Abstract
BCR-ABL fusion oncogene is the molecular hallmark of chronic myelogenous leukemia (CML), a condition characterized by a progression from a chronic to acute phase leukemia because of secondary genetic events, the nature of which remains largely unknown. Here, we report that the expression of the p210 BCR-ABL fusion protein leads to a down-regulation of BRCA1 protein, a gene product involved in the maintenance of genome integrity. BRCA1 protein is nearly undetectable in leukemia cells from patients with CML, both during the chronic phase and in blast crisis. Similarly, stable transfection-enforced expression of p210 protein in established hematopoietic cell lines leads to severe BRCA1 depletion. The lack of significant change in BRCA1 mRNA level in cells expressing p210 supports the hypothesis that the regulation of BRCA1 protein level occurs after transcription. It is abolished on exposure of the cells to STI571 and by mutation in the adenosine triphosphate (ATP) pocket of p210 and thus seems to require the tyrosine kinase activity of BCR-ABL. Cell lines expressing high levels of BCR-ABL display an increased rate of sister chromatid exchange and chromosome aberrations after ionizing radiation. These findings reveal a novel link between the oncoprotein BCR-ABL and the tumor-suppressor protein BRCA1.
Introduction
Chronic myelogenous leukemia (CML) is a clonal malignancy of the pluripotent hematopoietic stem cell, characterized by an uncontrolled proliferation of myeloid cells that harbor the Philadelphia chromosome (Ph1) carrying the specific t(9;22) translocation. The rearranged BCR and c-ABL genes yields a BCR-ABL-p210Kd fusion protein (p210) carrying constitutive tyrosine kinase (TK) activity.1 The major characteristics of the disease is a progression from an initial chronic phase toward an acute phase that remains of poor prognosis despite major responses obtained by the use of STI571. This progression is accompanied by an enhanced expression of p210 in leukemia cells, with evidence of additional genetic and chromosomal abnormalities, suggesting that the up-regulation of the tyrosine kinase activity brings about genetic instability in Ph1+ cells.
The precise molecular mechanisms underlying this genetic instability, which has been extensively studied in both human CML as well as in experimental cell transformation systems, remain undetermined. Several genetic alterations occurring during acute phase have been reported such as p53 mutation,2 shortening of telomeres,3 and methylation of the ABL promoter.4 The role of microsatellite instabilities remains controversial.5,6 However, in experimental settings, few genetic events have been linked to genome instability in cells expressing p210. An interaction between the xeroderma pigmentosum group B gene product and p2107 and a mutator phenotype associated with an increased expression of the DNA polymerase beta8 have been reported. We have previously reported that exogenous p210 expression in both murine and human hematopoietic cells leads to massive down-regulation of the double-strand break repair protein DNA-PKCs (DNA-dependent protein kinase catalytic subunit).9 This down-regulation that is reverted by tyrosine kinase and proteasome inhibitors is associated with a marked deficiency in DNA repair, suggesting that this phenomenon could play a role in determining the genetic instability associated with the occurrence of the blast crisis in CML. More recently alterations in the level of RAD51, a protein involved in homologous recombination repair, has been linked to drug resistance in p210-expressing cells.10
In the course of exploring various pathways of DNA repair in p210-expressing cells, we observed that BRCA1 protein level was also markedly decreased. The role of BRCA1 in the surveillance of genome integrity is recognized, although the precise biochemical activities involved in this activity have not yet been identified. The displacement of BRCA1 within the nucleus, after exposure of cells to DNA-damaging agents, suggests that BRCA1 moves to “sites of repair.” Several protein partners of BRCA1 are key players in DNA repair: Rad51,11 Rad50/MRE11/NBS,12 ATM,13 BLM,14 and FANCD.15 Furthermore, BRCA1 is a substrate for several DNA damage-activated kinases,13,16 suggesting that its activity is prone to regulation on DNA damage.16 In addition, the association of BRCA1 with RNA polymerase II is consistent with its contribution into transcription-coupled repair of oxidative lesions.17,18 The interaction of BRCA1 with Rad51 in mitotic and meiotic cells points more specifically toward a putative intervention in recombination processes. Finally, cells nullizygous for BRCA119 are extremely sensitive to double-strand breaks.20 Here, we report a marked down-regulation of BRCA1 level on expression of the p210 protein in several experimental models, as well as in primary leukemia cells from patients with CML.
Materials and methods
Cell lines and culture conditions
Murine Ba/F3 and human UT-7 cells. Murine Ba/F3 cell clones and human hematopoietic UT-7 cell clones stably expressing BCR-ABL p210 have been previously described.9 The Ba/F3 clone expressing tetracycline-inducible BCR-ABL has been previously described.21 The p210 fusion protein is expressed at low levels in UT-7/E8-1 clone and at high levels in UT-7/E8-2 and UT-7/9 clones. Except for the UT-7/9, UT-7 cells were cultured in the presence of recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF; 10 ng/mL; R&D Systems, Minneapolis, MN). The Ba/F3 cell line was cultured in the presence of 10% Walter and Elisa Hall Institute (WEHI)–conditioned medium as a source of interleukin 3 (IL-3).
The proteasome inhibitor lactacystin (Calbiochem, La Jolla, CA) was diluted in dimethyl sulfoxide (DMSO) and used at the final concentration of 5 μM. Cells were exposed to the proteasome inhibitor for 8 hours. The tyrosine kinase inhibitor STI571 (Novartis, Bale, Switzerland) was added to cell cultures at a final concentration of 5 μM. Cells were collected after 24 hours of exposure.
Human MO7e cells. Retroviral infectious particles were produced in 293-EBNA cells. The retrovirus-producing cell line 293 EBNA was maintained in Dulbecco modified Eagle medium (DMEM) medium with 10% fetal calf serum (SVF; Gibco, Life Technologies, France). Vesicular stomatitis virus G (VSV-G)–pseudotyped retroviruses were produced by transient transfection of 293 EBNA with 3 different constructs. The vector pN8-ϵ containing the VSV-G sequence and the vector pN8-ϵ containing the Moloney murine leukemia virus (Mo-MuLV) gag-pol sequences were kindly supplied by Dr J. Morgenstern (Millenium, Boston, MA). The third vector was the retroviral vector MIGR, carrying either BCR-ABL-IRES-eGFP or BCR-ABL/K1172R-IRES-eGFP encoding sequence for p210 kinase dead activity or the plasmid MIGR containing (green fluorescent protein variant) eGFP cDNA that was used as control. Transfection was performed using the Exgen reagent (Euromedex, Mondolsheim, France) according to the manufacturer's protocol. Supernatant containing infectious retroviral particles was recovered after 2 and 3 days of culture. Viral titers were determined by limiting dilution of the supernatants on NIH-3T3 cells, which were then analyzed on the basis of green fluorescent protein (GFP) fluorescence.
Retroviral infections
MO7e cell line was cultured in α minimum essential medium (αMEM) with 10% SVF (Gibco) supplemented with 10 ng/mL of GM-CSF (Novartis). Cells plated in 24 wells were mixed with supernatant containing infectious retroviral particles, using a multiplicity of infection of 10 viruses per cell, in the presence of hexadimethrine bromide (4 μg/mL) (Sigma, Saint Quentin, France). The cells were cultured for 24 hours and subjected to a second round of infection in the same conditions. GFP-expressing cells were sorted using FACSVantage (Becton Dickinson) 96 hours later and cultured in αMEM medium containing GM-CSF.
Primary CML samples and healthy controls
Light-density CML cells were obtained using Ficoll-Hypaque gradients from patients in chronic phase at diagnosis (1 patient) or during accelerated (2 patients) or blast phases (2 patients). Three samples from mobilized peripheral blood were used as controls. Cells were subjected to lymphocyte separation medium (Eurobio, Les Ulis, France). Cells were washed twice in cold phosphate-buffered saline (PBS), lysed by boiling in sodium dodecyl sulfate (SDS) sample buffer, and sonicated. Total protein concentration was determined by the Bradford method with the Bio-Rad protein assay (Bio-Rad, Hercules, CA).
Western blot analysis
Cells were washed twice with cold PBS, and extracts were prepared by the addition of lysis buffer (50 mM Tris (tris(hydroxymethyl)aminomethane)–HCl [pH 8], 120 mM NaCl, 5 mM ethylenediamine tetra-acetic acid [EDTA], 0.1% NP40, 1 mM phenylmethylsulfonid [PMSF], 50 mM NaF, 10 μg/mL leupeptin, 10 μg/mL aprotinin, 5 μg/mL pepstatin, 1 mM dithiothreitol [DTT], tablets inhibitor EDTA–free) (Roche) and sonicated. Total protein concentration was determined by the Bradford method with the Bio-Rad protein assay (Bio-Rad). Whole-cell extracts boiled in SDS sample buffer were fractionated through SDS–polyacrylamide gel electrophoresis (SDS-PAGE), 10% SDS-PAGE gel for β-actin, 5% for BRCA1 and BCR-ABL. The proteins were electroblotted in a liquid transfer system to nitrocellulose membranes (Schleicher and Schuell, Dassel Germany), and then subjected to a standard Western blot analysis protocol using a panel of antibodies human anti-BRCA1 (OP92; Oncogene Research Products, Darmstadt, Germany), mouse anti-Brca1 (GH118 kindly supplied by S. Ganesan from the Dana-Farber Cancer Institute, Boston, MA), anti-RAD51 (PC130; Oncogene Research Products), anti-ABL (OP19; Oncogene Research Products), antiphosphotyrosine kinase (PY20: sc-508; Santa Cruz, Santa Cruz, CA). Equivalent protein loading was controlled by β-actin expression using an anti–β-actin (A5316; Sigma). The secondary antibody used was a peroxidase-conjugated affinity pure goat antimouse or a peroxidase-conjugated affinity pure goat antirabbit (Jackson ImmunoResearch Laboratories, West Grove, PA). Immunoblots were visualized by enhanced chemiluminescence (ECL+; Amersham, Uppsala, Sweden).
Measurement of BRCA1 mRNA level by quantitative reverse transcription polymerase chain reaction (RT-PCR)
mRNA isolation and reverse transcription. Total RNA was isolated by a single step guanidinium isothiocyanate method using the QuickPrep Micro mRNA Purification Kit (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's instructions. First-strand cDNA was synthesized in a volume of 80 μL, with TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA). Total RNA (1 μg) was mixed with 10 × reverse transcriptase buffer, 5 mM MgCl2, 500 μM of each deoxynucleoside triphosphate (dNTP), 2.5 μM random hexamers, 32 units RNase inhibitor, and 4 units multiscribe reverse transcriptase. The cycling conditions were 10 minutes at 25°C, 30 minutes at 48°C, and 5 minutes at 95°C. Reactions in which reverse transcriptase was omitted served as controls.
Real-time PCR quantification.BRCA1 and an internal reference gene (β-2 microglobulin) cDNA were quantified using a fluorescence-based real-time detection method. PCR was performed with an ABI PRISM 7700 Sequence Detector and SYBR Green reagents (Applied Biosystems). The specificity of the nucleotide sequence chosen was confirmed by conducting basic local alignment search tool analyses; primer sequences used are available on request. We used 2.5 μL reverse transcriptase product for PCR in a final volume of 25 μL. For each PCR, a standard curve was produced, using 4 consecutive 1:10 dilutions of a positive sample. All samples were run in triplicate. The relative amounts of mRNA for each tested gene in the samples were calculated by comparison with standard curves. For each sample, results were normalized, using the β-2 microglobulin. The PCR reaction mixture consisted of 600 nM of each primer; 2.5 U AmpliTaq Gold polymerase; 200 μM each deoxyadenosine triphosphate (dATP), deoxycytidine triphosphate (dCTP), and deoxyguanosine triphosphate (dGTP); 400 μM deoxyuracil triphosphate (dUTP); 5.5 mM MgCl2; and 1 × SYBR Green Taqman Buffer, to a final volume of 25 μL (all reagents were from Applied Biosystems). Cycling conditions were 50°C for 10 seconds, 95°C for 10 minutes, followed by 46 cycles at 95°C for 15 seconds and 60°C for 1 minute.
Analysis of chromosomal aberrations and sister chromatin exchange (SCE)
BrdU (20 μM) was added to flasks. Twenty-four hours later cells were exposed to 2 Gy gamma irradiation; 24 hours after medium was removed, it was replaced with fresh medium. Colcemid was added at 0.1 μg/mL for 1.5 hours. Fixed preparations were dropped onto clean glass slides and stained with Hoecsht 33528. The slides were washed in running water for 2 minutes and incubated in Sorensen buffer (67 mM KH2PO4 + 67 mM Na2HPO4) for 1 minute. The wet slides were covered with coverslips, and metaphases were photographed immediately with a fluorescence microscope. Images were captured with a digital camera (Kodak Micromax 1400).
Assessment of DNA repair by fluorescence in situ hybridization (FISH)
Chromosome preparation was performed as previously described.22 Cells were incubated 24 hours after x-irradiation with phytohemagglutinin M (GIBCO/BRL, Grand Island, NY), and 1% bromodeoxyuridine (BrdU); Colcemid (0.1 μg/mL) was added 2 hours later. Cells were given hypotonic shock (KCl at 75 mM) for 20 minutes at 37°C, and slides with chromosomes in metaphase were prepared according to the standard methanol acetic acid (3:1, vol/vol) procedure. Slides were stained by Fluorescent Plus Giemsa, and whole chromosome 1 painting was performed with a specifically fluorescent-labeled probe (GIBCO/BRL). The slides were analyzed under a fluorescence microscope by visual scoring of translocations (200 cells were counted for each condition).
Results
BRCA1 protein is down-regulated by p210 both in murine and human cell lines
We have previously reported that expression of p210 in human or murine hematopoietic cells leads to a marked decrease in the level of DNA-PKCs that could possibly contribute to the defects in repair observed in these cells.9 This observation has led us to investigate the status of other gene products involved in various DNA repair pathways in cells expressing p210. Here, we report a drastic reduction of BRCA1 accumulation in human UT-7 cells expressing p210 protein (Figure 1A). The same differences in BRCA1 protein expression between parental and p210-expressing cells were observed using cells lysed by boiling in sodium dodecyl sulfate (SDS) sample buffer and sonicated to rule out differences because of a different association of BRCA1 with DNA that would be lost during BRCA1 protein extraction (data not shown). Three independent clones expressing increasing levels of p210 protein are compared with the parental UT-7/P cells. Whereas BRCA1 protein is readily detectable in the parental cells (lane 1), it decreases as the level of p210 increases (lanes 2-3), to become nearly undetectable in the clone with the highest levels of p210 (UT-7/9, lane 4). It is noteworthy that no change in the levels of endogenous c-ABL is observed. This mirror image correlation between the respective levels of accumulation of these 2 proteins suggests that p210 mediates a pathway leading to depletion of BRCA1 protein. This phenomenon is also observed in murine hematopoietic Ba/F3 cells expressing p210. BRCA1 is nearly undetectable in a Ba/F3 clone expressing p210 (Figure 1B lane 2) compared with the parental cell line (Figure 1B lane 1). Finally, straightforward evidence for this phenomenon is brought about by the massive depletion of BRCA1 observed in a clone of Ba/F3 in which p210 expression is conditionally activated by deprivation of doxycycline for 48 hours in a tetracycline-regulated promoter (Tetoff) setting (Figure 1C). In the presence of doxycycline these cells display low constitutive level of BCR-ABL relatively to endogenous c-ABL (BCR-ABL/c-ABL ratio = 0.6). Deprivation of doxycycline for 48 hours leads to a nearly 10-fold increase of BCR-ABL accumulation (BCR-ABL/c-ABL ratio = 5).
Western blot analysis showing BRCA1 and BCR-ABL expression in human and murine clones expressing variable levels of BCR-ABL. (A) Levels of BRCA1 and BCR-ABL protein were analyzed by immunoblotting 50 μg whole-cell extracts of UT-7 cell clones (lane 1, UT-7/P; lane 2, UT-7/E8-1; lane 3, UT-7/E8-2; lane 4, UT-7/9). (B) Levels of Brca1 and BCR-ABL protein were analyzed by immunoblotting 50 μg whole-cell extracts of murine Ba/F3 clones (lane 1, Ba/F3; lane 2, Ba/F3 210). (C) Levels of Brca1 and BCR-ABL protein were analyzed by immunoblotting 50 μg whole-cell extracts of Tetoff conditional cell line (lane 1, cells exposed to doxycycline; lane 2, no exposure of cells to doxycycline).
Western blot analysis showing BRCA1 and BCR-ABL expression in human and murine clones expressing variable levels of BCR-ABL. (A) Levels of BRCA1 and BCR-ABL protein were analyzed by immunoblotting 50 μg whole-cell extracts of UT-7 cell clones (lane 1, UT-7/P; lane 2, UT-7/E8-1; lane 3, UT-7/E8-2; lane 4, UT-7/9). (B) Levels of Brca1 and BCR-ABL protein were analyzed by immunoblotting 50 μg whole-cell extracts of murine Ba/F3 clones (lane 1, Ba/F3; lane 2, Ba/F3 210). (C) Levels of Brca1 and BCR-ABL protein were analyzed by immunoblotting 50 μg whole-cell extracts of Tetoff conditional cell line (lane 1, cells exposed to doxycycline; lane 2, no exposure of cells to doxycycline).
Down-regulation of BRCA1 is observed in primary CML samples
As an attempt to assess whether the down-regulation of BRCA1 associated with p210 expression in cultured cells could be extended to the actual pathologic condition of Ph1+ cells in patients with CML, the level of BRCA1 protein was examined in primary CML samples from patients at different stages of the disease. For this purpose, light-density peripheral blood mononucleated cells were purified by Ficoll-Hypaque gradients. One patient was at first in chronic phase of the disease, whereas the other 4 patients had been previously treated and were at the accelerated (n = 2) or acute (n = 2) phases. As shown in Figure 2, a decrease in BRCA1 level is observed in all patient cells displaying detectable levels of p210 protein, as compared with control cells (mobilized peripheral blood cells). This effect appears irrespective of the disease stage.
Western blot analysis showing BRCA1 and BCR-ABL expression in primary CML samples and in mobilized peripheral blood (PB) from autografts. Whole-cell extracts were immunoblotted with anti-BRCA1, anti–c-ABL, and anti–β-actin (lanes 1-3, PB from autografts; lanes 4-5, CML blast phase; lanes 6-7, CML accelerated phase; lane 8, CML chronic phase).
Western blot analysis showing BRCA1 and BCR-ABL expression in primary CML samples and in mobilized peripheral blood (PB) from autografts. Whole-cell extracts were immunoblotted with anti-BRCA1, anti–c-ABL, and anti–β-actin (lanes 1-3, PB from autografts; lanes 4-5, CML blast phase; lanes 6-7, CML accelerated phase; lane 8, CML chronic phase).
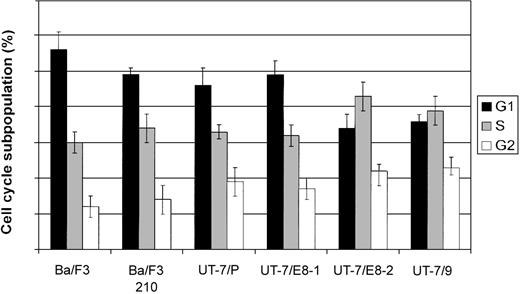
Level of BRCA1 protein does not reflect differences in cell cycle distribution
The level of BRCA1 protein is regulated throughout the cell cycle. It peaks in S and G2 phases with a localization in nuclear foci and decreases thereafter to reach a minimum in G1 phase in which it is diffusely dispersed in the nucleoplasm. The differential localization and phosphorylation of the protein throughout the cell cycle suggest that BRCA1 exerts its functions at specific stages of the cell cycle.23,24 The low abundance of BRCA1 in cells expressing high levels of p210 could indeed reflect an alteration of the cell cycle leading to an accumulation of cells in G1 phase. This situation does not seem to be the case because cell cycle distribution (Figure 3) and doubling time of BA/F3 and UT-7 clones appear to be minimally affected by the expression of p210 (data not shown). This result is somewhat unexpected, considering that the lack of BRCA1 is presumed to cause growth arrest both in vitro and in vivo.25
Cell cycle analysis of UT-7 cell clones expressing different levels of BCR-ABL. Exponentially growing cells were washed in PBS and stained with propidium iodide and submitted to fluorescence-activated cell sorter (FACS) analysis. Results shown are the averages of 3 independent experiments; error bars show 5% confidence intervals.
Cell cycle analysis of UT-7 cell clones expressing different levels of BCR-ABL. Exponentially growing cells were washed in PBS and stained with propidium iodide and submitted to fluorescence-activated cell sorter (FACS) analysis. Results shown are the averages of 3 independent experiments; error bars show 5% confidence intervals.
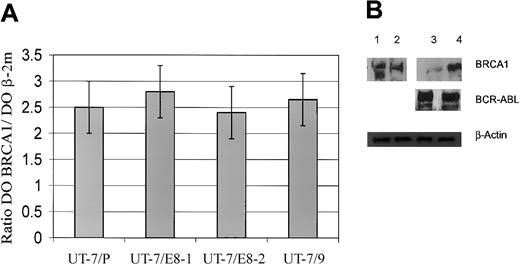
Down-regulation of BRCA1 is determined after transcription
To elucidate the molecular mechanism underlying the depletion of BRCA1 protein, the levels of BRCA1 mRNA accumulated in the parental and the p210 protein-expressing UT-7 cells (Figure 1A-B) were measured by quantitative PCR. As shown in Figure 4A, the levels of BRCA1 mRNA normalized to β2 microglobulin are not significantly different between the various cell lines and do not reflect the differences observed in protein levels. This finding leads to the conclusion that the down-regulation of BRCA1 protein must be determined after transcription, thus leaving 2 major mechanisms to account for the phenomenon: either p210 interferes with the translation of the BRCA1 mRNA (initiation or elongation) or it mediates a degradation process of the protein.
Expression of BRCA1 mRNA and Western blot analysis. (A) Relative expression of BRCA1 mRNA in UT-7 cell clones expressing different levels of BCR-ABL. Mean levels of expression of the BRCA1 gene, normalized to β-2 microglobulin gene, are displayed logarithmically on the y-axis. For clarity, only positive error bars (SD) are shown. For all categories, n = 3. (B) Western blot analysis showing the effects of proteasome inhibitor in UT-7 cell clones. Parental UT-7/P and BCR-ABL–expressing UT-7/9 cells were grown in recombinant human granulocyte macrophage colony-stimulating factor (rhGM-CSF) and exposed to proteasome inhibitor lactacystin 5 μM diluted in DMSO; similar volume of DMSO was added for 8 hours as a control. Whole-cell extracts were subjected to Western blot analysis using anti-BRCA1, anti–c-ABL (lane 1, UT-7/P + DMSO; lane 2, UT-7/P + lactacystin; lane 3, UT-7/9 + DMSO; lane 4, UT-7/9 + lactacystin).
Expression of BRCA1 mRNA and Western blot analysis. (A) Relative expression of BRCA1 mRNA in UT-7 cell clones expressing different levels of BCR-ABL. Mean levels of expression of the BRCA1 gene, normalized to β-2 microglobulin gene, are displayed logarithmically on the y-axis. For clarity, only positive error bars (SD) are shown. For all categories, n = 3. (B) Western blot analysis showing the effects of proteasome inhibitor in UT-7 cell clones. Parental UT-7/P and BCR-ABL–expressing UT-7/9 cells were grown in recombinant human granulocyte macrophage colony-stimulating factor (rhGM-CSF) and exposed to proteasome inhibitor lactacystin 5 μM diluted in DMSO; similar volume of DMSO was added for 8 hours as a control. Whole-cell extracts were subjected to Western blot analysis using anti-BRCA1, anti–c-ABL (lane 1, UT-7/P + DMSO; lane 2, UT-7/P + lactacystin; lane 3, UT-7/9 + DMSO; lane 4, UT-7/9 + lactacystin).
Ubiquitin-proteasome–mediated degradation has been hypothesized to account for the reduced level of DNA-PKCs9 as well as the low level of the cycline-dependent kinase inhibitor p2726 observed in p210-expressing cells. This hypothesis led us to explore the possibility that exposure to the proteasome inhibitor lactacystin will affect the accumulation of BRCA1 in the UT-7/9 clone expressing the highest level of p210. As shown on Figure 4B, incubation with lactacystin for 7 hours at a concentration of 5 μM leads to a marked increase of BRCA1 level (lane 4) compared with untreated cells (lane 3) without significant changes in the p210 level.
It should be noted that in these conditions lactacystin treatment for longer exposure results in a significant reduction of p210 accumulation, an effect previously reported in K-562 cells.27 This information provides an independent confirmation of the mirror image correlation between the respective levels of accumulation of BRCA1 and P210 proteins. However, it prevents from drawing a firm conclusion as to the role played by the proteasome in the status of BRCA1.
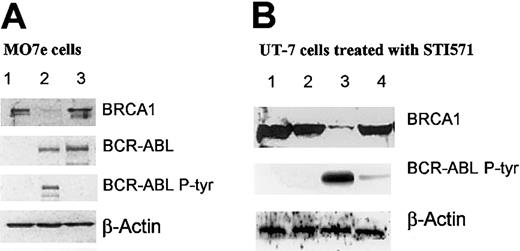
Tyrosine kinase activity of p210 mediates the effect on BRCA1 level
Two strategies were developed in an attempt to link the reduced level of BRCA1 to the tyrosine kinase activity of p210. The data presented in Figure 5A show that the depletion of BRCA1 observed in MO7e cells expressing the kinase-active p210 (lane 2) is not observed in cells expressing the kinase-defective mutant K1172R28 (lane 3). The kinase deficiency of the mutant is demonstrated by its impaired capacity to autophosphorylate on tyrosine as shown on Figure 5A by probing with antiphosphotyrosine antibody (lanes 3). It is noteworthy that wild-type and mutant proteins are expressed at the same level (lanes 3 and 2). This result strongly suggests that the tyrosine kinase activity of p210 plays a role in the down-regulation of BRCA1. This conclusion is supported by the observation that exposure of UT-7/9 cells expressing the wild-type p210 to the tyrosine kinase inhibitor STI571 does indeed restore the accumulation of BRCA1 to levels close to that observed in control UT-7/P cells (Figure 5B lane 4). This effect is observed after 24 hours of exposure to 5 μM STI571, a condition in which the autophosphorylation of p210 is strongly repressed. We have noted that exposure to lower concentration of the drug (1 μM) for shorter time (7-8 hours) does not affect the level of BRCA1, although the kinase activity is severely inhibited.
Western blot analyses. (A) Western blot analysis showing BRCA1 and BCR-ABL expression in different MO7e cells. Cell extracts (50 μg) were subjected to Western blot analysis using anti-BRCA1, anti–c-ABL, and antiphosphotyrosine (lane 1, MO7e MIGR EGFP; lane 2, MO7e MIGR BCR-ABL-IRES-eGFP; lane 3, MO7e MIGR BCR-ABL/1172-IRES-eGFP). (B) Western blot analysis showing the effects of STI571 (C-ABL tyrosine kinase inhibitor) in UT-7 cell clones. Parental UT-7/P and BCR-ABL–expressing UT-7/9 cells were grown in rhGM-CSF and exposed to STI571 5 μM for 24 hours. Whole-cell extracts were subjected to Western blot analysis using anti-BRCA1, anti–c-ABL, antiphosphotyrosine, and anti–β-actin (lane 1, UT-7/P control; lane 2, UT-7/P + STI571; lane 3, UT-7/9 control; lane 4, UT-7/9 + STI571).
Western blot analyses. (A) Western blot analysis showing BRCA1 and BCR-ABL expression in different MO7e cells. Cell extracts (50 μg) were subjected to Western blot analysis using anti-BRCA1, anti–c-ABL, and antiphosphotyrosine (lane 1, MO7e MIGR EGFP; lane 2, MO7e MIGR BCR-ABL-IRES-eGFP; lane 3, MO7e MIGR BCR-ABL/1172-IRES-eGFP). (B) Western blot analysis showing the effects of STI571 (C-ABL tyrosine kinase inhibitor) in UT-7 cell clones. Parental UT-7/P and BCR-ABL–expressing UT-7/9 cells were grown in rhGM-CSF and exposed to STI571 5 μM for 24 hours. Whole-cell extracts were subjected to Western blot analysis using anti-BRCA1, anti–c-ABL, antiphosphotyrosine, and anti–β-actin (lane 1, UT-7/P control; lane 2, UT-7/P + STI571; lane 3, UT-7/9 control; lane 4, UT-7/9 + STI571).
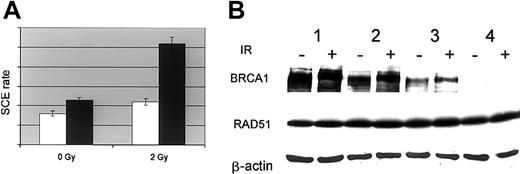
Increased frequency of sister chromatid exchange in BRCA1-deficient cells
Multiple evidences supporting the concept that BRCA1 plays a role in homologous recombination29 have prompted us to evaluate the effect of the down-regulation of BRCA1 on the frequency of sister chromatid exchange induced by exposure to ionizing radiation. As shown in Figure 6A, parental and p210-expressing cells display very similar constitutive frequency of SCE. However, exposure to 2 Gy more than doubles this frequency in p210-expressing cells and affects modestly the SCE in parental cells. These observations are in agreement with the report of an increased number of SCEs in patients with CML at blast crisis.30
SCE and BRCA1 and RAD51 accumulation. (A) Sister chromatid exchange rate in parental (UT-7/P, □) and BCR-ABL–expressing cells (UT-7/9, ▪). Cells were pulsed with 5′Brdu for 24 hours and dropped on slides after hypotonic shock. SCE was revealed by Hoechst 33528 staining. The SCE rate was expressed as the average of the number of exchanged chromatid adjusted to the individual cell ploidy; error bars show 5% confidence intervals. (B) BRCA1 and RAD51 accumulation in UT-7 cell clones expressing variable levels of BCR-ABL. UT-7 clones expressing different levels of BCR-ABL were irradiated (6 Gy; +) or not (–). Six hours after irradiation 50 μg whole-cell extracts were subjected to Western blot analysis using anti-BRCA1, anti-RAD51, and anti–β-actin (lane 1, UT-7/P; lane 2, UT-7/E8-1; lane 3, UT-7/E8-2; lane 4, UT-7/9).
SCE and BRCA1 and RAD51 accumulation. (A) Sister chromatid exchange rate in parental (UT-7/P, □) and BCR-ABL–expressing cells (UT-7/9, ▪). Cells were pulsed with 5′Brdu for 24 hours and dropped on slides after hypotonic shock. SCE was revealed by Hoechst 33528 staining. The SCE rate was expressed as the average of the number of exchanged chromatid adjusted to the individual cell ploidy; error bars show 5% confidence intervals. (B) BRCA1 and RAD51 accumulation in UT-7 cell clones expressing variable levels of BCR-ABL. UT-7 clones expressing different levels of BCR-ABL were irradiated (6 Gy; +) or not (–). Six hours after irradiation 50 μg whole-cell extracts were subjected to Western blot analysis using anti-BRCA1, anti-RAD51, and anti–β-actin (lane 1, UT-7/P; lane 2, UT-7/E8-1; lane 3, UT-7/E8-2; lane 4, UT-7/9).
It is tempting to correlate this high SCE frequency to the lack of BRCA1 leading to a defect in the proper repair of strand breaks by homology-directed repair as demonstrated in murine cells. An alternative explanation would attribute this effect to a putative up-regulation of RAD51. p210 has recently been reported to up-regulate the level of RAD51,10 resulting in an increased recombination repair activity. The examination of the accumulation of RAD51 in response to p210 expression in the UT-7 clones presented in Figure 6B shows no difference in the constitutive or the radiation-induced RAD51 accumulation between these cells.
The appropriate cell response to radiation is attested by the hyperphosphorylation of BRCA1 leading to an electrophoretic mobility shift.
DNA repair deregulation in BRCA1-deficient cells
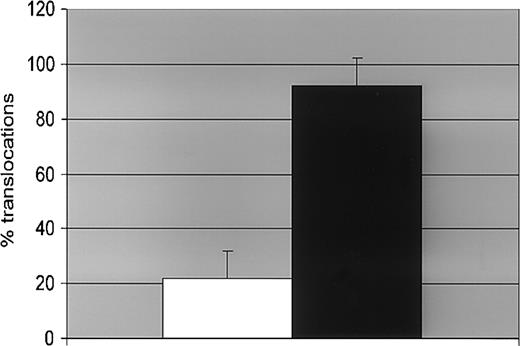
DNA damage repair capacity of UT-7/P cells (parental clone) and UT-7/9 cells (clone with high level of p210) was assessed by counting the chromosomal damages induced by ionizing radiation revealed by fluorescence in situ hybridization (FISH). Chromosome 1 was used as a standard, because of its size it is the most sensitive target for rearrangements. The data presented in Figure 7 illustrate deficiency in DNArepair induced by p210 in the UT-7/9 clone exposed to radiation by comparison with the control cell line UT-7/P. The respective frequencies of the major chromosomal aberrations observed in cells exposed to radiation illustrate quantitatively the deregulated DNA repair in p210-expressing cells (Figure 7). It is noteworthy that cells carrying mutant alleles of BRCA1 have been reported to display a high rate of rearrangement.
FISH analysis. Effect of BCR-ABL expression on DNA repair evaluated by FISH. Human parental UT-7/P (□) and BCR-ABL–expressing UT-7/9 (▪) cells were irradiated with 4 Gy and collected 24 hours later. A total of 200 metaphases were counted for each point. The figures represent the percentage of cells with chromosome 1 translocation.
FISH analysis. Effect of BCR-ABL expression on DNA repair evaluated by FISH. Human parental UT-7/P (□) and BCR-ABL–expressing UT-7/9 (▪) cells were irradiated with 4 Gy and collected 24 hours later. A total of 200 metaphases were counted for each point. The figures represent the percentage of cells with chromosome 1 translocation.
Discussion
Here, we report that the expression of p210 BCR-ABL fusion protein correlates with severe down-regulation of BRCA1 protein in human or murine hematopoietic cells. This depletion occurs both in cultured cell lines and in fresh leukemia cells from patients with CML and relies on the tyrosine kinase activity of p210.
The lack of significant change in BRCA1 mRNA level in cells expressing p210 supports the hypothesis that the regulation of BRCA1 protein level occurs after transcription. This regulation can be achieved at 2 levels: efficiency of translation of the BRCA1 mRNA or stability of the protein. There is evidence that p210 suppresses expression of C/EBPα through the induction of the RNA binding heterogeneous nuclear RNA-protein (hnRNP) E2 that inhibits the translation of C/EBPα mRNA by binding to the loop-forming 5′ untranslated open reading frame (5′uORF).31 Interestingly, BRCA1 is indeed translationally regulated by stable structures present in 5′uORF that can be potential targets for hnRNP E2 induced by p210.32 This hypothesis deserves experimental support. We have raised the possibility that BRCA1 protein level may be regulated by proteasome-mediated degradation.
Unfortunately, the experiments involving lactacystin exposure of the cells reported here do not allow us to draw firm conclusions as to the role played by the proteasome in the status of BRCA1 because long-term exposure (24 hours) to lactacystin induces a decrease in the accumulation of p210 itself. It is noteworthy that in breast tumor cells lactacystin does not protect BRCA1 against proteolytic degradation, whereas another inhibitor of the proteasome N-acetylleucylleucylnorleucinal (ALLN) is effective.33
Cells undergoing the p210-induced down-regulation of BRCA1 display an increased sensitivity to ionizing radiation in terms of clonogenic survival associated with an increased rate of SCE and chromosome aberrations.
The limited level of BRCA1 together with the down-regulation of other genes involved in DNA repair (DNA-PKCs, DNA polymerase β, XPB) might contribute to the genetic instability that eventually leads to blast crisis and, therefore, plays a role in the pathogenesis of CML. Major phenotypic manifestations that relate to the role of BRCA1 in cell survival and in maintenance of genetic stability could occur in cells in which it is present in limited amounts. The essential role of BRCA1 in cell survival was initially revealed by the lethality of Brca1null/null embryos that die early in development because of growth arrest believed to result from the activation of a p53-driven replication checkpoint that prevents the accumulation of damaged DNA.25,34 The limited success in establishment of BRCA1null/null cell lines in vitro from tumor tissues has been interpreted as a support for such a role. In this context, it is remarkable that the limited level of BRCA1 does not seem to affect adversely the growth capacities of the UT-7 or Ba/F3 cell lines expressing p210. This may reflect the capacity of p210-induced signaling to circumvent functional BRCA1 deficiency in these hematopoietic cells by avoiding a checkpoint control that is activated in cells lacking BRCA1 such as the p53-driven replication checkpoint.
Two reports have indicated a possible implication of p210 in the DNA repair processes. On the one hand, we have reported a down-regulation of DNA-PKC expression concomitant with increasing p210 protein levels.9 This effect was associated with major DNA repair deficiency on exposure to DNA-damaging agents. A down-regulation of DNA-PKCs was also observed in CD34+ cells from patients with CML at diagnosis, suggesting that this phenomenon constitutes an important feature of this disease. It is thus possible that the down-regulation of DNA-PKCs by p210 plays a role in determining one of the main characteristics of CML, which is the chromosome and genetic instability leading to the blast crisis. On the other hand, it has been recently reported that p210 controls the transcription, degradation, and phosphorylation of RAD51 a major component of the homologous recombination repair machinery.10 This regulation results in increased recombination repair and may account for the resistance of leukemic cells to cisplatin and mitomycin C. Our results do not confirm these data and rule out a modification of the RAD51 status as a cause of the increased SCE frequency in the cells studied here.
BRCA1 operates in the maintenance of genetic stability through its role both in DNA repair35 and centrosome duplication.36 Indeed, the BRCA1null/null cell line HCC-193720 carries multiple genetic alterations (aneuploidy, p53, phosphotase and tensin homolog deleted on chromosome 10 [PTEN] tumor suppressor inactivation) and displays deficiencies for repair of both double-stranded breaks37 or oxidative lesions.18
It would be tempting to speculate that limiting the level of BRCA1 resulting from p210 expression could indeed lead to an overall genetic instability manifested by a significant increase in SCE and chromosomal damages induced by ionizing radiation.
Radiation sensitivity of p210-expressing cells is still a matter of controversy. Increased radiation sensitivity in p210-expressing murine cells has been reported.38 The Ph1+ leukemic cell line, K-562, is resistant to DNA-damaging agents, including gamma-irradiation.39 Although the results reported in this work do not solve these discrepancies, they might have other implications. Indeed, the fact that the CML cells are both deficient in DNA repair and refractory to apoptosis may explain that chromosomal and genetic abnormalities can accumulate in leukemic cells and why this myeloproliferative disorder constantly progresses to a blast crisis.
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2002-10-3011.
Supported by a fellowship grant (S.J.) and a research grant (J.F.) from the Association de Recherche contre le Cancer (ARC); from fondation pour la recherche médicale (S.J.); from Ligue nationale contre le cancer (Ligue du Cher) et Ligue contre le cancer, section des Hauts de Seine; and from the Fondation de France (J.B.), from the ARC and comité de recherche clinique (CRC) of the Institut Gustave Roussy (A.G.T.).
E.D. and S.J. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Nazanine Modtjahedi for stimulating discussions and Lionel Tintignac and Serge Leibovitch for helpful advices. We thank Dr R. Van Etten and Dr W. Pear for the gift of the p210 K1172R mutant and MIGR-BCR-ABL constructs, respectively. The generous gift of STI571 by Novartis for in vitro experiments (Dr E. Buchdunger) is also gratefully acknowledged.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal