Abstract
The immunoglobulin variable heavy chain (IgVH) gene mutation status is an important prognostic factor in chronic lymphocytic leukemia (CLL), since cases with mutated VH genes show significantly longer survival than unmutated cases. Recently, we reported a preferential use of the VH3-21 gene in mutated CLL and showed that mutated VH3-21 cases had an inferior overall survival compared with other mutated CLL. In order to further characterize this subset, we performed VH gene analysis in 265 CLL cases and identified 31 VH3-21 cases (11.7%); 21 cases had mutated and 10 cases unmutated VH genes. Regardless of VH gene mutation status, a poor overall survival was found in the VH3-21 cases with a median survival of 83 months. These survival data confirm that VH3-21 cases do not fit into the general prognostic grouping of mutated and unmutated CLL. A large fraction of VH3-21 cases also demonstrated unique features with shorter lengths of the third complementarity determining region (CDR3) and CDR3s with highly homologous amino acid sequences. Furthermore, the VH3-21 cases showed a striking dominance of λ light chain expression, and analysis of the Igλ gene rearrangements revealed highly restricted use of the Vλ2-14/Jλ3 genes in the majority of cases. Taken together, our new findings strengthen the suggestion that VH3-21–using cases comprise a new CLL entity, irrespective of VH gene mutation status, and implicate that a common antigen epitope, perhaps of pathogenic significance, is recognized by the highly homologous VH3-21/Vλ2-14 Ig molecules expressed in individual tumors.
Introduction
In recent years, analysis of the immunoglobulin (Ig) genes has been of considerable interest in chronic lymphocytic leukemia (CLL), particularly in terms of somatic hypermutation status of the Ig variable heavy chain (VH) region and VH gene use.1-11 The hypermutation status of the VH genes has revealed 2 subsets of CLL, one with unmutated VH genes and one with somatically hypermutated VH genes.3,5,7,8 The hypothesis is that the CLL cells with mutated VH genes originate from B cells that have passed through the germinal center, whereas unmutated CLL cells derive from naive B cells. The clinical course of these 2 subsets has also been shown to differ with worse prognosis for patients with unmutated Ig genes.7-9,12 In a previous report from our group, we analyzed the VH gene mutation status in 119 CLL cases, in which 42% of the cases were found to be mutated and 58% unmutated.13 Survival analysis verified previous reports that mutated CLL cases had a significantly longer survival than unmutated cases. Furthermore, the prognostic importance of unmutated VH genes to identify CLL patients with poor outcome has also recently been confirmed in multivariate analyses.14,15
Biased VH gene use has been reported in CLL with overrepresentation of individual VH1 and VH4 gene family members.1,2,5,6,16-19 The VH1 gene family member, VH1-69, has previously been shown to be frequently used in CLL (12%-21%) and especially in unmutated CLL, while it has been found expressed in only 1.6% to 4.7% of normal peripheral blood CD5+ B cells.3,4,6,8,13,20,21 Several groups have demonstrated that the VH1-69 rearrangements in CLL display characteristic features, such as biased use of certain D and JH genes and significantly longer average length of the third complementarity determining region (CDR3), compared with normal B cells and other CLL cases in general,4,6,13,21,22 whereas one study could not find any differences in VH1-69 structure between CLL and normal B cells.20 Considering that the CDR3 is highly important in the antigen-binding site, the finding of molecular similarities in VH1-69–using CLL has led to the postulation of a possible antigenic component in the pathogenesis of this CLL subset.4,5,7,8,23
We could recently confirm the overexpression of the VH1-69 gene in unmutated CLL, but, additionally, a novel overrepresentation of the VH3-21 gene was revealed in the mutated group.13 Interestingly, these mutated VH3-21 cases showed a worse overall survival compared with the remaining mutated CLL patients. Also, the VH3-21 subset (n = 13) demonstrated shorter CDR3 lengths than normal B cells and other CLL cells in general, and clonal expression of λ light chains. We therefore suggested that CLL with mutated VH3-21 genes might comprise an additional entity of CLL and speculated upon a possible role for specific antigen(s) in the development of this subgroup.
To further characterize this VH3-21 subset, we have extended our series of CLL and identified 31 VH3-21-using cases in total, including cases from different parts of Sweden and from Finland. This group continued to show poor overall survival, regardless of mutation status, and analysis of the CDR3 showed remarkable similarities in both the nucleotide sequence and thus amino acid composition in a number of the VH3-21 cases. We have also analyzed the light chain V gene use, revealing a considerable restricted use of the Igλ light chain gene Vλ2-14 in the majority of the VH3-21 cases.
Patients, materials, and methods
Patients and materials
Tumor samples, collected from 265 patients with CLL, had been identified from the archives of frozen tissue specimens at the Departments of Pathology and Clinical Immunology (n = 154) at Uppsala University Hospital, the Department of Pathology (n = 51) at Umeå University Hospital, the Department of Hematology (n = 17) at Linköping University Hospital, the Department of Hematology (n = 7) at Huddinge University Hospital, Sweden, and the CLL outpatient clinic (n = 36) at Tampere University Hospital, Finland, between 1981 and 2001. Frozen tumor material was obtained mainly from peripheral blood and bone marrow, but also from lymph nodes and spleen. Morphologic classification was performed according to the World Health Organization (WHO) classification, based on morphology of sections and imprints and combined immunophenotyping.24 According to the Royal Marsden scoring system, the tumor cells typically expressed CD5, CD19, and CD23 and showed a weak expression of Ig.25 The median age at diagnosis was 65 years for the whole material, and the male-female ratio was 2:1. Survival data were available in 244 of 265 patients. Median follow-up alive was 65 months.
Polymerase chain reaction (PCR) amplification and nucleotide sequence analysis
High-molecular-weight DNA was prepared from fresh-frozen tumor material using standard protocols including proteinase K treatment. VH and VL gene family–specific PCR amplification was performed using specific VH/JH primers and VL/JL primers as previously described.26,27 To distinguish monoclonal PCR products from polyclonal, a single-strand conformational polymorphism analysis was performed using polyacrylamide gel or GenePhor system electrophoresis (Pharmacia Biotechnology, Uppsala, Sweden) as reported previously.28
In the majority of samples, clonal products from the VH gene PCR were sequenced directly using the BigDye Terminator Cycle Sequencing Reaction Kit (Perkin-Elmer, Foster City, CA). Cloning of the VH gene (n = 51) and VL gene (n = 33) PCR products was performed as previously described26 or using the Zero Blunt TOPO PCR cloning kit (Invitrogen, Groningen, The Netherlands). Briefly, in the latter method, the PCR products were ligated into the pCR-BluntII-TOPO vector, which was followed by transformation into competent Escherichia coli cells. On average, 5 colonies were selected randomly for sequencing using the BigDye Terminator Cycle Sequencing Reaction Kit. All sequence reactions were analyzed using an automated DNA sequencer (ABI 377; Applied Biosystems, Foster City, CA).
Sequencing of the germline VH3-21 gene
Granulocytes were obtained from one mutated VH3-21–positive patient (case 11, 2.6% mutations), and T cells were isolated from frozen tumor samples of 2 cases with mutated VH3-21 genes (case 12, 3.6% mutations and case 22, 2.7% mutations) and one unmutated VH3-21 case with a low number of mutations (case 1, 0.9% mutations) using a CD4 Positive Isolation Kit (Dynal, Oslo, Norway). Primers were designed to amplify the germline VH3-21 gene using the Primer3 software program (Whitehead Institute, Cambridge, MA); forward primer, 5′-ACGGTGGATGTGTGTGACAG and reverse intronic primer, 5′-TCCTCGGAACTTTCATCCTG. The PCR reaction was carried out in a total volume of 50 μL and included 200 μM deoxyribonucleoside triphosphates (dNTPs), 1.5 mM MgCL2, 0.125 μM primers, 5 μL 10 × gelatin, 5 μL 10 × PCR buffer, and 0.5 U PfuTurbo hotstart DNA polymerase (Stratagene, La Jolla, CA). The reaction was carried out as follows: denaturation at 95°C for 5 minutes; 45 cycles of 90 seconds at 95°C, 30 seconds at 60°C, and 80 seconds at 72°C; and a final cycle of 5 minutes at 72°C. The PCR products were cloned and sequenced using the Zero Blunt TOPO PCR cloning kit.
Analyses of Ig sequences
The sequences were aligned to Ig sequences from the Basic Local Alignment Search Tool (BLAST) database (National Center for Biotechnology Information, Bethesda, MD), the V-BASE database (Medical Research Council [MRC] Centre for Protein Engineering, Cambridge, United Kingdom), and the international ImMunoGeneTics database (IMGT, http://imgt.cines.fr:8104; initiator and coordinator: Marie-Paule Lefranc, Montpellier, France). VH and VL gene sequences deviating more than 2% from the corresponding germline gene were defined as mutated. The distribution of replacement (R) mutations and silent (S) mutations within the CDRs and framework regions (FRs) was assessed. A higher frequency of R mutations and a lower frequency of S mutations in the CDRs than in the FRs indicate that antigen selection may have occurred. A multinomial distribution model proposed by Lossos et al was used to determine the probability that hypermutations within the VH genes resulted from antigen selection.29 The mutated VH3-21 genes were analyzed for mutational hotspots, RGYW (where R is a purine [A or G], Y is a pyrimidine [C or T], and W is an A or T) and its inverse repeat WRCY.30 For D gene determination, a requirement of a minimum of 7 matching nucleotides was used. The length of the CDR3 of the Ig heavy chain (IgH) gene rearrangement was calculated according to the IMGT database starting from the first codon after the conserved cysteine (Cys) up to the position preceding the conserved phenylalanine (Phe) or tryptophan (Trp) of the JH gene segment. Using the nomenclature of the IMGT database the Vλ2-14 (IgBlast database) gene is named Vλ3-21, but to avoid confusion between the Vλ3-21 and VH3-21 gene we have chosen to name the VL genes according to the IgBlast database.
Statistical analyses
Kaplan-Meier survival analysis and log-rank test were performed to study the prognostic significance of VH gene mutations and VH3-21 gene use in CLL using the Statistica 6.0 software (Stat Soft, Tulsa, OK). Overall survival was calculated from the date of diagnosis until the last follow-up or death.
Results
VH gene analysis
A total of 312 IgH rearrangements were amplified and sequenced from 265 CLL cases, including 45 cases with 2 rearrangements and 1 case with 3 rearrangements. The sequence results from the first 119 cases have been published previously.13 Of the 265 cases, 110 (41.5%) showed somatically mutated VH genes, while 155 (58.5%) were unmutated. In the 46 cases who displayed more than 1 IgH rearrangement, 21 cases had only unmutated VH gene rearrangements (judged as unmutated) and 18 cases had only mutated VH gene rearrangements (judged as mutated), whereas 7 cases had 1 mutated and 1 unmutated VH gene rearrangement (judged as mutated). VH1-69 gene use was demonstrated in 46 (17.4%) of 265 analyzed cases, in which the majority displayed unmutated VH genes (45 of 46 cases). VH3-21 use was shown in 31 (11.7%) of 265 cases (Table 1); 21 of these were from southern/central Sweden (Uppsala, Linköping, and Stockholm), 7 from northern Sweden (Umeå), and 3 from Finland.
IgH and IgL gene rearrangement data from the 31 analyzed VH3-21 positive cases
Case no. . | VH homology, % . | D gene . | JH gene . | VH reading frame . | Vκ family† . | Vλ . | VL gene homology, % . | Jλ/κ . | VL gene reading frame* . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 99.1 | — | 6 | F | 2 | 2-14 | 98.4 | 3 | F |
| 2 | 98.2 | — | 6 | F | 2 & 4 | 2-14 | 97.2 | 3 | F |
| 3 | 96.9 | — | 6 | F | 1 | 2-14 & 2-11 | 98.4 & 98.7 | 3 & 3 | F & NF |
| 4 | 96.4 | — | 6 | F | 1 | 2-14 | 97.6 | 3 | F |
| 5 | 97.8 | — | 6 | F | Vκ2 A18 | — | 100 | 4 | NF |
| 6 | 95.9 | — | 6 | F | 3 | 2-14 | 98 | 3 | F |
| 7 | 95.9 | — | 6 | F | 2 | 2-14 | 98.8 | 3 | F |
| 8 | 97.3 | — | 6 | F | — | 2-14 | 96.9 | 3 | F |
| 9 | 96.4 | — | 6 | F | 1 & 4 | 2-14 | 98 | 3 | F |
| 10 | 96.4 | — | 6 | F | 1 & 3 | 2-14 | 89.4 | 3 | F |
| 11 | 97.3 | — | 6 | F | — | 2-14 | 98.4 | 3 | F |
| 12 | 96.4 | — | 6 | F | — | 2-14 | 99.6 | 3 | F |
| 13 | 99.1 | — | 6 | F | 3 & 4 | 2-14 | 98 | 3 | F |
| 14 | 99.6 | — | 6 | F | 2 & 4 | 2-14 | 98.4 | 3 | F |
| 15 | 95.9 | — | 6 | F | 4 | 2-14 | 98 | 3 | F |
| 16 | 96.4 | — | 6 | F | 1 | 2-14 | 98.4 | 3 | F |
| 17 | 99.1 | — | 6 | F | 4 | 2-14 | 98.4 | 3 | F |
| 18 | 100 | — | 6 | F | 1 | 2-14 | 99.2 | 3 | F |
| 19 | 95.1 | — | 6 | F | — | 2-14 | 98.4 | 3 | F |
| 20 | 95.9 | — | 6 | F | 3 | 3-4 | 96.3 | 3 | NF |
| 21 | 96.9 | — | 6 | F | 1 | 2-14 | 99.2 | 3 | F |
| 22 | 97.3 | 3-22 | 4 | F | 1 | 2-14 & 1-3 | 97.6 & 91.9 | 3 & 3 | F & NF |
| 23 | 95.1 | 5-5/5-18 | 3 | F | — | 2-14 | 97.6 | 3 | F |
| 24 | 97.3 | 6-13 | 4 | F | 1 | 2-14 | 98.4 | 3 | F |
| 25 | 94.2 | 2-2 | 3 | F | 3 | 2-14 | 98.8 | 1 | F |
| 26 | 96.4 | — | 4 | F | 1 | 2-14 & 1-17 | 98.8 & 97.6 | 3 & 1 | Both F |
| 27 | 100 | 6-19 | 4 | F | 1 | 2-11 | 100 | 1 | F |
| 28 | 100 | 6-13 | 6 | F | Vκ3 A27 & Vκ1 L12 | — | 100 & 100 | 1 & 1 | F & NF |
| 29 | 100 | 3-10 | 6 | F | Vκ3 L2 | — | 100 | 2 | F |
| 30 | 100 | 3-3 | 6 | F | Vκ1 L12 | — | 100 | 1 | F |
| 31 | 92.8 | 1-7 | 6 | NF | 2 & 5/6 | 1-4 | 98.6 | 1 | F |
Case no. . | VH homology, % . | D gene . | JH gene . | VH reading frame . | Vκ family† . | Vλ . | VL gene homology, % . | Jλ/κ . | VL gene reading frame* . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 99.1 | — | 6 | F | 2 | 2-14 | 98.4 | 3 | F |
| 2 | 98.2 | — | 6 | F | 2 & 4 | 2-14 | 97.2 | 3 | F |
| 3 | 96.9 | — | 6 | F | 1 | 2-14 & 2-11 | 98.4 & 98.7 | 3 & 3 | F & NF |
| 4 | 96.4 | — | 6 | F | 1 | 2-14 | 97.6 | 3 | F |
| 5 | 97.8 | — | 6 | F | Vκ2 A18 | — | 100 | 4 | NF |
| 6 | 95.9 | — | 6 | F | 3 | 2-14 | 98 | 3 | F |
| 7 | 95.9 | — | 6 | F | 2 | 2-14 | 98.8 | 3 | F |
| 8 | 97.3 | — | 6 | F | — | 2-14 | 96.9 | 3 | F |
| 9 | 96.4 | — | 6 | F | 1 & 4 | 2-14 | 98 | 3 | F |
| 10 | 96.4 | — | 6 | F | 1 & 3 | 2-14 | 89.4 | 3 | F |
| 11 | 97.3 | — | 6 | F | — | 2-14 | 98.4 | 3 | F |
| 12 | 96.4 | — | 6 | F | — | 2-14 | 99.6 | 3 | F |
| 13 | 99.1 | — | 6 | F | 3 & 4 | 2-14 | 98 | 3 | F |
| 14 | 99.6 | — | 6 | F | 2 & 4 | 2-14 | 98.4 | 3 | F |
| 15 | 95.9 | — | 6 | F | 4 | 2-14 | 98 | 3 | F |
| 16 | 96.4 | — | 6 | F | 1 | 2-14 | 98.4 | 3 | F |
| 17 | 99.1 | — | 6 | F | 4 | 2-14 | 98.4 | 3 | F |
| 18 | 100 | — | 6 | F | 1 | 2-14 | 99.2 | 3 | F |
| 19 | 95.1 | — | 6 | F | — | 2-14 | 98.4 | 3 | F |
| 20 | 95.9 | — | 6 | F | 3 | 3-4 | 96.3 | 3 | NF |
| 21 | 96.9 | — | 6 | F | 1 | 2-14 | 99.2 | 3 | F |
| 22 | 97.3 | 3-22 | 4 | F | 1 | 2-14 & 1-3 | 97.6 & 91.9 | 3 & 3 | F & NF |
| 23 | 95.1 | 5-5/5-18 | 3 | F | — | 2-14 | 97.6 | 3 | F |
| 24 | 97.3 | 6-13 | 4 | F | 1 | 2-14 | 98.4 | 3 | F |
| 25 | 94.2 | 2-2 | 3 | F | 3 | 2-14 | 98.8 | 1 | F |
| 26 | 96.4 | — | 4 | F | 1 | 2-14 & 1-17 | 98.8 & 97.6 | 3 & 1 | Both F |
| 27 | 100 | 6-19 | 4 | F | 1 | 2-11 | 100 | 1 | F |
| 28 | 100 | 6-13 | 6 | F | Vκ3 A27 & Vκ1 L12 | — | 100 & 100 | 1 & 1 | F & NF |
| 29 | 100 | 3-10 | 6 | F | Vκ3 L2 | — | 100 | 2 | F |
| 30 | 100 | 3-3 | 6 | F | Vκ1 L12 | — | 100 | 1 | F |
| 31 | 92.8 | 1-7 | 6 | NF | 2 & 5/6 | 1-4 | 98.6 | 1 | F |
— represents no D gene identified or no Vκ/λ family amplified.
F indicates functional; NF, nonfunctional.
The Vκ PCR products were sequenced only in cases 5 and 28 to 30.
Characterization of VH3-21 gene rearrangements
Somatic hypermutation (< 98% homology) was demonstrated in 21 (68%) of 31 VH3-21 cases, whereas the remaining 10 cases (32%) were considered unmutated. In the mutated cases, the range of mutations varied from 2.2% to 7.2% with a median of 3.6%. One mutated VH3-21–using case (case 31) displayed double rearrangements with one functional VH4-61 gene rearrangement but a nonfunctional VH3-21 gene rearrangement with a stop codon introduced in the CDR3. The D gene use was identified in only approximately one third of the cases (10/31) with no single D gene overrepresented. The JH6 gene was found in the majority of the VH3-21 cases (81%).
The mutational hotspots, RGYW and its inverse repeat WRCY, accounted for 26.7% of the nucleotides of the VH3-21 gene. In the mutated VH3-21 cases, 46% of the identified mutations occurred in these regions. A multinomial model by Lossos et al was used to test for evidence of antigen selection in the mutated VH3-21 cases, in which 13 of 21 mutated cases showed evidence for antigen selection in the FRs and/or CDR3s29 ; 3 cases showed significant excess of R mutations in the CDRs, 2 of which also had significant scarcity of R mutations in FRs, whereas another 10 mutated cases demonstrated significant scarcity of R mutations in FRs alone.
The mean length of the CDR3 in the VH3-21 cases (n = 31) was considerably shorter (10.5 codons) compared with VH1-69 rearrangements (21.3 codons) and CLL rearrangements in general (14.0 codons). For the mutated VH3-21 (n = 21) cases the mean length was 9.1 codons. The mean length of the VH3 family in the whole material was 12.6 codons. The most frequently used members of this family VH3-30 (n = 15), VH3-23 (n = 13), VH3-7 (n = 10), and VH3-33 (n = 10) showed mean codon lengths of 12.4, 10.8, 11.2, and 12.4, respectively. Following cluster analysis of the CDR3 in the VH3-21 cases we found that 7 of the cases had an identical CDR3 amino acid sequence and 5 others differed by only one amino acid from these 7 cases (Figure 1). The conserved amino acid structures consisted of an Ala-Arg-Asp-Ala-Asn-Gly-Met-Asp-Val (ARDANGMDV) motif.
Amino acid sequence in the VH3-21–using cases. Of 31 cases, 30 are shown, since the VH3-21 gene rearrangement in case 31 was nonfunctional with a stop codon in the CDR3. Sequence homology is shown to the germline VH3-21 gene. Homology to the Ala-Arg-Asp-Ala-Asn-Gly-Met-Asp-Val (ARDANGMDV) CDR3 sequence is also shown. Dots indicate homology with the consensus sequence. A dash indicates that an amino acid is missing compared to the germline sequence. Case 17 displayed an extra amino acid insertion that could not be found in the germline sequence or the remaining sequences.
Amino acid sequence in the VH3-21–using cases. Of 31 cases, 30 are shown, since the VH3-21 gene rearrangement in case 31 was nonfunctional with a stop codon in the CDR3. Sequence homology is shown to the germline VH3-21 gene. Homology to the Ala-Arg-Asp-Ala-Asn-Gly-Met-Asp-Val (ARDANGMDV) CDR3 sequence is also shown. Dots indicate homology with the consensus sequence. A dash indicates that an amino acid is missing compared to the germline sequence. Case 17 displayed an extra amino acid insertion that could not be found in the germline sequence or the remaining sequences.
Analysis of the germline VH3-21 gene in 4 cases
Using VH3-21–specific primers, the germline gene was amplified from granulocyte DNA from 1 mutated VH3-21 case and from T-cell DNA from 2 mutated and 1 unmutated (with 0.9% mutations) VH3-21 case. The PCR products were cloned and 10 colonies were sequenced per sample, all of which showed 100% homology to the VH3-21 germline gene.
Analysis of the IgL gene rearrangements
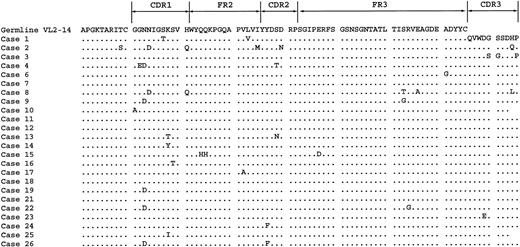
Of the VH3-21 cases, 28 showed clonal λ light chain expression, whereas 3 cases expressed κ light chains. PCR analysis of the Ig light chain rearrangements (κ/λ) was performed in all 31 VH3-21 cases (Table 1). Clonal Vκ PCR products were detected in 26 cases, but were sequenced only in κ-expressing cases (nos. 28, 29, and 30) and in one λ-expressing case (case 5) in which only a VK product was obtained. Clonal Igλ PCR products were subcloned and sequenced in 27 of 28 cases. Sequence analysis revealed that the Vλ2-14 gene was used in 24 of 27 cases (Figure 2). In addition, cases 20, 27, and 31 used the Vλ3-4, Vλ2-11, and Vλ1-4 gene, respectively. Of the Vλ2-14–using cases, 3 were also found to rearrange Vλ2-11 (case 3), Vλ1-3 (case 22), and Vλ1-17 (case 26). Of the 27 cases sequenced, 19 cases showed more than 98% homology to their respective germline gene. The most frequently used Jλ gene was found to be Jλ3 (83%), while the remaining 5 cases used the Jλ1 gene. With the exception of case 25, all Vλ2-14 cases used the Jλ3 gene and, with the exception of case 10, showed between 96.9% and 99% germline homology. The CDR3 of the light chains analyzed showed little variability.
Amino acid sequence of the Vλ2-14 gene in the VH3-21–using cases. Sequence homology is shown to the germline Vλ2-14 gene. Homology to the Gln-Val-Trp-Asp-Gly-Ser-Ser-Asp-His-Pro (QVWDGSSDHP) CDR3 sequence is also shown. Dots indicate homology with the consensus sequence.
Amino acid sequence of the Vλ2-14 gene in the VH3-21–using cases. Sequence homology is shown to the germline Vλ2-14 gene. Homology to the Gln-Val-Trp-Asp-Gly-Ser-Ser-Asp-His-Pro (QVWDGSSDHP) CDR3 sequence is also shown. Dots indicate homology with the consensus sequence.
Survival analysis
Survival curves were plotted for 244 CLL patients and a significant difference was found between the mutated and unmutated CLL. The median survival for mutated cases (n = 100) was 120 months (interquartile range, 64-185 months) compared with 70 months (interquartile range, 38-109 months) for the unmutated cases (n = 144) (P < .001). The mutated VH3-21 cases (n = 21) showed a median survival of 72 months, which was significantly different from the remaining mutated CLL cases (146 months, P = .018) but similar to the unmutated group (70 months). All VH3-21 cases (mutated and unmutated, n = 31) showed a median survival of 83 months (interquartile range, 59-110 months; Figure 3). When excluding the VH3-21 cases, the median survival for the mutated (n = 79) and unmutated (n = 134) group was 137 months and 64 months, respectively. The median age at diagnosis was 64 years for VH3-21 cases, 67 years for the mutated cases, and 63 years for the unmutated cases. Using the Mann-Whitney U test no significant difference between the diagnostic ages was indicated.
Kaplan-Meier curve of the CLL patient material investigated in the study showing mutated (n = 79), unmutated (n = 134), and VH3-21 cases (n = 31) with a median survival of 137, 64, and 83 months, respectively.
Kaplan-Meier curve of the CLL patient material investigated in the study showing mutated (n = 79), unmutated (n = 134), and VH3-21 cases (n = 31) with a median survival of 137, 64, and 83 months, respectively.
Discussion
The prognostic role of VH gene somatic hypermutation status in CLL is now well established, since CLL cases with mutated VH genes have a more favorable prognosis than cases with unmutated VH genes.7-9,12-15 However, we recently identified a group of patients with mutated VH3-21 genes that showed a worse outcome compared with other mutated CLL cases,13 and suggested that they may constitute a new subset of CLL. In this follow-up study, VH gene analysis was performed in an extended number of VH3-21 cases (n = 31) and showed that two thirds of patients had mutated VH genes, whereas one third were unmutated. Taken as a group, survival analysis of the VH3-21 cases revealed a median survival (83 months) similar to the unmutated CLLs (64 months) (Figure 3). The poor overall survival was also observed when analyzing only the mutated VH3-21 cases (72 months). From a prognostic point of view, these data clearly show that VH3-21 cases represent a distinct subgroup within CLL, as they do not fit the traditional pattern of mutated or unmutated grouping. Our finding highlights that use of an individual VH gene (ie, VH3-21) can have prognostic impact in CLL, which is further supported by a recent study showing that VH3-21 and VH3-23 use predict poor outcome in CLL.31
The overall mutation rate in the hypermutated VH3-21 cases was lower (median, 3.6%) than CLL in general (median, 5.4%) and close to the cutoff level of 2%. Nollet et al have recently questioned our finding that CLL with mutated VH3-21 use constitutes a new CLL subset.32 They suggested that a different cutoff level, for example 4%, would change most of the mutated VH3-21 cases to unmutated cases that already have a known worse outcome.32 However, the analysis of the mutational hotspots RGYW and its inverse repeat WRCY in the VH3-21 cases revealed that 46% of the mutations occurred in these motifs, which accounted for only 27% of the VH3-21 gene sequence, that is, half of the mutations occurred in approximately one quarter of the VH gene region. These hotspots are known to be targeted by the hypermutational mechanism and therefore provide evidence that the leukemia precursor cells most probably have undergone a germinal center reaction.30 In 3 mutated VH3-21–using cases, we amplified the germline VH3-21 gene, showing that the germline VH3-21 sequences were 100% identical with the published VH3-21 sequence. This result shows that the mutations identified in the VH3-21 rearrangements do not represent polymorphisms and that the rearrangements have gone through somatic hypermutation. Additionally, analysis of the mutated VH3-21 cases using the multinomial model proposed by Lossos et al indicated that 13 of 21 cases showed evidence of antigen selection.29 Taken together, these latter findings have convinced us that the mutated VH3-21 cases are truly hypermutated. Furthermore, we analyzed one case that was classified as unmutated (using the 2% border) but contained 2 mutations compared with the germline sequence, and showed that the unrearranged VH3-21 gene in T cells from this patient was 100% homologous to the published germline sequence. Whether these low numbers of mutations indicate involvement of the hypermutation mechanism is currently unknown, but our finding raises the discussion of the use of the 2% border to indicate germinal center experience.
In contrast to what could be expected in the majority of CLLs, a striking preference for λ light chain expression was demonstrated in the VH3-21 group (28 of 31 cases). This led us to further investigate the Igλ gene rearrangements by nucleotide sequencing. Interestingly, the VH3-21 cases demonstrated a highly restricted use of one particular light chain gene, Vλ2-14 (24 of 31 analyzed cases). These Vλ2-14 rearrangements also showed similarities of the CDR3 and the majority used the Jλ3 gene (Figure 2). The finding of combined use of one certain IgH and IgL gene indicates that these genes have been preferentially rearranged in this subset of CLL patients. This phenomenon has been previously described in IgG+ CLL showing restricted VH and Vκ family use; although the number was small with only 2 cases with a specific VH4 (VH4-18) gene rearranging with the same Vκ gene (VκO2).23
The VH3-21–rearranged cases also displayed shorter CDR3s than average (10.5 codons), especially among the mutated cases (9.1 codons), whereas the VH1-69 cases showed longer CDR3s (21.3 codons) compared with CLL in general (14.0 codons). Although the VH3-21 gene rearrangements were shorter than average CLL VH gene rearrangements, other members of the VH3 gene family also had short CDR3s (VH3-23 and VH3-07). The reason behind the shorter CDR3s in VH3-21 cases is unknown, but one hypothesis could be that these gene rearrangements have undergone VHDJH recombination in a certain fashion that allows deletion of parts or most of the D gene segment, thus making them short. High sequence homology was observed in the CDR3s of several of the VH3-21 cases, in which 7 cases showed identical CDR3s and additionally 5 cases showed only one amino acid difference from the 7 cases (Figure 1). In total, 11 cases demonstrated a short and more-or-less identical amino acid motif of the CDR3 (Ala-Arg-Asp-Ala-Asn-Gly-Met-Asp-Val [ARDANGMDV]). These specific CDR3 features combined with the biased use of the VH3-21/Vλ2-14 genes implicate that the CLL tumors are expressing highly similar Ig structures on their surfaces. Considering the importance of the variable regions of both the heavy and light Ig chains in the formation of the antigen-binding site of the Ig molecule and in particular the CDR3,33 these data indicate a common antigenic binding site being expressed by these individual CLL tumors, which could be a consequence of selection by a common antigen. Sparse data are available about the VH3-21 rearrangement in lymphocytes in the healthy population, but the VH3-21 gene has been implicated in the production of rheumatoid factors in rheumatoid arthritis.34
We and others have shown that the rearrangements of VH1-69–using cases showed longer CDR3s and preferentially used D and JH genes,4,6,13,22 which have been suggested to predispose VH1-69–positive B cells for neoplastic transformation by yet unidentified antigens. In contrast to the VH3-21–using cases, our VH1-69–expressing cases displayed high variability in the CDR3s (data not shown), and, thus, different antibody specificity for various epitopes on the antigen surface cannot be ruled out. However, the highly homologous structures observed in a number of the VH3-21 cases are indicative of a common epitope being recognized by a high proportion of these Ig structures and therefore make them unique in the CLL repertoire.
In summary, we present data on a cohort of CLL patients, using the rearranged VH3-21 gene, that as a group show poor overall prognosis, irrespective of mutation status, and do not fit the recently postulated classification of mutated/unmutated CLL. That these patients were found in all geographic regions studied also emphasizes the significance of our data. Additionally, a high proportion of these cases coexpresses the Vλ2-14 light chain gene and demonstrates remarkable similarities in the CDR3s, indicating a common epitope being recognized by these tumors. Further studies will be necessary to additionally characterize this subset of CLL cases and to identify the putative antigenic binding specificity of these unique CLL patient tumors.
Prepublished online as Blood First Edition Paper, February 13, 2003; DOI 10.1182/blood-2002-11-3485.
Supported by grants from the Swedish Cancer Society, Lion's Cancer Research Foundation, Umeå University, Uppsala University, the research foundation of the Department of Oncology at Uppsala University, Sweden, and Tampere University Research Fund, Finland.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are grateful to Anita Lindström for skillful technical assistance.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal