, of which there were 28 samples: duplicated cylinders from B-cell chronic lymphocytic leukemia (B-CLL; 2 cases), follicular lymphoma (FCL; 2 cases), mantle cell lymphoma (MCL; 2 cases), diffuse large B-cell lymphoma (DLBCL; 2 cases), Burkitt lymphoma (BL; 2 cases, EBV-positive and -negative), peripheral T-cell lymphoma (PTCL; 2 cases, EBV-positive and -negative), and anaplastic large-cell lymphoma (ALCL; 2 cases).
Immunohistochemistry
TMA blocks were sectioned at a thickness of 3 μm and dried for 16 hours at 56°C before being dewaxed in xylene and rehydrated through a graded ethanol series and washed with phosphate-buffered saline. Antigen retrieval was achieved by heat treatment in a pressure-cooker for 2 minutes in 10 mM citrate buffer (pH 6.5). Before staining the sections, endogenous peroxidase was blocked.
Immunohistochemical staining was performed on these sections using 27 different antibodies (Abs), described in Table1. After incubation, immunodetection was performed with the LSAB Visualization System (DAKO, Glostrup, Denmark) employing diaminobenzidine chromogen as substrate. Sections were counterstained with hematoxylin.
Antibodies used in the analyses, indicating source, dilution, threshold, and pattern of reactivity used, and positive controls
| Protein . | Clone . | Source . | Dilution . | Reactivity . | Threshold . | Internal control . |
|---|---|---|---|---|---|---|
| Ki67 | M1B1 | DAKO | 1:100 | High/low | >50% positive H/RS cells | Proliferating cells |
| Bcl2 | 124 | DAKO | 1:25 | High/low | >50% positive H/RS cells with strong expression | Small lymphocytes |
| Bax | POLYCLONAL | Santa Cruz | 1:1000 | Positive/negative | >10% positive H/RS cells | Benign B lymphocytes |
| Bcl-XL | 2H12 | ZYMED | 1:10 | High/low | >50% positive H/RS cells with strong expression | HL-derived cell lines |
| Mcl1 | POLYCLONAL | DAKO | 1:100 | High/low | >50% positive H/RS cells with strong expression | Proliferating cells |
| Survivin | POLYCLONAL | RD Systems | 1:1500 | High/low | >50% positive H/RS cells with strong expression | HL-derived cell lines |
| p65/RelA | F-6 (p65) | Santa Cruz | 1:2000 | Positive/negative | Nuclear expression in H/RS cells | HL-derived cell lines |
| Cyclin A | 6E6 | Novocastra | 1:100 | Positive/negative | >10% positive H/RS cells | Proliferating cells (G2/M) |
| Cyclin B1 | 7A9 | Novocastra | 1:25 | Positive/negative | >10% positive H/RS cells | Proliferating cells (G2/M) |
| Cyclin D1 | DCS-6 | DAKO | 1:100 | Positive/negative | Any positive H/RS cells | Macrophages and endothelial cells |
| Cyclin D3 | DCS-22 | Novocastra | 1:10 | Positive/negative | >10% positive H/RS cells | Proliferating cells |
| Cyclin E | 13A3 | Novocastra | 1:10 | Positive/negative | >10% positive H/RS cells | TMA controls, proliferating cells |
| CDK1 | 1 | Transduction Lab | 1:1500 | Positive/negative | >10% positive H/RS cells | TMA controls, proliferating cells |
| CDK2 | 8D4 | NeoMarkers | 1:500 | Positive/negative | >10% positive H/RS cells | TMA controls, proliferating cells |
| CDK6 | K6.83 | Chemicon | 1:10 | Positive/negative | >10% positive H/RS cells | TMA controls |
| Bcl6 | PG-B6p | DAKO | 1:10 | Positive/negative | >10% positive H/RS cells | CG B cells and B-cell lymphomas |
| SKP2 | 1G12E9 | ZYMED | 1:10 | Positive/negative | >50% positive H/RS cells | Proliferating cells |
| STAT1 | C-136 | Santa Cruz | 1:50 | Positive/negative | Nuclear expression in H/RS cells | Reactive lymphocytes and macrophages |
| STAT3 | F-2 | Santa Cruz | 1:500 | Positive/negative | Nuclear expression in H/RS cells | Reactive lymphocytes and macrophages |
| PTEN | 28H6 | Novocastra | 1:500 | Positive/negative | Expression in H/RS cells similar to benign cells | Normal cells |
| P53 | DO-7 | Novocastra | 1:50 | High/low | >80% positive H/RS cells | Scattered GC cells |
| P21 | EA10 | Oncogene | 1:50 | High/low | >50% positive H/RS cells | Scattered GC cells |
| P16 | POLICLONAL | Santa Cruz | 1:50 | High/low | Expression in H/RS cells similar to benign cells | Normal cells |
| P27 | 57 | Transduction Lab | 1:1000 | High/low | >50% positive H/RS cells | Resting lymphoid cells |
| Hdm2 | IF2 (Mdm2) | Oncogen | 1:10 | High/low | >50% positive H/RS cells | TMA internal controls, macrophages, and endothelial cells |
| Rb | G3-245 | BD PharMingen | 1:250 | High/low | >50% positive H/RS cells | Proliferating cells |
| EBV-LMP | CS1-4 | Novocastra | 1:250 | Positive/negative | Cytoplasmic and membranous expression in H/RS | EBV-positive TMA controls |
| Protein . | Clone . | Source . | Dilution . | Reactivity . | Threshold . | Internal control . |
|---|---|---|---|---|---|---|
| Ki67 | M1B1 | DAKO | 1:100 | High/low | >50% positive H/RS cells | Proliferating cells |
| Bcl2 | 124 | DAKO | 1:25 | High/low | >50% positive H/RS cells with strong expression | Small lymphocytes |
| Bax | POLYCLONAL | Santa Cruz | 1:1000 | Positive/negative | >10% positive H/RS cells | Benign B lymphocytes |
| Bcl-XL | 2H12 | ZYMED | 1:10 | High/low | >50% positive H/RS cells with strong expression | HL-derived cell lines |
| Mcl1 | POLYCLONAL | DAKO | 1:100 | High/low | >50% positive H/RS cells with strong expression | Proliferating cells |
| Survivin | POLYCLONAL | RD Systems | 1:1500 | High/low | >50% positive H/RS cells with strong expression | HL-derived cell lines |
| p65/RelA | F-6 (p65) | Santa Cruz | 1:2000 | Positive/negative | Nuclear expression in H/RS cells | HL-derived cell lines |
| Cyclin A | 6E6 | Novocastra | 1:100 | Positive/negative | >10% positive H/RS cells | Proliferating cells (G2/M) |
| Cyclin B1 | 7A9 | Novocastra | 1:25 | Positive/negative | >10% positive H/RS cells | Proliferating cells (G2/M) |
| Cyclin D1 | DCS-6 | DAKO | 1:100 | Positive/negative | Any positive H/RS cells | Macrophages and endothelial cells |
| Cyclin D3 | DCS-22 | Novocastra | 1:10 | Positive/negative | >10% positive H/RS cells | Proliferating cells |
| Cyclin E | 13A3 | Novocastra | 1:10 | Positive/negative | >10% positive H/RS cells | TMA controls, proliferating cells |
| CDK1 | 1 | Transduction Lab | 1:1500 | Positive/negative | >10% positive H/RS cells | TMA controls, proliferating cells |
| CDK2 | 8D4 | NeoMarkers | 1:500 | Positive/negative | >10% positive H/RS cells | TMA controls, proliferating cells |
| CDK6 | K6.83 | Chemicon | 1:10 | Positive/negative | >10% positive H/RS cells | TMA controls |
| Bcl6 | PG-B6p | DAKO | 1:10 | Positive/negative | >10% positive H/RS cells | CG B cells and B-cell lymphomas |
| SKP2 | 1G12E9 | ZYMED | 1:10 | Positive/negative | >50% positive H/RS cells | Proliferating cells |
| STAT1 | C-136 | Santa Cruz | 1:50 | Positive/negative | Nuclear expression in H/RS cells | Reactive lymphocytes and macrophages |
| STAT3 | F-2 | Santa Cruz | 1:500 | Positive/negative | Nuclear expression in H/RS cells | Reactive lymphocytes and macrophages |
| PTEN | 28H6 | Novocastra | 1:500 | Positive/negative | Expression in H/RS cells similar to benign cells | Normal cells |
| P53 | DO-7 | Novocastra | 1:50 | High/low | >80% positive H/RS cells | Scattered GC cells |
| P21 | EA10 | Oncogene | 1:50 | High/low | >50% positive H/RS cells | Scattered GC cells |
| P16 | POLICLONAL | Santa Cruz | 1:50 | High/low | Expression in H/RS cells similar to benign cells | Normal cells |
| P27 | 57 | Transduction Lab | 1:1000 | High/low | >50% positive H/RS cells | Resting lymphoid cells |
| Hdm2 | IF2 (Mdm2) | Oncogen | 1:10 | High/low | >50% positive H/RS cells | TMA internal controls, macrophages, and endothelial cells |
| Rb | G3-245 | BD PharMingen | 1:250 | High/low | >50% positive H/RS cells | Proliferating cells |
| EBV-LMP | CS1-4 | Novocastra | 1:250 | Positive/negative | Cytoplasmic and membranous expression in H/RS | EBV-positive TMA controls |
The staining of the TMA sections was evaluated by 2 different pathologists (J.F.G. and F.I.C.), using uniform criteria. In order to guarantee the reproducibility of this method, we decided to employ straightforward and clear-cut criteria. Briefly, the pattern of staining for each Ab was recorded as positive or negative, and high or low expression, taking into account the expression in H/RS cells and different cutoffs for each marker (Table 1).
Some exceptions to these rules were as follows: (1) Cytoplasmic STAT1, STAT3, and NF-κB expression can generally be found in normal lymphoid cells and in H/RS cells. In this study, we considered positive cases only when nuclear expression in the tumoral cells could be seen without difficulty, indicating the activated form of these proteins.9 (2) p53 nuclear overexpression is a well-known phenomenon in HL, so we considered cases with high expression when more than 80% of tumoral cells were strongly positive. (3) Bcl2 and other antiapoptoic regulators (Bcl-XL, survivin) overexpression was considered only when more than 50% of H/RS cells strongly expressed the protein. Cases with only faint cytoplasmic expression in some tumoral cells were classified as negative. (4) Cyclin D1 expression is considered abnormal in lymphoid cells, being positive only in tumoral cells from MCL, multiple myeloma (MM), and some prolymphocytic leukemia (PLL). Therefore, those HL cases displaying any kind of expression in H/RS cells were considered to be positive.
Although the reactivity of most of the antibodies used have been validated in different previous studies, we cannot fully exclude that undescribed reactivities of the antibodies or the technique used here may affect some of the results.
In situ detection of apoptosis and EBER in situ hybridization
Apoptosis was detected using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Intergen, Oxford, United Kingdom), based on the TdT-mediated biotin-dUTP nicked-end labeling (TUNEL) methodology. Briefly, TMA sections were pretreated with proteinase K for 15 minutes, and then incubated with TdT enzyme for 1 hour at 37°C and afterward with antidigoxigenin peroxidase conjugate for 1 hour. Color was developed with 3, 3′ diaminobenzidine tetrahydrochloride (10 minutes) and counterstained with hematoxylin.
EBV was detected by in situ hybridization with fluorescein-conjugated Epstein-Barr virus (EBER) peptide nucleic acid (PNA) probe (DAKO). This probe is complementary to the 2 nuclear EBER RNAs encoded by the Epstein-Barr virus. TMA sections were pretreated with proteinase K for 10 minutes at 37°C, and then incubated with EBER PNA probe for 1.5 hours at 55°C in a humid chamber and afterward washed with Stringent Wash Solution for 25 minutes at 55°C in a water bath with shaking. Detection was performed with anti–fluorescein isothiocyanate (FITC) antibody (clone DAK-FITC4, from DAKO) diluted at 1:100. After incubation, immunodetection was performed with biotinylated anti–mouse immunoglobulins, followed by peroxidase-labeled streptavidin (LSAB Visualization System, DAKO) with diaminobenzidine chromogen as substrate. Sections were counterstained with hematoxylin.
Validation of the technique
The reproducibility of the results thus obtained was confirmed by comparing them with those from whole sections in 42 randomly selected cases, stained using the same procedures for some of the Abs and probes used in this study, and for other markers routinely in use in HL that have variable and well-known expression in H/RS cells. Specifically, we evaluated the expression of CD20, CD30, CD15, LMP, Bcl2, Bcl6, p53, PTEN, p16, and EBER-ISH.
Statistical study
The Pearson chi-square test and Fisher exact test were used where appropriate to establish whether there were any relationships between the different markers included in this study. Differences were considered to be significant for values of P < .05.
Survival analyses were performed of all the classical HL cases for which clinical information was available (approximately 70%). Actuarial survival curves, in terms of overall survival (OS), were plotted using the Kaplan and Meier method. Statistical significance of associations between individual variables and OS was determined by using the log-rank test. The Cox proportional hazard univariate analysis was also performed independently for each variable, estimating values of the relative risk (RR), chi-square, and P. All the statistical analyses were performed using the software package SPSS (Chicago, IL).
Results
With this design, 89.3% individual core biopsies were found to include identifiable H/RS cells. As each TMA included 2 different core cylinders from the same patient, this resulted in 95.3% of cases being considered as having TMA results that could be evaluated (Figure1). An analysis comparing both core samples from the same cases showed a concordance of 87%.
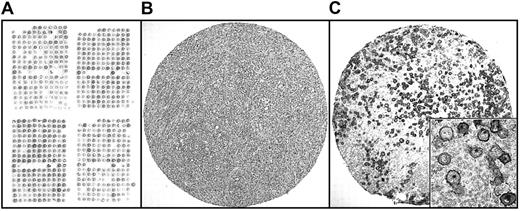
TMA design.
(A) Distribution of core cylinders in TMAs. Preservation of morphology, antigen preservation, and representativity of these samples is shown in one of these cylinders, corresponding to nodular sclerosis HL, stained for H/E (B) and CD30 (C). Original magnification, panels B-C, × 40; panel C inset, × 600.
TMA design.
(A) Distribution of core cylinders in TMAs. Preservation of morphology, antigen preservation, and representativity of these samples is shown in one of these cylinders, corresponding to nodular sclerosis HL, stained for H/E (B) and CD30 (C). Original magnification, panels B-C, × 40; panel C inset, × 600.
To validate the chosen approach, the results obtained from the staining of 10 different Abs and probes using TMAs were compared in a series of 42 randomly selected cases with those obtained in the analysis of whole sections. These proved to be highly reproducible (Table 2). The overall concordance was 93.8%, a figure that accords with previously reported results for other tumoral models,27 where concordances of more than 90% have been recorded when comparing results from the use of TMAs with whole sections.
Evaluation of IHC results in whole sections by comparison with TMA results: number of concordant cases/number of evaluable cases
| . | Concordant results between whole sections and TMA cores (%) . |
|---|---|
| CD20 | 32/39 (82.5) |
| CD30 | 38/38 (100) |
| CD15 | 36/39 (92.3) |
| BCL2 | 35/39 (89.8) |
| BCL6 | 37/41 (90.2) |
| P53 | 39/39 (100) |
| P16 | 30/37 (81.8) |
| PTEN | 35/35 (100) |
| LMP | 37/37 (100) |
| EBER | 41/41 (100) |
| Overall concordance | 93.5% |
| . | Concordant results between whole sections and TMA cores (%) . |
|---|---|
| CD20 | 32/39 (82.5) |
| CD30 | 38/38 (100) |
| CD15 | 36/39 (92.3) |
| BCL2 | 35/39 (89.8) |
| BCL6 | 37/41 (90.2) |
| P53 | 39/39 (100) |
| P16 | 30/37 (81.8) |
| PTEN | 35/35 (100) |
| LMP | 37/37 (100) |
| EBER | 41/41 (100) |
| Overall concordance | 93.5% |
Percentage concordance is shown in brackets. All the cases correspond to classical HL samples.
Immunohistochemistry
Threshold reactivity was defined for each Ab according to the characteristics of the staining, giving priority to the ease of reproducibility of results. Internal control was provided in each case by the reactivity of accompanying lymphocytes and macrophages, or the cylinders included in the TMA for control purposes. Figure2 shows the common pattern of expression for each Ab, and a summary of the results is shown in Table3.
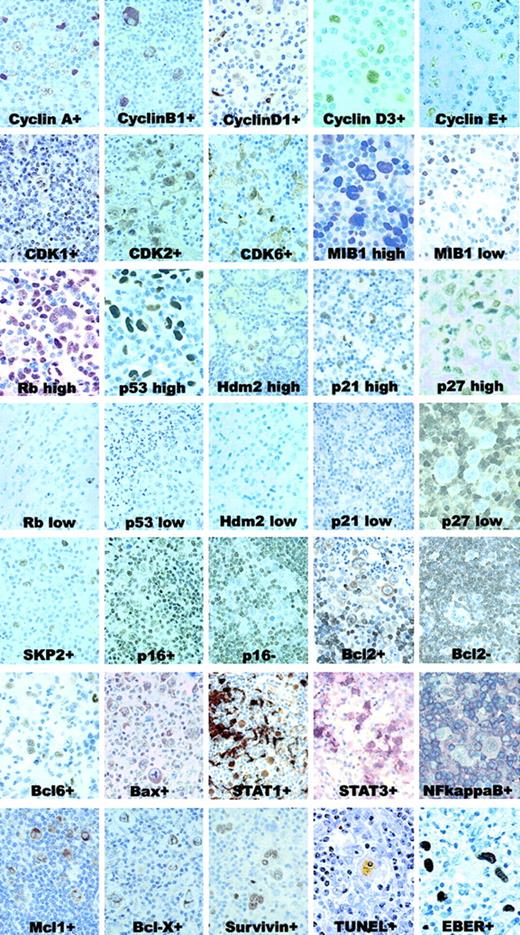
IHC and ISH patterns.
Samples with positive and negative (or high vs low expression) for each marker are shown. All these cases are representative of classical HL, with either nodular sclerosis or mixed cellularity. Original magnification for cyclins, MIB1, Rb, p53, p27, p65/RelA, Bcl6, TUNEL, and EBER, × 1000; remaining panels, × 600.
IHC and ISH patterns.
Samples with positive and negative (or high vs low expression) for each marker are shown. All these cases are representative of classical HL, with either nodular sclerosis or mixed cellularity. Original magnification for cyclins, MIB1, Rb, p53, p27, p65/RelA, Bcl6, TUNEL, and EBER, × 1000; remaining panels, × 600.
IHC and ISH results, indicating number of positive cases
| Marker . | Positive cases (%) . |
|---|---|
| PTEN | 258/258 (100) |
| Bax | 259/266 (97.4) |
| Cyclin A | 262/271 (96.7) |
| Cyclin B1 | 239/257 (93) |
| Cyclin E | 231/257 (89.9) |
| Survivin | 234/262 (89.3) |
| STAT1 | 236/267 (88.4) |
| CDK1 | 269/264 (86.7) |
| SKP2 | 226/268 (84.3) |
| CDK2 | 223/265 (83.8) |
| P21 | 218/270 (80.7) |
| TUNEL | 169/210 (80.5) |
| P65/Re1A | 196/257 (76.3) |
| P16 | 189/259 (73) |
| Hdm2 | 171/244 (70.1) |
| STAT3 | 145/261 (55.6) |
| M1B1 | 121/265 (45.7) |
| Rb | 102/262 (38.9) |
| CyclinD3 | 103/267 (38.6) |
| EBER | 98/256 (38.3) |
| LMP | 96/252 (38.1) |
| Bcl2 | 61/264 (23.1) |
| P27 | 59/256 (23.0) |
| CDK6 | 54/262 (20.6) |
| Bcl-XL | 50/260 (19.2) |
| P53 | 41/265 (15.5) |
| Bcl6 | 24/254 (9.4) |
| Mcl1 | 18/263 (6.8) |
| Cyclin D1 | 14/270 (5.2) |
| Marker . | Positive cases (%) . |
|---|---|
| PTEN | 258/258 (100) |
| Bax | 259/266 (97.4) |
| Cyclin A | 262/271 (96.7) |
| Cyclin B1 | 239/257 (93) |
| Cyclin E | 231/257 (89.9) |
| Survivin | 234/262 (89.3) |
| STAT1 | 236/267 (88.4) |
| CDK1 | 269/264 (86.7) |
| SKP2 | 226/268 (84.3) |
| CDK2 | 223/265 (83.8) |
| P21 | 218/270 (80.7) |
| TUNEL | 169/210 (80.5) |
| P65/Re1A | 196/257 (76.3) |
| P16 | 189/259 (73) |
| Hdm2 | 171/244 (70.1) |
| STAT3 | 145/261 (55.6) |
| M1B1 | 121/265 (45.7) |
| Rb | 102/262 (38.9) |
| CyclinD3 | 103/267 (38.6) |
| EBER | 98/256 (38.3) |
| LMP | 96/252 (38.1) |
| Bcl2 | 61/264 (23.1) |
| P27 | 59/256 (23.0) |
| CDK6 | 54/262 (20.6) |
| Bcl-XL | 50/260 (19.2) |
| P53 | 41/265 (15.5) |
| Bcl6 | 24/254 (9.4) |
| Mcl1 | 18/263 (6.8) |
| Cyclin D1 | 14/270 (5.2) |
The different markers are ordered by the percentage of positive cases in the series (additional unreviewed information will be available on our website athttp://bioinfo.cnio.es/data/Hodgkin2003/).
As part of the validation process, the immunohistochemical expression of latent membrane protein (LMP) from EBV virus was analyzed and the results compared with those of EBER-ISH. A concordance of nearly 100% was found, thus confirming the specificity of this system.
Additionally, the different control samples illustrated the expression pattern for the proteins analyzed and provided internal controls for EBER-ISH and TUNEL techniques. As expected, proliferating cells in normal tissue samples showed expression of MIB1, cyclins A, B1, and D3, CDK1 and 2, SKP2, and Rb, but were negative for p27, whose expression is opposed to proliferation. Tumor suppressor genes such as PTEN and p16 were expressed in most cells in normal tissues, whereas only scattered lymphoid cells in the germinal centers were p53- or Hdm2-positive. The levels of cyclin D1 and CDK6 were undetectable in normal lymphoid cells. STAT1 and STAT3 expression was associated with proliferating cells, markedly in EBV-infected tissues such as those with infectious mononucleosis. Cytoplasmic p65/RelA expression could be seen in most lymphocytes, but nuclear protein was undetectable in normal lymphoid cells. The pattern of expression of Bcl2, Bax, Bcl-XL, survivin, and Bcl6 in lymphoid tissue has been described elsewhere.28 29
In the HL-derived cell lines, overexpression and/or loss of some oncogenes, tumor suppressor genes, STAT proteins, and p65/RelA have been previously described.7,30 31 Our results from the TMAs confirmed these observations and also revealed the frequent expression of PTEN, cyclins A, B1, and E, CDK1, 2, and 6, and Rb. Finally, the analysis of the different lymphoma samples included in these TMAs demonstrated the pattern of expression for each protein, provided internal controls for markers such as cyclin D1 in MCLs or Bcl2 and Bcl6 in FCLs or DLBCLs, and also delineated the differences between low-growth fraction lymphomas (indolent lymphomas, such as CLL, FCL, or MCL, showing low-level expression of cyclins, p53, and CDKs and expressing CDKIs) and high-growth fraction lymphomas (aggressive lymphomas, such as DLBCL, BL, ALCL, and PTCL, with high-level expression of cyclins and CDKs, frequent expression of p53, loss of some CDKIs, and generally expressing Bcl6).
Considering these results together, it seems that H/RS cells express cyclins and CDKs involved in G1/S transition (cyclin D, cyclin E1, CDK2, CDK6), G2/M transition (cyclin A, cyclin B1, CDK1), and molecules involved in apoptosis control (p65/RelA, Bcl2, Bcl-XL, survivin, and Bax). At the same time, they exhibit markers suggestive of dysfunction of the major tumor suppressor pathways, such as high p53 and Hdm2 staining, frequent loss of p27KIP1 and also p16INK4A in a number of cases, with increased expression of SKP2.
Some of the most striking findings were the overexpression of cyclin E, CDK2, and CDK6, and the increased nuclear expression of p65/RelA, STAT1, and STAT3.
Thus, in 89.9% of cases, H/RS cells showed stronger cyclin E staining than that observed in the reactive germinal centers included for control within the TMA. Similar findings have been observed for CDK2, the normal partner of cyclin E, whose overexpression could be detected in 83.8% of samples. Lower percentages of positive cases were observed for CDK6 (21%), cyclin D3 (39%), and cyclin D1 (5%). A high degree of expression of cyclin A and cyclin B1 was also observed in nearly all cases (97% and 93%, respectively).
In this series, nuclear p65/RelA could also be demonstrated in 76.3% of cases, with a clear difference from the results obtained in reactive germinal centers, where nuclear p65/RelA was only rarely detected. Survivin expression was found in 89.3% of cases.
Nuclear STAT1 and STAT3 staining of H/RS cells was detectable in up to 88% and 56% of samples, respectively. In this case there was also a marked difference in comparison with the rarity of either or both transcription factors in reactive lymphocytes, with the sole exception of infectious mononucleosis control cases.
Cyclin D1, besides its expected presence in endothelial cells and macrophages, was unexpectedly found in H/RS cells in 5% of samples. Nuclear PTEN was demonstrable in all cases, in reactive and tumoral cells. Bcl6 was observed in the majority of NLPHL (67%), and also in a small proportion (7.3%) of classical HL cases.
Classical HL versus lymphocyte-predominant HL
The comparison of the expression profiling of classical HL with NLPHL revealed differences in the profile between the 2 types of HL. These differences were significant even though few NLPHL cases were included. Thus, NLPHL cases were more frequently negative for EBV than were classical HL cases (0% positive cases vs 39.5%,P = .020), p65/RelA (37.5% vs 77.5%,P = .020), CDK2 (37.5% vs 85.2%, P = .003), p16 (37.5% vs 74.1%, P = .035), and Bcl2 (0% vs 23.9%,P = .048), and they were commonly positive for Bcl6 (66.7% vs 7.3%, P = .000). (P values are those associated with calculated Fisher exact test statistics.)
Relationship between different markers
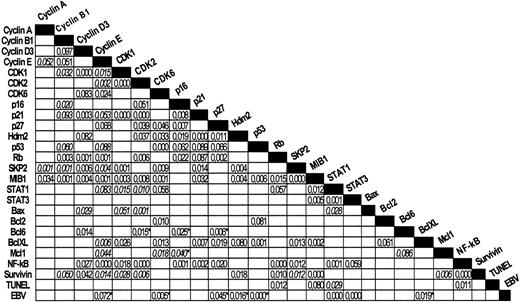
The relationships between the different markers are illustrated in Figure 3. Statistical analysis using the Pearson and Fisher exact tests revealed a large number of significant associations between different characters (see also additional information).
Chi-square tests.
Most significant results are shown. P values of Pearson (nonitalicized) or Fisher exact tests (italicized) are indicated. Inverse relationships between pairs of markers are indicated with asterisks.
Chi-square tests.
Most significant results are shown. P values of Pearson (nonitalicized) or Fisher exact tests (italicized) are indicated. Inverse relationships between pairs of markers are indicated with asterisks.
All NPLHL cases were EBV-negative. In classical HL, the presence of EBV was more frequent in mixed cellularity (MC) types than in other subtypes (64.6% vs 20.2%, P = .000). The presence of EBV in this series appears to be directly associated with STAT1 and STAT3 expression, while there is an inverse relation with cyclin E, CDK6, p27, p53, Hdm2, and Bcl-XL. This confirms the role that EBV plays in this disease. It is of particular note that EBV presence is strongly associated with STAT1 and STAT3 expression.
One of the most striking associations is that established between MIB1 and apoptosis. In addition, growth fraction, as detected by MIB1, is associated with a large set of markers including STAT1 and STAT3, CDK1, CDK2, CDK6, SKP2, cyclins B1, D3, and E, Hdm2, p53, p21, Rb, Bcl-XL, and survivin. The strongest relationships were found for SKP2 and STAT3.
Apoptosis rate, as measured with TUNEL, was related with nuclear p65/RelA, Rb, and STAT1 expression. At the same time, both MIB1 and TUNEL were closely related.
Cyclin E appears to have a more ubiquitous relationship, being directly related with cyclins A and B1, MIB1, p21, p27, Rb, SKP2, STAT1, STAT3, CDK1, CDK2, CDK6, and p65/RelA. Of particular relevance is the strong relationship between cyclin E and p65/RelA identified here.
NF-κB (p65/RelA) is also one of the most widely related markers, as shown by 2 × 2 contingency analyses, since in this series its expression is associated with p21, p16, p27, Rb, cyclins E and D3, CDK1, CDK2, SKP2, STAT1, STAT3, survivin, and TUNEL.
Lastly, Bcl6 expression is strongly opposed to p27 in this series. This finding is reminiscent of what may be observed in reactive germinal centers. At the same time, cyclin D3 is simultaneously expressed with Bcl6.
Relationship with survival probability
The survival analyses, restricted to classical HL cases, were done in those cases for which a complete clinical follow-up was available. The results of survival analyses using Kaplan-Meier and Cox regression models are shown in Table 4. Shorter OS was significantly related with high apoptotic index (TUNEL), high proliferative index (MIB1), and overexpression of Bcl2, p53, Bcl-XL, and Bax. Trends were also observed for low p21 and high STAT3 expression, but they were not statistically significant (see also additional information).
Biologic markers related with patient's outcome
| . | Kaplan-Meier P . | Cox regression model . | ||
|---|---|---|---|---|
| Chi-square . | RR . | P . | ||
| Bcl2 | .0041 | 8.41 | 2.83 | .004 |
| MIB1 | .0307 | 4.67 | 2.28 | .031 |
| p53 | .0232 | 5.15 | 2.45 | .023 |
| Bcl-xL | .0121 | 6.28 | 2.47 | .012 |
| Bax | .0130 | 6.16 | 4.05 | .013 |
| TUNEL | .0296 | 4.72 | 6.78 | .030 |
| STAT3 | .0623 | — | — | .068 |
| p21 | .0842 | — | — | .061 |
| . | Kaplan-Meier P . | Cox regression model . | ||
|---|---|---|---|---|
| Chi-square . | RR . | P . | ||
| Bcl2 | .0041 | 8.41 | 2.83 | .004 |
| MIB1 | .0307 | 4.67 | 2.28 | .031 |
| p53 | .0232 | 5.15 | 2.45 | .023 |
| Bcl-xL | .0121 | 6.28 | 2.47 | .012 |
| Bax | .0130 | 6.16 | 4.05 | .013 |
| TUNEL | .0296 | 4.72 | 6.78 | .030 |
| STAT3 | .0623 | — | — | .068 |
| p21 | .0842 | — | — | .061 |
— indicates data not needed.
Discussion
Experimental models and observations in viral-induced cell transformation have shown the synergistic effect of concurrent and multiple tumor suppressor pathway inactivation. Examples of this are provided by viral E6 and E7 HPV proteins binding p53 and Rb,32,33 and EBV-mediated B-cell transformation through p53 and Rb binding by EBNA5 protein.34-36 Observations performed in different experimental models have proved difficult to transfer and confirm in human tumors, where the simultaneous analysis of different suppressor pathways requires multiple genes to be explored with an array of techniques. The availability of monoclonal Abs for paraffin-embedded tissue and the development of TMA now make it possible to explore this matter in tissue samples from human tumors, even in the case of HL, where the low proportion of tumoral cells requires the use of highly sensitive techniques.
HL is exceptional among malignant neoplasms in that the tissue samples involved consist mainly of reactive lymphocytes, plasma cells, and often fibrous stroma, containing only a limited number of the putative neoplastic cell, the H/RS cell. Thus, in a majority of cases H/RS cells represent less than 10% of the lymph node cell population, making in situ techniques such as IHC and ISH the most suitable methods for analyzing protein, DNA, and RNA expression, as they allow the visualization and identification of this expression selectively in the neoplastic cells.
TMA allows simultaneous analysis of several proteins in a large series of patients, thus revealing the complex interactions between diverse pathways and genes, and, in turn, the clinical relevance of multiple biologic parameters. Different groups in several types of human tumors have previously demonstrated the suitability of this technique. To our knowledge, a single limited study24 has previously provided evidence of the feasibility of this tool in HL, based solely on the expression of a single protein.
In this paper we have shown the adequacy of TMA in the analysis of heterogeneous tumors such as HL, made a comparative study of results from analyses of whole tissue sections, and demonstrated the suitability of this design and the chosen conditions (two 1-mm diameter cylinders per sample). Detection of EBV LMP by IHC versus EBER by ISH has also shown a high concordance rate, highlighting the accuracy of the technique. Since TMA technology is a population-level research tool, and is not intended for making clinical diagnoses of individual cases; the general picture provided by these experiments more than compensates for the information probably lost concerning some of the cylinders and cases.
The checkpoint regulating transition from G1 to S is frequently disrupted in cancer, and the mechanism of this regulation is complex. There are 2 cyclin/CDK complexes, cyclin D/CDK4 or CDK6 and cyclinE/CDK2, that phosphorylate the retinoblastoma gene product, Rb, alter its ability to associate with other cellular proteins, such as E2F, and activate the transcription of several genes required for S progression. The kinase activities of CDK4/cyclin D and CDK2/cyclin E convert Rb to the hyperphosphorylated state with loss of binding activity. G1 CDK activity is positively regulated by growth factors and inhibited by a variety of physiologic signals from the cell microenvironment.37
This series shows that H/RS cells display, in most cases, deregulation of the genes involved in the G1/S and G2/M checkpoints, and inactivation of the tumor suppressor pathways defined by p14ARF-p53-p21WAF1, p16INK4a-Rb, and p27KIP1. Thus, the tumoral cells show almost constant Hdm2 overexpression, which has been found in previous studies to be associated with the presence of alternative transcripts of Hdm2 lacking the adhesion to p14ARF, its inhibitory protein.7 Hdm2 overexpression is considered to be a mechanism that leads to p53 inactivation, binding it and concealing its transcriptional activity.38 Although alterations in the p53 gene are the most common genetic alteration found in human cancer (> 50%), the incidence of p53 mutations is significantly lower in HL than in other neoplasias.39 40 The understanding of the activity of these proteins in HL may explain the unusual low frequency of p53 mutations in this tumor with overexpression of the protein and defective p53 function.
The p16INK4a-Rb pathway is also presumably inactivated in a large fraction of these samples as the result of p16INK4aloss (due to promoter region methylation or deletion) and/or cyclin D overexpression. Results obtained here concerning p16 loss differ slightly from those previously reported, probably as a consequence of the higher sensitivity of the techniques or the larger size of the series employed in the current study.
In the majority of cases, p27KIP1 protein is also lost, probably as a consequence of increased degradation mediated by SKP2. SKP2 protein acts as a ubiquitin ligase for p27KIP1, and most cases show overexpression of this protein in H/RS cells. Indeed, consistent with this interpretation, there is an inverse relationship between p27KIP1 and SKP2 expression in HL tumors.
At the same time, the tumoral cells are characterized by the increased expression in a large proportion of cases of cyclins and CDKs involved in G1/S and G2/M transition, such as cyclins D, A, B1, and E, CDK2, and CDK6. A striking finding here is the observation that 90% and 84% of cases show overexpression of cyclin E and CDK2, respectively, which differs strongly from the observations in reactive lymphoid tissue and other NHLs. Cyclin E and CDK2 both form a complex, which is negatively regulated through the interaction with p27KIP1, and whose balance determines the cell-cycle progression to the S phase. This almost universal finding in H/RS could be related with the increased growth fraction seen in these cells, as already described.41 Additionally, deregulated cyclin E increases chromosomal instability and polyploidy,42 affecting those processes involved in the faithful duplication and segregation of chromosomes. These are findings that could be considered to be the hallmark of HL cells.
Consistent with altered mitotic checkpoint control, the H/RS cells also express elevated protein levels of the mitotic regulatory proteins cyclin A, cyclin B1, and CDK1 (cdc2). The cyclin B/CDK1 complex (also called mitosis-promoting factor, MPF) is the primary regulator of transition from G2 to M phase, being involved in chromosome condensation, nuclear membrane breakdown, and spindle formation. The transition from G2 to M phases and the proper transition during mitosis depend on cyclin A and B proteosomal degradation.43 For example, onset of mitosis is regulated by the activation of cyclin B1/CDK1 and this event is controlled at several levels, ensuring that chromosome segregation does not occur in the case of unreplicated or damaged DNA, or misaligned chromosomes. The altered expression of these G2/M cyclins and CDKs in H/RS cells could depend on inappropriate ubiquitination/degradation, and also could, at least partially, explain the frequent alterations in cytokinesis and the morphologic abnormalities found in these cells, leading to the formation of the characteristic multinucleated cells.
H/RS cells also have defective regulation of apoptosis, as shown by the association of Bcl2 (23%), Bcl-XL (19%), and survivin (89%) overexpression, loss of Bax expression (3%), and increased nuclear expression of p65/RelA (76%). These findings are consistent with those of previous studies9,44,45 suggesting that Bcl2, Bcl-XL, and NF-κB activation are critical mechanisms involved in the resistance to apoptosis by H/RS cells, at least in some cases as a consequence of the transforming capacity of LMP1-EBV protein.46 The frequent expression of survivin in the H/RS cells in HL has not been previously described, and may represent an additional mechanism for evading apoptosis in the G2/M checkpoint. The relevance of Bcl2 and Bcl-XL overexpression is underlined by the shortened OS of these patients. This result is similar to observations in DLBCL, and confirms previous observations in HL.47 In this context it is worth mentioning that the disease in cases of HL with marked overexpression of p53 proteins, as observed in 15% of cases, is more aggressive and has a shortened OS. Here we have chosen this high threshold of p53 expression, similar to that found to reflect p53 mutations in other lymphoid tumors.
H/RS cells harbor clonally rearranged and somatically mutated immunoglobulin genes, indicating their derivation in most cases from germinal center B cells,1 but lack B-cell receptor expression. Although germinal center B cells that lack a functional high-affinity Ab undergo apoptosis within the germinal center, under physiologic conditions2 the H/RS cells escape programmed cell death (apoptosis), and instead proliferate and disseminate. The concurrent alterations in cell-cycle regulatory pathways observed in this series thus explain the insensitivity of H/RS cells to inhibitory signals, and the high growth fraction of these cells.
The analysis of the relationships among the proteins studied yielded some interesting findings. EBV-positive cases have a characteristic profile distinguished by the overexpression of STAT1 and STAT3 by the H/RS cells, and down-regulation of the expression of p27, Hdm2, p53, cyclin E, CDK6, and Bcl-XL, which seems to confirm that EBV presence in H/RS is not just an innocent bystander, but also plays a role in the progression of the disease and the survival of tumoral cells. STAT1 and STAT3 also seem to play a determinant role, since their expression in H/RS cells is significantly associated with the presence of increased levels of CDK1, CDK2, and CDK6, Rb, NF-κB, and Bax, and also is associated with higher proliferative index. Previous studies analyzing the expression profile in EBV-positive cases at the RNA level34 also revealed a specific pattern, but showed some differences from our results at the protein level. These findings provide convincing evidence that EBV exploits the normal program of B cell-cycle activation as part of its strategy.
Another key transcription factor revealed by this study as playing an important role in HL is NF-κB, as has been observed previously in HL-derived cells48,49 and tumors.9,10 This transcription factor regulates the expression of numerous genes regulating critical functions in the immune system, inflammatory responses involved in the control of cell proliferation and apoptosis. This analysis proves that the presence of NF-κB and its activation, as reflected by the nuclear expression, is related to changes in the expression of a set of proteins that play a role in the control of cell-cycle progression, apoptosis, and gene transcription, such as p21, p16, p27, Rb, cyclins E and D3, CDK1, CDK2, SKP2, STAT1, STAT3, and survivin. It is noteworthy that there is a strong relation observed with survivin (P < .000), a member of the inhibitor of apoptosis (IAP) family, which has been described as a target of NF-κB.50 Although the association with Bcl-XL (another NF-κB target) is not statistically significant, the majority of cases express high levels of both proteins.
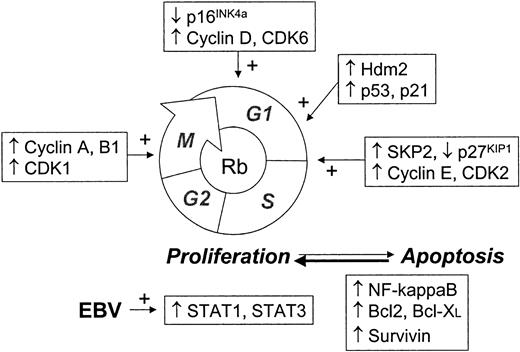
High MIB1 expression is associated here with the increased expression of Rb, p21, p53, Hdm2, cyclins A, B1, D3, and E, CDK1, CDK2, and CDK6, SKP2, STAT1, STAT3, and Bcl-XL. Thus it appears that the increased growth fraction, typical of Hodgkin cells, is the consequence of deregulation of multiple genes, significantly including cyclins and CDK, transcription factors and SKP2, a key gene in protein degradation mediated by ubiquitin. The complexity of the changes in cell-cycle regulation in H/RS cells is shown in Figure4.
Scheme of cell cycle showing main checkpoints commonly deregulated in HL tumors.
In spite of the small number of NLPHL cases included here, this study also reveals that these cases have a distinctive profile (Bcl6+, EBV-, p65/RelA-, p16-, Bcl2-, and CDK2-), different from that seen in classical HL, and not only attributable to the presence of EBV.
Clinical correlation also sheds further light on the relevance of these findings. Several of these regulator proteins seem to be associated with patient outcome. Thus, a shorter survival in this series is related with Bcl2, p53, Bcl-XL, Bax, and high proliferative and apoptotic indexes. Some of these prognostic markers had been previously noted in the literature.5,45,51 52 These findings have been shown to be statistically significant using Kaplan-Meier and Cox regression analyses and restricted to classical HL cases. Nevertheless, a more comprehensive analysis of these and other markers is currently being performed with the aim of incorporating biologic variables into the clinical prognosis scoring system.
In summary, TMA studies with a panel of Abs for cell-cycle markers show an unexpected constellation of abnormalities for the expression of oncogenes and cell-cycle control molecules, thus confirming the complexity of the changes involved in the malignant transformation in HL. Variations in the expression profiling reflect the histologic subtyping and account for some of the clinical variability observed in this disease.
We are indebted to Laura Cereceda (CNIO Tumor Bank) and L. Garcı́a (Hematology, H. Ramón y Cajal, Madrid) for their excellent assistance with data managing, and to R. Pajares and M. J. Acuña for their expertise and excellent technical assistance with TMA technology and immunohistochemical assays. We also thank all the participants of the Spanish Hodgkin Lymphoma Study Group for their cooperation.
Other members of the Spanish Hodgkin Lymphoma Study Group are as follows: P. Domı́nguez and C. Jara (FHA, Alcorcón), R. Quibén and L. Borbolla (H Móstoles, Madrid), C. Grande (H 12 Octubre, Madrid), J. Garcı́a (H Ramón y Cajal, Madrid), A. Castaño and P. Sáncjez-Godoy (H Servero Ochoa, Leganés), R. Martı́nez (HUC San Carlos, Madrid), J. Menárguez and P. Sabı́n (H Gregorio Marañón, Madrid), J. González-Carrero and C. Poderós (H Xeral-Cies, Vigo), Ll. Font (H Verge de la Cinta, Tortosa), M. A. Cruz (H Virgen de la Salud, Toledo), M. Llanos (HU Canarias), C. Morante (H Cabueñes, Gijón), E. Conde (HM De Valdecilla, Santander), M. F. Fresno and C. Rayón (HC de Asturias, Oviedo), R. Garcı́a (HCU Salamanca), J. Guma (H Sant Joan, Reus), P. Gonzalvo (HC de Jarrio, Coaña), G. Fernández (H Alvarez Buyllas, Mieres), J. Forteza and J. L. Bello (F Med Santiago de Compostela), and J. R. Méndez (H Valle de Nalón, Asturias).
Prepublished online as Blood First Edition Paper, September 12, 2002; DOI 10.1182/blood-2002-04-1128.
A complete list of the members of the Spanish Hodgkin Lymphoma Study Group appears in “.”
Supported by grants from the Comunidad Autónoma de Madrid (08.1/0028.1/2000) and the Ministerio de Ciencia y Tecnologia (1FD97-0431 and SAF2001-0060), Spain. F.I.C. is supported by a grant from the Madrid City Council and the CNIO.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Juan F. Garcı́a, Molecular Pathology Program, Centro Nacional de Investigaciones Oncológicas, C/ Melchor Fernández Almagro 3, E-28029 Madrid, Spain; e-mail:jfgarcia@cnio.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal