A prenatal origin of translocations associated with pediatric leukemia has been demonstrated for MLL-AF41 in infant leukemia, TEL-AML12,3 in common acute lymphocytic leukemia (cALL), and AML1-ETO4 in acute myeloid leukemia (AML). We investigated whether AML-associated translocations PML-RARA and CBFB-MYHII could arise before birth. PML-RARA arises from the t(15;17) translocation,5 characteristic of acute promyelocytic leukemia (APL)6 (AML FAB subtype M3), while CBFB-MYHII arises from inv(16)(p13q22)7 and t(16;16).8
Diagnostic samples from 2 t(15;17) and 2 inv(16) cases were obtained with informed consent and ethics committee approval from patients enrolled in the Northern California Childhood Leukemia Study (NCCLS) and from the Children's Oncology Group AML cell bank. Corresponding Guthrie cards (neonatal blood spots) for patients were obtained from a central repository maintained by the Genetic Diseases Branch of the California Department of Health Sciences. We obtained genomic break-points from patients by multiplex long-distance polymerase chain reaction (PCR) using eLONGase DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA), according to manufacturers' instructions, and sequenced the translocation junctions. For each PML-RARA case, 10 multiplex reactions were set up, containing 2 PML primers from bcr3 (intron 3; 1447 bp) or bcr1 (intron 6; 1056 bp), in combination with 1 of 5 RARA primers. For each CBFB-MYHII sample, 6 individual PCR reactions were set up using 1 of 6 CBFB primers (targeted to intron 5; 16 359 bp) in combination with the single MYHII primer (targeted to intron 11; 370 bp). Primer sequences are available on request.
We established nested or seminested PCR assays for each clonotypic sequence prior to analysis of the corresponding Guthrie cards for the presence of these sequences. Assay specificity was confirmed by testing patient-specific DNA as well as nonpatient DNA. Assay sensitivity was determined using serial dilutions of patient-specific DNA. Amplification was performed with Ampdirect buffers (Rockland Immunochemicals, Gilbertsville, PA) and eLONGase enzyme, according to the manufacturer's protocol. Prior to translocation-specific PCR, each Guthrie card was tested for capacity to support PCR amplification of a normal gene, NAD(P)H:quinone oxidoreductase (NQO1), as previously described.2
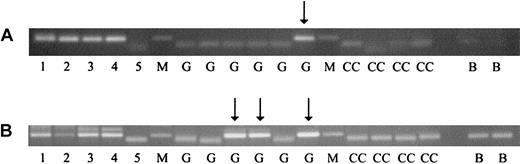
One of the t(15;17) cases (no. P1), aged 10.7 years, generated a positive PCR result in 2 separate assays (bands observed in 1 segment of 8 and 3 segments of 6 analyzed) (Figure 1). Of the 2 inv(16) cases examined, one (no. P5), aged 9.4 years, gave a positive result, in one segment of 14 analyzed in 2 separate assays (Figure 1). For both positive cases, PCR products were sequenced and confirmed as identical to the sequences obtained from the respective patient samples at diagnosis, verifying the in utero origination of the leukemic clonal translocations. The postnatal latencies observed in this study (10.7 and 9.4 years) are among the most protracted demonstrated by backtracking AML to birth, the longest such latency (12 years) having been reported for an AML case with t(8;21) and an AML1-ETO fusion gene.4 The latency period may reflect postnatal persistence of translocation-positive, quiescent multipotential cells, which, upon later recruitment into the myeloid differentiation pathway, acquire additional secondary changes necessary for leukemia. Animal models of PML-RARA and CBFB-MYHII support a multistep progression from fusion gene acquisition to development of these leukemias.9,10
PCR analysis of clonotypic genomic inv(16) (CBFB-MYHII)and t(15;17) (PML-RARA)sequences in neonatal Guthrie cards of leukemic patients. For analysis of Guthrie cards, 1/16 pieces were placed directly in first-round PCR reactions (50 μL), and for each individual, we tested at least 8 such segments (1/2 of a 1.5-cm spot). Second-round amplification (25 μL) was performed with 1 μL from the first-round PCR reactions. PCR conditions included 2 preincubation steps of 80°C for 15 minutes and 94°C for 4 minutes, followed by 40 amplification cycles of 94°C for 30 seconds, 58°C for 1 minute, 72°C for 1 minute, and, finally, a 72°C extension step for 7 minutes. Results shown are from the second round of nested PCR analysis. Lane markers: M indicates molecular weight marker. Lanes 1-5 show 1:10 dilutions of patient diagnostic DNA with lane 1 being 10 ng/μL and lane 5, 1 pg/μL DNA. G indicates patient Guthrie card samples; CC, control Guthrie card samples (from a healthy nonaffected individual); B, blank (no DNA sample). Arrows indicate PCR products from patient Guthrie segments. (A) CBFB-MYHII case no. P5: a single patient Guthrie segment yielded a PCR product from 6 analyzed. (B) PML-RARA case no. P1: 3 patient Guthrie segments yielded positive PCR products from 6 analyzed.
PCR analysis of clonotypic genomic inv(16) (CBFB-MYHII)and t(15;17) (PML-RARA)sequences in neonatal Guthrie cards of leukemic patients. For analysis of Guthrie cards, 1/16 pieces were placed directly in first-round PCR reactions (50 μL), and for each individual, we tested at least 8 such segments (1/2 of a 1.5-cm spot). Second-round amplification (25 μL) was performed with 1 μL from the first-round PCR reactions. PCR conditions included 2 preincubation steps of 80°C for 15 minutes and 94°C for 4 minutes, followed by 40 amplification cycles of 94°C for 30 seconds, 58°C for 1 minute, 72°C for 1 minute, and, finally, a 72°C extension step for 7 minutes. Results shown are from the second round of nested PCR analysis. Lane markers: M indicates molecular weight marker. Lanes 1-5 show 1:10 dilutions of patient diagnostic DNA with lane 1 being 10 ng/μL and lane 5, 1 pg/μL DNA. G indicates patient Guthrie card samples; CC, control Guthrie card samples (from a healthy nonaffected individual); B, blank (no DNA sample). Arrows indicate PCR products from patient Guthrie segments. (A) CBFB-MYHII case no. P5: a single patient Guthrie segment yielded a PCR product from 6 analyzed. (B) PML-RARA case no. P1: 3 patient Guthrie segments yielded positive PCR products from 6 analyzed.
A prenatal origin has therefore been demonstrated for 2 more translocations, PML-RARA and CBFB-MYHII, associated with childhood AML, suggesting that a wide diversity of childhood leukemias originate prenatally.
Supported by National Institutes of Health grants R01 CA89032, R01 ES09137, P42 ES04705, and P30 ES01896, to J.W., P.B., and M.S., respectively.
We thank physicians and hospitals in California who contributed diagnostic material to this study. We thank the California Department of Health Services for access to Guthrie cards.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal