Abstract
To get insight into the regulation of human interleukin-12 (IL-12) synthesis, we determined the chromatin organization of the IL-12(p35) promoter region. First, we determined positioning of nucleosomes within the IL-12(p35) promoter using the indirect end-labeling technique in the THP-1 monocytic cell line. On stimulation with bacterial lipopolysaccharide (LPS) and interferon-γ (IFN-γ), hypersensitivity to digestion with DNase I, micrococcal nuclease, and specific restriction enzymes was detected in the region encompassing nucleotide (nt) –310 to –160, indicating selective inducible chromatin remodeling involving disruption of a single nucleosome (named nuc-2). Using p35 promoter deletion mutants and reporter gene assays, we demonstrated that the –396/–241 region contained critical cis-acting elements. Within this latter region, we characterized physically and functionally 2 Sp1-binding sites, which were acting as key regulatory elements for both basal and LPS/IFN-γ–inducible p35 gene expression: Sp1#1 lies within the remodeled nuc-2 region and Sp1#2 is located in the nucleosome-free region immediately upstream of nuc-2. Finally, we extended the chromatin structure analysis to dendritic cells (DCs) derived from human monocytes and observed the same nucleosomal organization and remodeling as in the THP-1 cell line. Moreover, we found that in DCs, LPS and IFN-γ synergized in the induction of nucleosomal remodeling and that chromatin remodeling at the p35 locus immediately preceded IL-12(p35) mRNA synthesis. Taken together, our results demonstrate that IL-12(p35) gene activation in the course of DC maturation involves selective and rapid remodeling of a single positioned nucleosome within a region of the promoter containing critical Sp1-binding sites.
Introduction
Interleukin-12 (IL-12) represents an essential link between innate and adaptive immunity. Indeed, IL-12 is produced during inflammatory reactions and promotes the differentiation of type 1 helper T lymphocytes and effector cytotoxic T cells, which are critical for the eradication of intracellular pathogens and tumors.1 The induction of IL-12 synthesis is complex because it depends on the activation of 2 different genes encoding the subunits p35 and p40, which assemble to form the bioactive IL-12(p70) heterodimer.2 Transcriptional activation of the IL-12(p40) gene by microbial products and interferon-γ (IFN-γ) was shown to require the recruitment of various members of the CCAAT/enhancer-binding protein (C/EBP), nuclear factor-κB (NF-κB), ets (ets-2, PU-1), and interferon regulatory factor (IRF-1, IFN consensus sequence-binding protein [ICSBP]) families.3-6 Although evidence suggests that induction of IL-12(p35) mRNA is critical for the synthesis of bioactive IL-12(p70),7-9 regulation of IL-12(p35) gene transcription remains poorly characterized. Indeed, regulation of the p35 subunit seems very complex. In both murine and human cells, IL-12(p35) transcription can be initiated from different sites,10,11 thereby leading to the synthesis of various p35 isoforms. Recently, determination of important cis-acting DNA elements and trans-acting factors regulating the expression of the murine p35 gene has been reported: IRF-1 was shown to be involved in IFN-γ–dependent exon 2 promoter activity,12 and a κB site in exon 1 promoter was shown to be required for c-Rel-dependent induction of IL-12 by microbial products.13 Finally, posttranslational modifications are also likely to play an important role.14
The 5′-flanking region of the human p35 gene has been cloned and 2 distinct initiation sites (in Epstein-Barr virus [EBV]–transformed cell lines and primary monocytes) have been described.10 Apart from this report, the human p35 promoter remains completely uncharacterized.
In eukaryotes, genomic DNA is incorporated into chromatin, which consists of assembled histone octamer cores (nucleosomes). The ability of chromatin to modulate the access of specific transcription factors to DNA is now well established. Chromatin remodeling plays an important role in the regulation of immune functions by controlling the expression of a growing number of key cytokine genes.15 Specific modifications in the chromatin structure of some of these genes can occur during development and then persist in the differentiated cell. For example, during peripheral differentiation of naive CD4+ T cells into Th2 cells, the IL-4/IL-5/ IL-13 locus becomes accessible for transcription, whereas the IFN-γ locus is remodeled in Th1 cells.15-17 In other cytokine genes, precise changes in nucleosomal organization can occur in response to specific stimulus. In the case of the IFN-β gene, remodeling takes place after the recruitment of an enhanceosome to a nucleosome-free region.18 In contrast, at the murine IL-12(p40) locus, the major cis-acting elements (c-Rel and c/EBP sites) are located into a region corresponding to a positioned nucleosome.19 On activation, this nucleosome is disrupted, allowing transcription factors to bind to their recognition sites.
Herein, we considered a possible role for chromatin structure in the regulation of human IL-12(p35) gene expression. For this purpose, we first mapped the positions of nucleosomes within the endogenous IL-12(p35) promoter region in the THP-1 monocytic cell line. A single nucleosome, located in a region of the promoter where we identified critical Sp1-binding sites, was found to be specifically remodeled/disrupted on transcriptional activation with bacterial lipopolysaccharide (LPS) and IFN-γ. Importantly, the same chromatin remodeling was detected in dendritic cells (DCs), which represent a major source of IL-12(p70) in the course of the immune response.20
Materials and methods
Cells and reagents
THP-1 cells and monocyte-derived DCs were grown in RPMI 1640 (Biowhittaker Europe, Verviers, Belgium) supplemented with 2 mM l-glutamine (Biowhittaker), gentamicin (20 μg/mL; Schering Plough, Kenilworth, NJ), 50 μM 2-mercaptoethanol, 1% nonessential amino acids, and 10% fetal bovine serum (Biowhittaker and Hyclone, Logan, Utah, respectively). DCs were generated from peripheral blood mononuclear cells (PBMCs), as described by Romani et al.21 Briefly, PBMCs were resuspended in culture medium and allowed to adhere onto 75-cm2 flasks. After 2 hours at 37°C, nonadherent cells were removed and adherent cells were cultured in 20 mL medium containing granulocyte-macrophage colony-stimulating factor (GM-CSF; 800 U/mL, LEUCOMAX; Schering-Plough) and IL-4 (200 U/mL, kindly provided by K. Thielemans; VUB, Brussels, Belgium). Every 2 days, 800 U GM-CSF and 200 U IL-4 were added. After 6 days of culture, nonadherent cells corresponding to the DC-enriched fraction were harvested, washed, and used for subsequent experiments. Drosophila SL2 cells (LGC Promochem, Teddington, United Kingdom) were grown in Drosophila SFM medium (Invitrogen, Merelbeke, Belgium). The RAW 264.7 murine macrophage cell line (LGC Promochem) was maintained in Dulbecco modified Eagle medium (DMEM), supplemented with 10% fetal calf serum (FCS), nonessential amino acids, 2 mM glutamine, and penicillin/streptomycin. LPS from Escherichia coli (0128: B12) was obtained from Sigma-Aldrich (Bornem, Belgium). Recombinant human and murine IFN-γ were purchased from Biosource Europe (Nivelles, Belgium) and Roche Diagnostics (Brussels, Belgium), respectively.
Cloning and construction of luciferase reporter gene vectors
A 3.9-kb fragment of the IL-12(p35) gene (nucleotide [nt] –2606/+1308) was amplified by polymerase chain reaction (PCR) from human genomic DNA and subsequently cloned into the pGL2-BASIC vector (Promega, Leiden, The Netherlands) as a KpnI-HindIII insert. From this plasmid, we amplified by PCR a 1186-bp fragment (–1121/+66; corresponding to nt positions 490 through 1676 of the AF050083 clone), which was cloned into the pGL3-BASIC vector (Promega) to generate the luciferase reporter plasmid referred to as p35-lucWT. To this end, KpnI and BglII restriction sites were introduced into the 5′ and 3′ PCR primers, respectively. The PCR fragment was then cloned into pGL3-BASIC as a KpnI-BglII fragment. Deletion mutants of the 5′-flanking regions (–648/+66, –447/+66, –396/+66, –241/+66, and –131/+66) were generated using the same strategy. The –1121/+66 construct was used as a template for mutagenesis by the QuickChange Site-Directed Mutagenesis Method (Stratagene, La Jolla, CA). All constructs were fully resequenced prior to use.
Nuclease digestion of purified nuclei
The indirect end-labeling technique was performed as previously described22,23 with minor modifications. In brief, cell nuclei were resuspended in one of 3 buffers depending on nuclease digestion: buffer A (10 mM Tris [tris(hydroxymethyl)aminomethane], pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.3 M sucrose) for DNase I, buffer A supplemented with 10 mM CaCl2 for micrococcal nuclease (MNase), and buffer RE (recommended New England Biolabs [NEB], Beverly, MA) supplemented with 100 μg/mL bovine serum albumin [BSA] and 0.1 mM phenylmethylsulfonyl fluoride [PMSF]) for restriction enzymes. Sodium butyrate was added to all buffers at a final concentration of 1 mM. Nuclei were digested for 10 minutes on ice for DNase I, 20 minutes at 22°C for MNase, and 30 minutes at recommended temperature for restriction enzymes. Reactions were stopped by the addition of 2× proteinase K buffer (100 mM Tris, pH 7.5, 200 mM NaCl, 2 mM EDTA [ethylenediaminetetraacetic acid], 1% sodium dodecyl sulfate [SDS]). Following proteinase K and RNase treatment, genomic DNA underwent phenol/chloroform and chloroform extractions and was resuspended in sterile water after ethanol precipitation.
Purified DNA (15-30 μg) was incubated overnight at 37°C with an excess of EcoRI. Samples were then analyzed by electrophoresis on a 1.5% agarose gel. Size markers were generated by digestion of the IL-12 (p35) 3.9-kb plasmid with EcoRI and one of the following enzymes: BamHI (marker a, nt –1079), ApaI (marker b, nt –793), ScaI (marker c, nt –451), BstXI (marker d, nt –298), NaeI (marker e, nt –141), PstI (marker f, nt +64), BglII (marker g, nt +576), and XbaI (marker h, nt +865). These markers were mixed together and added to 15 or 30 μg genomic DNA. Samples were then transferred and hybridized with a [32P]dCTP-radiolabeled probe spanning nt –1610 to nt –890 from the IL-12(p35) promoter.
EMSAs
Nuclear extracts were prepared by a rapid method described by Osborn et al.24 Electrophoretic mobility shift assays (EMSAs) were performed as previously described.25 For supershift assays, polyclonal antibodies against Sp1 (PEP-2), Sp3 (D-20), or MEF-2 (used as a control; Santa Cruz Biochemicals, Santa Cruz, CA) were added to the binding-reaction mixture.
Transient transfection and luciferase assays
RAW 264.7 cells and Drosophila SL-2 cells were transfected using FuGENE-6 (Roche Diagnostics) according to the manufacturer's protocol. RAW cells were transfected using 2 μg reporter plasmid, 3 μL FuGENE-6 reagent, and as an internal control plasmid, 20 ng pRL-TK, in which a cDNA encoding Renilla luciferase is under the control of the herpes simplex virus thymidine kinase promoter region (Promega). If required, 16 hours after transfection, cells were pretreated with IFN-γ (100 U/mL, Roche Diagnostics) for 8 hours and then incubated with LPS (1 μg/mL) for 24 hours. Forty-eight hours after transfection, cells were then harvested and promoter activities were analyzed using the Dual Luciferase Reporter Assay System (Promega). Promoter activities were then normalized to Renilla luciferase activities.
Drosophila SL-2 cells were transfected with 1 μg of the p35-lucWT plasmid and increasing amounts of pPac (empty vector) or pPacSp1 (Sp1 expression vector). The latter plasmids were a kind gift from Dr G. Suske (Philipps-Universitat, Marburg, Germany). Luciferase activities were analyzed 48 hours after transfection.
RNA purification and real-time RT-PCR
Total RNA was extracted using a MagNa Pure LC RNA Isolation Kit (Roche Diagnostics) according to the manufacturer's instructions. Reverse transcription (RT) and real-time PCR reactions were then carried out using LightCycler-RNA Master Hybridization Probes (one-step procedure) on a LightCycler apparatus (Roche Diagnostics). The primers, standards, and real-time PCR conditions have been previously reported.26 The sequences of the oligonucleotides used for amplification of the IL-12(p35) and β-actin genes, respectively, were as follows: sense primers, 5′-CTCCTGGACCACCTCAGTTTG-3′ and 5′-GGATGCAGGAAGGAGATCACTG-3′; antisense primers, 5′-GGTGAAGGCATGGGAACATT-3′ and 5′-CGATCCACACGGAGTACTTG-3′; probes, 5′-(6-Fam)CCAGAAACCTCCCCGTGGCCA(Tamra)(phosphate)-3′ and 5′-(6-Fam)CCCTGGCACCCAGCACAATG(Tamra)(phosphate)-3′. Primers and probe for IL-12(p35) are located within exon 2 and therefore do not discriminate between the different p35 isoforms described.10
Results
Inducible alteration of DNase I hypersensitivity within the IL-12(p35) promoter region
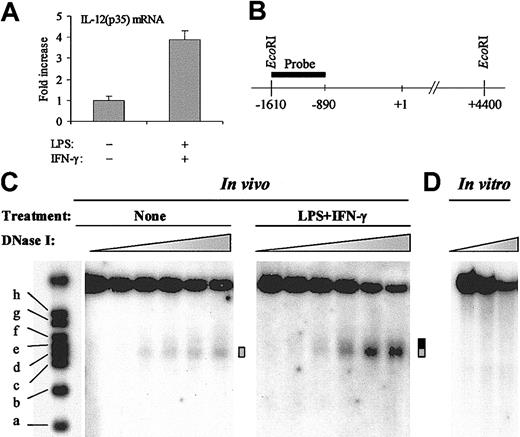
In eukaryotes, chromatin is increasingly recognized as an important modulator of transcriptional regulatory mechanisms.15 To study the chromatin structure of the IL-12(p35) promoter region, we used the human monocytic cell line THP-1 that displayed inducible expression of the p35 gene on stimulation with the combination LPS plus IFN-γ (4- to 5-fold induction; Figure 1A). The sensitivity of a particular nucleosomal DNA region to nuclease digestion reflects the DNA accessibility and the chromatin structure of this region.
Inducible DNase I hypersensitivity within the IL-12(p35) promoter. (A) Induction of IL-12(p35) mRNA in THP-1 cells. THP-1 cells were left untreated or were treated with LPS (1 μg/mL) and IFN-γ (100 U/mL) for 6 hours. Total RNA was reverse transcribed followed by real-time PCR with either IL-12(p35) or β-actin primers, using a one-step procedure. IL-12(p35) mRNA levels were normalized using β-actin mRNA levels and compared with unstimulated conditions. (B) Schematic representation of the probe used to map the nuclease-hypersensitive sites within the p35 promoter by the indirect end-labeling technique. Purified nuclei were treated with limiting concentrations of different nucleases (DNase I, MNase, or restriction enzymes). After isolation of genomic DNA, we performed overnight EcoRI digestion and analyzed the double-restriction products by Southern blotting using the indirect end-labeling technique. The probe spans nucleotides –1615 to –894 and is abutting the EcoRI restriction site. In vitro cleavage of genomic DNA by EcoRI yields an approximate 6-kb fragment. (C) In these experiments, THP-1 cells were either left untreated or treated with LPS (1 μg/mL) and IFN-γ (100 U/mL) for 4 to 6 hours. Nuclei were then purified and digested with increasing concentrations of DNase I (ranging from 0 to 50 U/mL). After purification of genomic DNA and in vitro digestion with EcoRI, DNA samples were analyzed by Southern blotting. Size markers are described in “Materials and methods.” The DNase I-hypersensitive site is indicated by a gray box. Increase in hypersensitivity on LPS/IFN-γ stimulation is indicated by a black extension of the gray box. (D) As a control, purified genomic DNA from THP-1 cells was digested in vitro with increasing concentrations of DNase I (ranging from 0.078 to 0.3125 U/mL). The results are representative of at least 3 independent experiments.
Inducible DNase I hypersensitivity within the IL-12(p35) promoter. (A) Induction of IL-12(p35) mRNA in THP-1 cells. THP-1 cells were left untreated or were treated with LPS (1 μg/mL) and IFN-γ (100 U/mL) for 6 hours. Total RNA was reverse transcribed followed by real-time PCR with either IL-12(p35) or β-actin primers, using a one-step procedure. IL-12(p35) mRNA levels were normalized using β-actin mRNA levels and compared with unstimulated conditions. (B) Schematic representation of the probe used to map the nuclease-hypersensitive sites within the p35 promoter by the indirect end-labeling technique. Purified nuclei were treated with limiting concentrations of different nucleases (DNase I, MNase, or restriction enzymes). After isolation of genomic DNA, we performed overnight EcoRI digestion and analyzed the double-restriction products by Southern blotting using the indirect end-labeling technique. The probe spans nucleotides –1615 to –894 and is abutting the EcoRI restriction site. In vitro cleavage of genomic DNA by EcoRI yields an approximate 6-kb fragment. (C) In these experiments, THP-1 cells were either left untreated or treated with LPS (1 μg/mL) and IFN-γ (100 U/mL) for 4 to 6 hours. Nuclei were then purified and digested with increasing concentrations of DNase I (ranging from 0 to 50 U/mL). After purification of genomic DNA and in vitro digestion with EcoRI, DNA samples were analyzed by Southern blotting. Size markers are described in “Materials and methods.” The DNase I-hypersensitive site is indicated by a gray box. Increase in hypersensitivity on LPS/IFN-γ stimulation is indicated by a black extension of the gray box. (D) As a control, purified genomic DNA from THP-1 cells was digested in vitro with increasing concentrations of DNase I (ranging from 0.078 to 0.3125 U/mL). The results are representative of at least 3 independent experiments.
To assess the presence of nuclease-hypersensitive sites, we performed indirect end-labeling experiments. In these experiments, THP-1 cells were either left untreated or treated with LPS/IFN-γ for 4 to 6 hours. Nuclei were then purified and digested by increasing concentrations of DNase I. After isolation of genomic DNA, we performed in vitro EcoRI digestion and analyzed the double restriction products by Southern blotting with a probe spanning nucleotides –1615 to –894 (Figure 1B). As shown in Figure 1C, in untreated conditions, a band gradually appeared in a DNase I concentration-dependent manner. Using the molecular weight markers, we mapped this DNase I-hypersensitive site to nt –420 to –280 (Figure 1C ▦). Importantly, on stimulation with LPS plus IFN-γ, the hypersensitive band appeared at lower DNase I concentrations and reached higher intensity than in untreated cells. Moreover, the site became larger at the expense of its 3′ boundary, which moved from nt –280 to nt –150 (Figure 1C ▦/▪).
To demonstrate that the DNase I hypersensitivity observed was a consequence of chromatin organization and not secondary to sequence-directed cleavage preference by DNase I, we performed control digestion of naked genomic DNA in vitro with increasing concentrations of DNase I. No preferential cutting was observed in conditions that generated levels of digestion comparable to those obtained in the in vivo experiments (Figure 1D).
Precise nucleosome positioning in the IL-12(p35) promoter region and chromatin remodeling associated with transcriptional activation
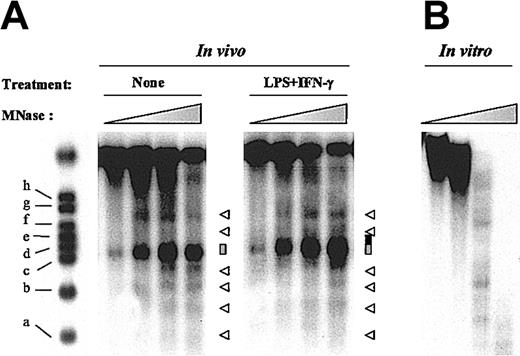
To confirm our results with another nuclease, we next performed indirect end-labeling experiments with MNase. This nuclease preferentially digests nucleosome-free DNA including linker domains between nucleosomes. As shown in Figure 2A, these experiments revealed a discrete banding pattern (indicated by arrowheads) corresponding to an approximate 160-bp nucleosome ladder, a result consistent with precise nucleosome positioning in this chromatin region. Moreover, we detected a major MNase-hypersensitive site (indicated by a gray box) spanning nt –450 to nt –320 and thus mapping to the same region as the DNase I-hypersensitive site. Remarkably, when the same region was examined after LPS plus IFN-γ treatment, similar results were obtained except that, as described for DNase I, we observed a displacement of the 3′ boundary of the MNase-hypersensitive site up to the adjacent linker region (marked by a black extension of the gray box; Figure 2A). In vitro digestion of naked DNA showed that MNase exhibited some sequence-dependent cleavage preference (Figure 2B). However, the MNase-hypersensitive site was noted only in vivo, supporting the hypothesis that it was secondary to chromatin organization.
Nucleosome positioning and inducible MNase hypersensitivity within the IL-12(p35) promoter. (A) THP-1 cells were either incubated with medium alone or stimulated with LPS (1 μg/mL) and IFN-γ (100 U/mL) for 4 hours. Nuclei were purified and digested with increasing concentrations of MNase, ranging from 0.015 to 0.12 U/mL. The major MNase-hypersensitive site is indicated by a gray box. Other MNase cutting sites are marked by arrowheads. Increase in MNase hypersensitivity on LPS/IFN-γ stimulation is indicated by a black extension of the gray box. (B) As a control, purified genomic DNA from THP-1 cells was digested in vitro with increasing concentrations of MNase (ranging from 0.00078 to 0.05 U/mL).
Nucleosome positioning and inducible MNase hypersensitivity within the IL-12(p35) promoter. (A) THP-1 cells were either incubated with medium alone or stimulated with LPS (1 μg/mL) and IFN-γ (100 U/mL) for 4 hours. Nuclei were purified and digested with increasing concentrations of MNase, ranging from 0.015 to 0.12 U/mL. The major MNase-hypersensitive site is indicated by a gray box. Other MNase cutting sites are marked by arrowheads. Increase in MNase hypersensitivity on LPS/IFN-γ stimulation is indicated by a black extension of the gray box. (B) As a control, purified genomic DNA from THP-1 cells was digested in vitro with increasing concentrations of MNase (ranging from 0.00078 to 0.05 U/mL).
Taken together, our results obtained with both DNase I and MNase demonstrate a selective stimulation-dependent chromatin remodeling in a region of the p35 locus located about 200 bp upstream of the transcription start site.
Chromatin remodeling within the p35 locus is restricted to a single nucleosome
To map more precisely the remodeled region, we next performed chromatin digestion with restriction enzymes. In these experiments, isolated nuclei from control or LPS/IFN-γ–stimulated THP-1 cells were digested with 11 different restriction endonucleases cutting at 22 distinct sites within the p35 promoter region spanning nt –793 to nt +117 (Figure 3; when several restriction sites for the same enzyme are found within the region under study, symbols are used [°, *, and $] to differentiate them). In untreated cells, strongest digestions were observed for NspI (nt –405), PvuII (nt –356), and BsaHI (nt –325), suggesting high accessibility within this region. Importantly, the region encompassed by these cutting sites overlapped the DNase I- and MNase-hypersensitive sites identified previously. After LPS/IFN-γ treatment, the intensities of digestion were unchanged compared with those observed in untreated cells except for 4 enzymes all located between size markers d and e, namely BstXI, (band °, nt –298), AlwNI (nt –256), BanI (band °, nt –185), and SmaI (nt –178). For these 4 enzymes, increased intensity of the bands (2.2-, 2.9-, 1.6-, and 1.5-fold increase, respectively) was observed after LPS/IFN-γ stimulation compared with the untreated conditions. Because digestion by several enzymes located outside of the remodeled region was either unchanged (ApaI, nt –451; NspI, nt –405; HaeII, nt –363 and –158; PvuII, nt –356; BsaHI, nt –325; NaeI, nt –141 and +69; SacII, nt +22; and ScaI, +99) or decreased (ScaI, nt –451 and PstI, nt +64) after LPS plus IFN-γ treatment, the enhanced digestion observed with BstXI, AlwNI, BanI, and SmaI is likely to be reflective of a true increase in accessibility. Moreover, BstXI and BanI cut at other sites (bands * [nt +117] and $ [nt +456] for BstXI, * [nt +571] and $ [nt +980 and +1303] for BanI) at the same rate in untreated and treated conditions, providing excellent internal controls in the experiment. The remodeled region delineated by these 4 enzymes closely matched the remodeled region predicted by the DNase I and MNase experiments. Because digestion with enzymes cutting just upstream (BsaHI, nt –325) and downstream (HaeII, nt –158) remained unchanged after LPS plus IFN-γ treatment, we more precisely mapped the boundaries of the region undergoing chromatin remodeling on IL-12(p35) induction between nt –325 to –298 in 5′and nt –178 and –158 in 3′. The size of this region, ranging from 120 to 167 bp, corresponds to the length of DNA protected by a nucleosome core, suggesting the selective remodeling of this nucleosome on p35 transcriptional activation.
Pattern of restriction enzyme digestion of chromatin within the IL-12(p35) promoter region. Nuclei from untreated (–) or LPS/IFN-γ–treated (+) THP-1 cells were digested with the indicated restriction enzymes. After purification of genomic DNA and in vitro digestion with EcoRI, DNA samples were examined using Southern blotting and the indirect end-labeling technique. Molecular weight markers are described in “Material and methods.” When several restriction sites for the same enzyme are found within the region under study, symbols are used (°, *, and $) to differentiate them. Fold induction refers to the ratio between stimulated and untreated samples after quantification of the bands with a PhosphorImager.
Pattern of restriction enzyme digestion of chromatin within the IL-12(p35) promoter region. Nuclei from untreated (–) or LPS/IFN-γ–treated (+) THP-1 cells were digested with the indicated restriction enzymes. After purification of genomic DNA and in vitro digestion with EcoRI, DNA samples were examined using Southern blotting and the indirect end-labeling technique. Molecular weight markers are described in “Material and methods.” When several restriction sites for the same enzyme are found within the region under study, symbols are used (°, *, and $) to differentiate them. Fold induction refers to the ratio between stimulated and untreated samples after quantification of the bands with a PhosphorImager.
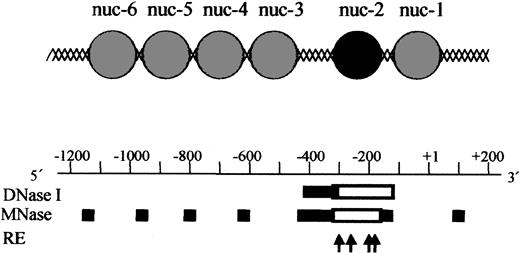
Based on our chromatin structure analysis of the p35 locus using DNase I, Mnase, and restriction enzymes, we established a tentative map of nucleosomes positioning in the region (Figure 4). The IL-12(p35) promoter contains an array of precisely positioned nucleosomes. Because MNase preferentially digests DNA in linker regions between nucleosomes, it is possible to locate nucleosomes within the p35 region. Four nucleosomes (named nuc-3 to nuc-6 from 3′ to 5′) are located between nt –1100 and nt –450. A major nuclease-hypersensitive site is located downstream of nuc-3. Because nuclease-hypersensitive sites usually indicate the presence of nonnucleosomal DNA in vivo, our results indicate that this region is likely to be nucleosome free. Remarkably, the region immediately downstream of this hypersensitive site is resistant to nuclease digestion in basal conditions, but becomes sensitive to all nucleases tested after stimulation with LPS plus IFN-γ. This region spans about nt –310/–168 or 152 nucleotides, which corresponds approximately to the length of DNA protected by a nucleosome core. Therefore, our results strongly suggest that a nucleosome (named nuc-2) is present in this location in basal conditions and selectively remodeled on transcriptional activation. The region downstream of nuc-2 contains one precisely positioned nucleosome referred to as nuc-1 and located on the transcription initiation site (Figure 4). Because assembly of a DNA region into chromatin generally represses transcription, these experiments strongly suggest that the region between nt –410 and –310 could harbor constitutively accessible cis-acting elements, whereas accessibility to the region between nt –310 and –160 could be regulated through nucleosomal remodeling.
Model for the chromatin organization within the human IL-12(p35) promoter region. A tentative assignment of nucleosome positions in this region based on nuclease digestion is shown and aligned with the nuclease-hypersensitive sites. Sites of cutting by DNase I and MNase are depicted by solid bars for basal conditions. Because MNase preferentially digests DNA in linker regions between nucleosomes, it is possible to locate nucleosomes within the p35 region. The DNA domain between nuc-2 and nuc-3 is strongly hypersensitive to DNase I and MNase cleavage. It is therefore likely to represent a nucleosome-free region. Empty boxes indicate increase in DNase I and MNase hypersensitivities on LPS/IFN-γ stimulation. The 4 arrows refer to the cutting sites of BstXI (nt –298), AlwNI (nt –256), BanI (nt –185), and SmaI (nt –178), which become more accessible on activation. Taken together, these results strongly suggest that nuc-2 (black circle) is selectively remodeled on p35 transcriptional activation.
Model for the chromatin organization within the human IL-12(p35) promoter region. A tentative assignment of nucleosome positions in this region based on nuclease digestion is shown and aligned with the nuclease-hypersensitive sites. Sites of cutting by DNase I and MNase are depicted by solid bars for basal conditions. Because MNase preferentially digests DNA in linker regions between nucleosomes, it is possible to locate nucleosomes within the p35 region. The DNA domain between nuc-2 and nuc-3 is strongly hypersensitive to DNase I and MNase cleavage. It is therefore likely to represent a nucleosome-free region. Empty boxes indicate increase in DNase I and MNase hypersensitivities on LPS/IFN-γ stimulation. The 4 arrows refer to the cutting sites of BstXI (nt –298), AlwNI (nt –256), BanI (nt –185), and SmaI (nt –178), which become more accessible on activation. Taken together, these results strongly suggest that nuc-2 (black circle) is selectively remodeled on p35 transcriptional activation.
The IL-12(p35) promoter region harbors critical cis-acting elements
Because nuclease-hypersensitive sites are frequently associated with regulatory regions of genes (such as promoters and enhancers),15 the presence of the open chromatin configuration and of the nucleosomal remodeling we identified in vivo prompted us to examine the potential regulatory role of the underlying DNA element in p35 transcription.
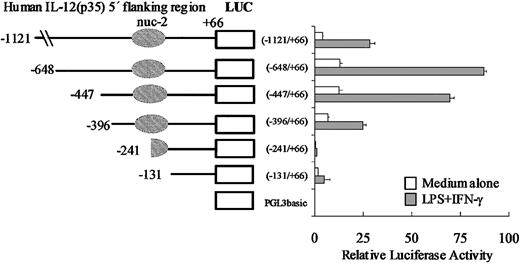
We first assessed the promoter activity of the p35 region under study and cloned a 1.2-kb region (nt –1121/+66) upstream of the luciferase reporter gene (into pGL3-BASIC). This reporter plasmid, referred to as p35-lucWT, was transiently transfected into the monocytic cell line THP-1. We observed that in unstimulated cells, the 1.2-kb region conferred a 100-fold increase of luciferase activity compared with the empty vector pGL3-BASIC (data not shown). However, despite many attempts, we were unable to reproducibly induce expression of the reporter construct p35-lucWT by stimulation with LPS plus IFN-γ regardless of the method of transfection or kinetics of stimulation (data not shown). Nevertheless, because the induction of the endogenous gene was rather low in THP-1 cells (4- to 5-fold; Figure 1A), the low transfection efficiency (5%-10%) we obtained in these cells might have been limiting to detect the inducibility of the p35 reporter construct by LPS plus IFN-γ. Therefore, another monocytic cell line (the murine RAW 264.7 cell line) was also tested and found to be an appropriate system in which to study LPS/IFN-γ–stimulated p35 gene expression. As shown in Figure 5, we observed that in unstimulated cells, the –1121/+66 fragment conferred a 20-fold increase of luciferase activity compared with the empty vector pGL3-BASIC. Activation with IFN-γ and LPS led to a 5- to 10-fold increase in luciferase activity. As a control, the activity of the TK promoter did not increase significantly (data not shown).
Promoter activity of the human IL-12(p35) gene and 5′ deletion analysis of the promoter region. A series of luciferase (LUC) reporter constructs (□) were generated using the pGL3basic vector and different 5′-flanking fragments of the p35 gene. Positions of the 5′ ends of the various fragments and the region protected by nuc-2 are indicated. RAW 264.7 cells were transiently transfected with 2 μg of the reporter plasmids and 20 ng pRL-TK as an internal control plasmid. RAW 264.7 cells were then left untreated (□) or treated with IFN-γ and LPS (▦). Luciferase activities (Firefly and Renilla) were measured in cellular extracts. Promoter activities were normalized using Renilla luciferase activities. Each point is the mean ± SD of triplicate transfections performed in the same experiment. A representative experiment of 3 independent transfections is shown.
Promoter activity of the human IL-12(p35) gene and 5′ deletion analysis of the promoter region. A series of luciferase (LUC) reporter constructs (□) were generated using the pGL3basic vector and different 5′-flanking fragments of the p35 gene. Positions of the 5′ ends of the various fragments and the region protected by nuc-2 are indicated. RAW 264.7 cells were transiently transfected with 2 μg of the reporter plasmids and 20 ng pRL-TK as an internal control plasmid. RAW 264.7 cells were then left untreated (□) or treated with IFN-γ and LPS (▦). Luciferase activities (Firefly and Renilla) were measured in cellular extracts. Promoter activities were normalized using Renilla luciferase activities. Each point is the mean ± SD of triplicate transfections performed in the same experiment. A representative experiment of 3 independent transfections is shown.
To gain additional insight into p35 gene regulation, we generated a series of p35-luciferase reporter constructs containing 5′ deletions within the p35 promoter region. The 6 resulting plasmids were transiently transfected into untreated or IFN-γ/LPS-stimulated RAW 264.7 cells. As shown in Figure 5, our 5′ deletion analysis of the p35 promoter revealed that deletion from –1121 to –648 was associated with a 2- to 3-fold increase of luciferase activities both in unstimulated and in IFN-γ/LPS-stimulated cells, suggesting that some repressor elements might be located in this region. Strikingly, the 155-bp deletion from nt –396 to nt –241 greatly reduced (by 8- and by 30-fold) the basal and the IFN-γ/LPS-stimulated luciferase activities, respectively, suggesting that cis-acting elements, critical for both basal and inducible p35 transcriptional activities, are located within this –396/–241 region. Remarkably, this region overlaps part of the remodeled region corresponding to nuc-2 and the nucleosome-free region immediately upstream.
Sp proteins bind to 2 sites in the –396/–241 p35 promoter region
Because deletion of the –396/–241 region caused a profound loss of basal and inducible human IL-12(p35) promoter activity (Figure 5), we decided to analyze the nucleotide sequence of this region for the presence of known transcription factor–binding sites (Figure 6A). Underlined nucleotides in the p35 sequence correspond to the nucleosome-free region (from nt –410 to –310), whereas bold nucleotides correspond to the region of the remodeled nucleosome nuc-2 (from nt –310 to –160). Both regions represent a CpG island containing several GC boxes with close homology to the Sp1 consensus sequence. We therefore designed 2 double-stranded oligonucleotides, designated Sp1#1wt (nt –301/–274) and Sp1#2wt (nt –360/–334) encompassing the potential Sp1#1 and Sp1#2 sites, respectively (Figure 6B). These oligonucleotides were radiolabeled and tested in EMSAs for DNA-protein interactions with nuclear extracts from monocyte-derived DCs (Figure 7A). With both Sp1 probes, one major retarded complex was observed, although the binding affinity to Sp1#2 site was much lower than to the Sp1#1 site (Figure 7A). Similar results were obtained with nuclear extracts from THP-1 cells and with affinity-purified human Sp1 protein (data not shown). To evaluate the sequence specificity of the binding to the Sp1 probes, we performed competition EMSAs using increasing concentrations of different unlabeled double-stranded competitor oligonucleotides (Figure 7A). Protein binding in the major retarded complex was shown to be sequence specific because its formation was inhibited by competition with an excess of the unlabeled homologous Sp1#1wt and Sp1#2wt oligonucleotides but not by the same molar excesses of mutated versions of these oligonucleotides (Sp1#1mut and Sp1#2mut) or of heterologous oligonucleotides of unrelated sequence (data not shown). An Sp1 consensus oligonucleotide (Sp1 cons) inhibited the formation of the retarded complex to the same extent (or even at lower concentration for Sp1#2) than the homologous oligonucleotide. In contrast, this complex was not competed by a mutated version of the Sp1 consensus oligonucleotide (Sp1mut) containing a GG>TT substitution, thereby demonstrating the specificity of the major retarded complex to the Sp1 site. The absence of competition by the mutated homologous oligonucleotides (Sp1#1mut and Sp1#2mut) demonstrated that the selected mutations (substitution of the GG cores with the dinucleotides TT) abolished Sp1 binding to the Sp1#1 and Sp1#2 sites. Moreover, we confirmed the lack of Sp binding to the probes corresponding to the mutated p35 Sp1 sites #1 and #2 (data not shown).
Analysis of the nucleotide sequence of the p35 promoter. (A) Nucleotide sequence of the human IL-12(p35) proximal promoter region (accession no. AF050083). Nucleotides corresponding to the nuclease-hypersensitive site in untreated cells are underlined. Nucleotides encompassed by nuc-2 appear in bold. The putative Sp1-binding sites (Sp1#1 and Sp1#2, indicated by boxes) were identified by computer analysis (MatInspector V2.2). Nucleotide +1 denotes the S1 initiation site in monocytes as determined by Hayes et al.10 A putative TATA box (–32) is underlined. (B) Mutagenesis of the p35 Sp1-binding sites. The 2 oligonucleotides used as probes in EMSA (named Sp1#1wt and Sp1#2wt) encompass several putative binding sites for Sp1 (these sites are indicated by underlined nucleotides). The recognition core sequence 5′-GG-3′ of each potential Sp1 site is indicated by a thicker bar. Nucleotides that differ from the Sp1 consensus sequence are marked by an asterisk. For Sp1#1mut and Sp1#2mut oligonucleotides, only the bases that are changed compared with the wt sequence are indicated (substitution of the GC cores with the dinucleotide TT). The Sp1 consensus DNA-binding site is shown.
Analysis of the nucleotide sequence of the p35 promoter. (A) Nucleotide sequence of the human IL-12(p35) proximal promoter region (accession no. AF050083). Nucleotides corresponding to the nuclease-hypersensitive site in untreated cells are underlined. Nucleotides encompassed by nuc-2 appear in bold. The putative Sp1-binding sites (Sp1#1 and Sp1#2, indicated by boxes) were identified by computer analysis (MatInspector V2.2). Nucleotide +1 denotes the S1 initiation site in monocytes as determined by Hayes et al.10 A putative TATA box (–32) is underlined. (B) Mutagenesis of the p35 Sp1-binding sites. The 2 oligonucleotides used as probes in EMSA (named Sp1#1wt and Sp1#2wt) encompass several putative binding sites for Sp1 (these sites are indicated by underlined nucleotides). The recognition core sequence 5′-GG-3′ of each potential Sp1 site is indicated by a thicker bar. Nucleotides that differ from the Sp1 consensus sequence are marked by an asterisk. For Sp1#1mut and Sp1#2mut oligonucleotides, only the bases that are changed compared with the wt sequence are indicated (substitution of the GC cores with the dinucleotide TT). The Sp1 consensus DNA-binding site is shown.
Physical and functional characterization of 2 Sp1-binding sites within the IL-12(p35) promoter. (A) Competition assays. Nuclear extracts (10 μg protein) were prepared from DCs and incubated with [32P]-radiolabeled Sp1#1wt and Sp1#2wt probes in the absence of competitor or in the presence of increasing concentrations (5- to 80-fold molar excess) of the indicated unlabeled competitors. The figure shows only the specific retarded complex. (B) The major retarded complex is predominantly composed of Sp1. Supershift assays were performed by incubating Sp1#1wt and Sp1#2wt probes with DC nuclear extracts in the absence of antibody, or in the presence of anti-Sp1, anti-Sp3, or as a control, anti–MEF-2 antibodies. (C) Effect of the mutations in the Sp1 sites on IL-12(p35) gene expression. Unstimulated and IFN-γ/LPS-stimulated RAW 264.7 cells were transiently transfected with 2 μg p35-lucWT, p35-mutSp1#1, or p35-mutSp1#2 plasmids and 20 ng pRL-TK as an internal control plasmid. Promoter activities were normalized using Renilla luciferase activities. Values represent the means ± SD of triplicate samples performed in the same experiment. A representative experiment of 3 independent experiments is shown. RLU indicates relative light units. (D) Ectopic expression of Sp1 transcription factor up-regulates p35 promoter activity. SL2 cells were transiently cotransfected with p35-lucWT (1 μg) and increasing amounts of pPacSp1. To maintain the same amount of transfected DNA and to avoid squelching artifacts, the different amounts of plasmid DNA were complemented to 1.6 μg total DNA by using the empty pPac plasmid. Results are presented as histograms indicating the induction by Sp1 (in fold) with respect to the activity of the p35-lucWT in the absence of Sp1, which was assigned a value of 1. Values represent the means ± SD of triplicate transfection performed in the same experiment. A representative experiment of 4 independent experiments is shown.
Physical and functional characterization of 2 Sp1-binding sites within the IL-12(p35) promoter. (A) Competition assays. Nuclear extracts (10 μg protein) were prepared from DCs and incubated with [32P]-radiolabeled Sp1#1wt and Sp1#2wt probes in the absence of competitor or in the presence of increasing concentrations (5- to 80-fold molar excess) of the indicated unlabeled competitors. The figure shows only the specific retarded complex. (B) The major retarded complex is predominantly composed of Sp1. Supershift assays were performed by incubating Sp1#1wt and Sp1#2wt probes with DC nuclear extracts in the absence of antibody, or in the presence of anti-Sp1, anti-Sp3, or as a control, anti–MEF-2 antibodies. (C) Effect of the mutations in the Sp1 sites on IL-12(p35) gene expression. Unstimulated and IFN-γ/LPS-stimulated RAW 264.7 cells were transiently transfected with 2 μg p35-lucWT, p35-mutSp1#1, or p35-mutSp1#2 plasmids and 20 ng pRL-TK as an internal control plasmid. Promoter activities were normalized using Renilla luciferase activities. Values represent the means ± SD of triplicate samples performed in the same experiment. A representative experiment of 3 independent experiments is shown. RLU indicates relative light units. (D) Ectopic expression of Sp1 transcription factor up-regulates p35 promoter activity. SL2 cells were transiently cotransfected with p35-lucWT (1 μg) and increasing amounts of pPacSp1. To maintain the same amount of transfected DNA and to avoid squelching artifacts, the different amounts of plasmid DNA were complemented to 1.6 μg total DNA by using the empty pPac plasmid. Results are presented as histograms indicating the induction by Sp1 (in fold) with respect to the activity of the p35-lucWT in the absence of Sp1, which was assigned a value of 1. Values represent the means ± SD of triplicate transfection performed in the same experiment. A representative experiment of 4 independent experiments is shown.
To identify directly the factors present in the retarded complex, we performed supershift assays using specific antibodies directed against Sp1 and Sp3. For both the Sp1#1wt and Sp1#2wt probes, addition of the anti-Sp1 antibody interfered with the formation of the complex and generated a supershift complex (Figure 7B). The anti-Sp3 antibody generated a very weak supershift and only slightly decreased the intensity of the complex. When both anti-Sp1 and anti-Sp3 antibodies were included in the binding reaction, a further reduction of the retarded complex was noted. No supershifted complex was observed with a control antibody against MEF-2 (Figure 7B) or when the anti-Sp antibodies were added to the probe alone (data not shown), thus indicating the specificity of the protein-antibody interactions. Overall, these results demonstrate that the transcription factor Sp1 interacts with 2 Sp sites, which lie immediately upstream (Sp1#2) and within (Sp1#1) the p35 promoter region protected by nuc-2. We report a 2-bp point mutation that abrogates Sp binding to these sites.
The Sp1 sites are critical for both basal and LPS/IFN-γ–inducible p35 gene expression
To examine the functional role of the Sp1-binding sites in the transcriptional activity of the p35 promoter, the same 2-bp mutation described was introduced in each Sp1 site individually by site-directed mutagenesis in the context of the –1121/+66 construct. The mutated plasmids were designated p35mutSp1#1 and p35mutSp1#2, respectively. Significant reduction of basal promoter activity in transiently transfected RAW 264.7 cells was observed by mutation of the Sp1#1 site (5-fold reduction) and of the Sp1#2 site (2.5-fold reduction; Figure 7C), indicating that both Sp sites are required for optimal basal p35 promoter activity. Importantly, induction of the p35 promoter activity in response to IFN-γ/LPS stimulation was completely abrogated by mutation of the Sp1#1 site and significantly reduced as a consequence of the Sp1#2 mutation. These results are consistent with the binding of a transcriptional activator, such as Sp1, to both Sp sites. Thus, both Sp1 sites act as key cis-acting elements for regulating the inducibility of the p35 promoter activity by IFN-γ/LPS.
To further examine the role of Sp1 in the regulation of the p35 promoter, we studied the effect of Sp1 overexpression on luciferase activity. For this purpose, we used Drosophila SL2 cells, which lack endogenous Sp factors. As seen in Figure 7D, the IL-12(p35) promoter activity was up-regulated in a dose-dependent manner by ectopically expressed Sp1 protein (up to 37-fold).
Taken together, our functional studies demonstrate a positive regulatory role of the Sp sites Sp1#1 and Sp1#2 in both basal and IFN-γ/LPS-inducible p35 promoter-driven gene expression. Importantly, the Sp1#1 site, found to be the most important both physically and functionally, precisely lies within the nuc-2 region undergoing the selective stimulation-dependent chromatin remodeling. Furthermore, The Sp1#2 site is located in the nucleosome-free region immediately upstream of nuc-2.
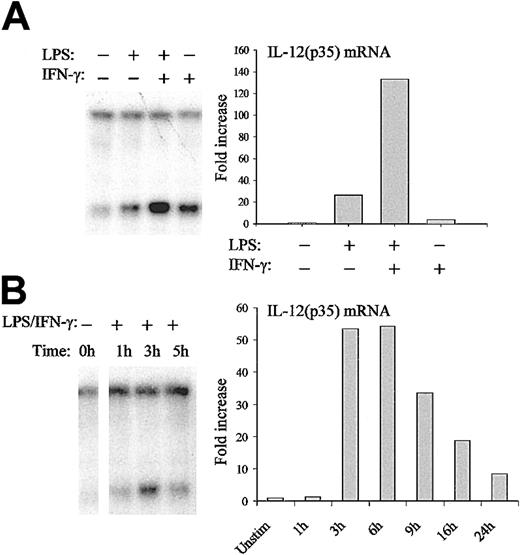
Regulation of IL-12(p35) chromatin remodeling in DCs
DCs represent the major source of IL-12.27 It is therefore important to validate our results on chromatin organization of the IL-12(p35) promoter in these primary cells. Moreover, the use of DCs also allows the study of the relative contribution of different stimuli such as LPS and IFN-γ that act synergistically on IL-12(p35) induction.28 Indirect end-labeling experiments using DNase I confirmed the hypersensitive site observed in THP-1 cells. Moreover, similar remodeling was also observed in DCs on stimulation with LPS and IFN-γ (data not shown). For further studies, we used the BstXI (cutting site, nt –298) restriction enzyme that cuts very close to the Sp1#1 site. As seen in the right panel of Figure 8A, LPS and IFN-γ acted synergistically on IL-12(p35) mRNA expression in DCs. Importantly, both stimuli also contributed to chromatin opening and their combination acted synergistically in the remodeling (Figure 8A left panel). We then looked at the kinetics of chromatin remodeling and IL-12(p35) mRNA synthesis in DCs. As seen in Figure 8B, mRNA synthesis started 1 to 3 hours after LPS/IFN-γ stimulation, peaked at 6 hours, and rapidly decreased thereafter. In contrast, chromatin remodeling started already 1 hour after stimulation and reached a peak after 3 hours. We therefore conclude that chromatin remodeling at the p35 locus is a highly regulated and transient event, which precedes mRNA synthesis.
Regulation of nuc-2 remodeling in DCs. (A) DCs were either incubated with medium alone or stimulated with LPS (1 μg/mL) or IFN-γ (100 U/mL) or both for 3 hours. Nuclei were then isolated and digested with BstXI. After purification of genomic DNA and in vitro digestion with EcoRI, samples were analyzed by Southern blotting and indirect end-labeling (left panel). IL-12(p35) mRNA levels were determined by real-time RT-PCR (right panel). For this purpose, DCs were incubated with medium alone or stimulated with LPS (1 μg/mL) or IFN-γ (100 U/mL) or both for 6 hours. IL-12(p35) mRNA levels were normalized using β-actin mRNA levels and were compared with unstimulated conditions. (B) DCs were either incubated with medium alone or stimulated with LPS (1 μg/mL) and IFN-γ (100 U/mL) for the indicated periods of time. DNA accessibility in chromatin within the p35 locus (left panel) and IL-12(p35) mRNA levels (right panel) were assessed as described in panel A. The results are representative of at least 3 independent experiments performed with different donors.
Regulation of nuc-2 remodeling in DCs. (A) DCs were either incubated with medium alone or stimulated with LPS (1 μg/mL) or IFN-γ (100 U/mL) or both for 3 hours. Nuclei were then isolated and digested with BstXI. After purification of genomic DNA and in vitro digestion with EcoRI, samples were analyzed by Southern blotting and indirect end-labeling (left panel). IL-12(p35) mRNA levels were determined by real-time RT-PCR (right panel). For this purpose, DCs were incubated with medium alone or stimulated with LPS (1 μg/mL) or IFN-γ (100 U/mL) or both for 6 hours. IL-12(p35) mRNA levels were normalized using β-actin mRNA levels and were compared with unstimulated conditions. (B) DCs were either incubated with medium alone or stimulated with LPS (1 μg/mL) and IFN-γ (100 U/mL) for the indicated periods of time. DNA accessibility in chromatin within the p35 locus (left panel) and IL-12(p35) mRNA levels (right panel) were assessed as described in panel A. The results are representative of at least 3 independent experiments performed with different donors.
Discussion
In this study, we have investigated the chromatin environment of the human IL-12(p35) promoter. We were able to demonstrate precise nucleosomal positioning within this region. Strikingly, on activation of the gene by LPS/IFN-γ, a single nucleosome, nuc-2 located in the region –310/–160, was selectively remodeled or disrupted. The determination of the chromatin environment was performed in the monocytic cell line THP-1. Stimulation of these cells by LPS/IFN-γ led to a 4- to 5-fold induction of p35 mRNA synthesis (Figure 1A), which is rather low compared with the levels obtained in primary cells such as DCs (Figure 8A). During the initiation of an immune response, DCs represent the major source of IL-12. We therefore used these primary cells to confirm our observations in a more relevant model and to further investigate the regulation of nuc-2 remodeling.
One important aspect of IL-12(p35) and (p40) regulation is the synergistic activation by LPS and other bacterial compounds with IFN-γ.28 Here we demonstrate that IFN-γ treatment of human DCs clearly induces chromatin remodeling at the p35 promoter and acts synergistically with LPS for maximal opening/remodeling. This result suggests that one of the major actions of IFN-γ on p35 induction could be the control of the accessibility of the promoter to transcription factors. Interestingly, no such effect of IFN-γ was observed at the p40 locus in murine macrophages.19 Results obtained on chromatin remodeling at the murine p40 and human p35 promoters suggest differential regulation of these 2 genes and involvement of different signaling pathways. However, to address this issue, it would be required to simultaneously study chromatin organization of both genes in the same system and experimental settings.
In this report, we show that nuc-2 remodeling is rapid, transient, and precedes p35 mRNA synthesis, implying that chromatin remodeling might be a prerequisite for high levels of gene expression. Moreover, IFN-γ readily induces chromatin remodeling in the IL-12(p35) gene promoter without activating its transcription. These results imply that, although nuc-2 remodeling might be necessary, it is not sufficient for full expression.
We assessed p35 promoter activity and by using 5′ deletion mutants and reporter gene assays, we showed that response to LPS/IFN-γ stimulation required a region located between nt –396 and –241. Within this region, we identified 2 binding sites for the transcription factor Sp1. We demonstrated that these 2 Sp1 sites were critical for both basal and inducible p35 promoter activities. Sp1 is generally considered to be a ubiquitous transcription factor, constitutively expressed in various cell types29 including DCs (data not shown). It is therefore surprising that an Sp1 element might be the target of a signal transduction pathway involved in the induction of the p35 gene. However, several lines of evidence have suggested that Sp1 contributes to the inducible transcription of a few other genes including the IL-1β,30 p15INK4B,31 tumor necrosis factor receptor II (TNFR-II),32 IL-6,33 IL-10,34 and TNF-α35 genes. The Sp1 sites we identified in the p35 promoter could constitute another example for the role of Sp1 in inducible transcription. This hypothesis is currently under further investigation in our laboratory. Inducible posttranslational modifications of Sp1, such as phosphorylation,36 may account for these observations. Alternatively, Sp1 binding might be critical for the assembly of a functional enhanceosome on further stimulation.37 Indeed, inducible transcription factors were recently shown to be involved in the regulation of the IL-12(p35) gene in mice.13,38 It is likely that human IL-12(p35) promoter activity depends on a coordinate recruitment of constitutive as well as inducible transcription factors, such as members of the NF-κB, IRF, or stat families.
Unlike stably transfected plasmids or episomal constructs, chromatin structure of transiently transfected plasmids is usually incomplete or aberrant and may vary with the transfection method used. Our transient transfection experiments might therefore not fully assess the involvement of chromatin remodeling in the regulation of the p35 promoter activity. It will be critical to determine the respective role of the different Sp1 sites identified here in their native chromatin context. Indeed, the Sp1#1 site precisely lies within the nuc-2 region. It is noteworthy that we identified a third Sp1 site (Sp1#3) of weaker affinity, which is also located in the remodeled nuc-2 region (data not shown). Sp1 was shown to be able to access nucleosomal DNA; however, its affinity for nucleosomal GC boxes is greatly reduced compared with naked DNA.39 Our results suggest that the Sp1#1 site is inaccessible in resting cells and that the selective stimulation-dependent remodeling of nuc-2 leads to unmasking of this critical cis-element. A similar mechanism could be involved in the inducible recruitment of Sp1 to the TNF-α promoter, which was described on activation of J774 cells.35 In contrast, the Sp1#2 site is located in the nucleosome-free region immediately upstream of nuc-2 and might therefore be constitutively accessible. Occupancy of the Sp1#2 site might be required for the active positioning of nuc-2 as suggested for other promoters such as the HIV-1 5′ long terminal repeat.23,40 Alternatively, the Sp1#2 site might be required for nuc-2 remodeling through recruitment of chromatin remodeling complexes. Indeed, Sp1 is able to recruit CBP/p300 complexes, which possess acetyltransferase activities,41 and a recent study reported the interaction of Sp1 with the mammalian SWI/SNF-like chromatin remodeling BAF complex.42 The mechanisms involved in the selective stimulation-dependent remodeling of nuc-2 in the p35 promoter remains to be elucidated and could implicate the transcription factor Sp1 or a still-unidentified protein binding close to the region occupied by nuc-2. Interestingly, at the monocyte chemoattractant protein 1 (MCP-1) and regulated on activation normal T cell expressed and secreted (CCL5; RANTES) promoters, IFN-γ was found to induce H4 acetylation facilitating NF-κB recruitment,43 whereas BRG1, the essential adenosine triphosphatase (ATPase) subunit of the BAF complex, is involved in IFN-γ-induced class II transactivator (CIITA) expression.44
Finally, together with recent data from Saccani et al,45 showing the role of histone H3 phosphorylation in activated DCs, our findings provide evidence that selective and regulated changes in the chromatin environment of key cytokine genes are involved in the process of DC maturation, a critical step in the induction of the immune response.46
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2002-09-2851.
Supported by the Centre de Recherche Interuniversitaire en Vaccinologie sponsored by the Région Wallonne and SmithKline Beecham Biologicals (Belgium), by the NEOVAC program of the 5th framework of the European Commission (contract no. QLK2-CT-1999-00429), by the Fonds National de la Recherche Scientifique (FNRS, Belgium), the Télévie-Program, the Université Libre de Bruxelles (ULB, ARC program no. 98/03-224), the International Brachet Stiftung (IBS), the CGRI-INSERM Cooperation, the Région Wallonne-Commission Européenne FEDER, and the Theyskens-Mineur Foundation. S.G. is a research fellow of the FNRS. C.V.L. is Maître de Recherches of the FNRS. D.D. is supported by a postdoctoral fellowship from the Région Wallonne (grant no. 991/4202) and S.N. is a fellow of the FNRS-Télévie.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



![Figure 7. Physical and functional characterization of 2 Sp1-binding sites within the IL-12(p35) promoter. (A) Competition assays. Nuclear extracts (10 μg protein) were prepared from DCs and incubated with [32P]-radiolabeled Sp1#1wt and Sp1#2wt probes in the absence of competitor or in the presence of increasing concentrations (5- to 80-fold molar excess) of the indicated unlabeled competitors. The figure shows only the specific retarded complex. (B) The major retarded complex is predominantly composed of Sp1. Supershift assays were performed by incubating Sp1#1wt and Sp1#2wt probes with DC nuclear extracts in the absence of antibody, or in the presence of anti-Sp1, anti-Sp3, or as a control, anti–MEF-2 antibodies. (C) Effect of the mutations in the Sp1 sites on IL-12(p35) gene expression. Unstimulated and IFN-γ/LPS-stimulated RAW 264.7 cells were transiently transfected with 2 μg p35-lucWT, p35-mutSp1#1, or p35-mutSp1#2 plasmids and 20 ng pRL-TK as an internal control plasmid. Promoter activities were normalized using Renilla luciferase activities. Values represent the means ± SD of triplicate samples performed in the same experiment. A representative experiment of 3 independent experiments is shown. RLU indicates relative light units. (D) Ectopic expression of Sp1 transcription factor up-regulates p35 promoter activity. SL2 cells were transiently cotransfected with p35-lucWT (1 μg) and increasing amounts of pPacSp1. To maintain the same amount of transfected DNA and to avoid squelching artifacts, the different amounts of plasmid DNA were complemented to 1.6 μg total DNA by using the empty pPac plasmid. Results are presented as histograms indicating the induction by Sp1 (in fold) with respect to the activity of the p35-lucWT in the absence of Sp1, which was assigned a value of 1. Values represent the means ± SD of triplicate transfection performed in the same experiment. A representative experiment of 4 independent experiments is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/12/10.1182_blood-2002-09-2851/6/m_h81234446007.jpeg?Expires=1769094090&Signature=rV7h11lskrek40XNU4EuvDj9CKchrZEmuwaGfmE1qNXNxhhKuHYyaZcIKyCMPt4HrXn7KShIMAzaGm8ZMGFV6GSfvJW0j68BlJoEs3uY4iYVAbSKDPqkqPI7ExBxO~QO8k7tTBNlviUP~JVhmV8xa12Zy0vazAf9cjvjPnQcf61Z8dQ4ZJK-Y9IiA~DoeJFXm3KTcjfOrIL~qqKrLNfN7LMQTdq0LDagl5y4kJM7IX-pjzKMid5znJKfZxPAqDGUOnPP3wmSL9-mS6loUuUNzNgJbEJyisQVLoKZ7hjx9q0CvNaiwTb1JRstRH22qheQLxr3oTz6sR-~ql-qY2AWRQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal