Abstract
Rearrangement of T-cell receptor (TCR) and immunoglobulin genes by a common V(D)J recombination machinery is regulated by developmentally specific chromatin changes at the target locus, a process associated with transcription. At the TCRβ locus, the Eβ enhancer and the Dβ1 promoter regulate germline transcription originating near the TCR Dβ1 gene segment. The Dβ1 promoter contains 3 GC-rich motifs that bind a common set of nuclear proteins from pro–T-cell lines. Mutations that diminish the binding of nuclear proteins also diminish the activity of the Dβ1 promoter in transcriptional reporter assays. Using a yeast one-hybrid approach, 3 Krüppel-like factors—KLF3, KLF5, and KLF6—and a novel zinc finger protein were identified in a thymus library, all of which bound the GC-rich motif in a sequence-specific manner. Of these genes, KLF5 mRNA was expressed in a restricted manner in lymphoid cells and tissues, with highest expression in pro–T-cell lines and Rag-deficient thymocytes. Antibody supershift studies and chromatin immunoprecipitation assay confirmed that KLF5 bound the Dβ1 promoter. In reporter gene assays, KLF5 but not KLF6 efficiently transactivated the Dβ1 promoter, whereas a dominant-negative KLF5 construct inhibited reporter expression. These data suggest that reiterated GC motifs contribute to germline TCRβ transcription through binding of KLF5 and other Krüppel family members and that restricted expression of KLF5 may contribute to lineage-specific regulation of germline TCRβ transcription.
Introduction
The adaptive immune system in mammals requires a vast repertoire of individual specificities in antigen receptor genes, far beyond what could be directly encoded in the genome. Diversity is created by the assembly of variable region gene segments in immunoglobulin and T-cell–receptor genes by V(D)J recombination.1,2 This process is now understood to involve the coupled, site-specific cleavage of DNA at conserved recombination signal sequences flanking the borders of the V, D, and J elements by RAG-1 and RAG-2 proteins. Processing and rejoining of signal and coding ends occurs by the nonhomologous end-joining DNA repair pathway that is expressed in all cells.3, 4, 5, 6 Antigen receptor genes are rearranged in a highly ordered fashion with respect to lineage and developmental stage through locus-specific processes that modify the structure of chromatin to provide “accessibility” to, or otherwise promote assembly of, the recombinase complex.7, 8, 9, 10 This modification of chromatin is accompanied by transcription of the unrearranged (germline) locus. Regulation of germline transcription by cis-acting enhancer and promoter elements is closely linked with the regulation of recombination.11, 12, 13, 14, 15, 16 Numerous studies now demonstrate the importance of these cis regulatory elements in controlling the rearrangement of antigen receptor loci, whether as chromosomally integrated substrates or transgenic mouse lines or by mutation of such elements in endogenous loci using gene targeting in mice.9,17, 18, 19, 20, 21, 22, 23
Rearrangement of the TCR β-chain locus is among the earliest steps in the development of αβ T cells and is required for generation of this lineage. As with other antigen receptor genes, rearrangement is preceded by transcription of the locus and includes transcripts from individual V genes, along with transcripts that encompass each of the duplicated DJC complexes.8,24,25 Germline transcription of the DJC portion of the locus is controlled by its enhancer/locus control region element, Eβ, and occurs at high levels at the pro–T-cell stage 2.4,26 Gene-targeting studies demonstrate that Eβ is required for the rearrangement of TCRβ genes and the development of T cells, and, in the absence of Eβ, germline transcription occurs at a substantially reduced level.10,17,18 Germline transcripts are initiated separately near the Dβ1 and Dβ2 gene segments, near regions with high sequence conservation between human and mouse.27,28 The Dβ1 promoter has been characterized, extending approximately 350 bp upstream of Dβ1. This promoter directs the efficient transcription of reporter genes in pro–T-cell lines in the context of the Eβ enhancer and in more mature T- and B-cell lines in the context of the Eμ enhancer.24,25 Dβ1 promoter function is required for efficient rearrangement involving this element.22,23
Through knockout, overexpression, and ectopic expression studies, members of several classes of transcription factors have been identified that regulate T-cell development. These include basic HLH family members such as E2A, HEB, Id3, and HES-129,30 ; zinc finger proteins such as Ikaros family members and GATA-329, 30, 31 ; Wnt pathway transcription factors TCF-1 and LEF-129,31,32 ; and Notch family members. Although several targets of these transcription factors have been identified among T-cell–specific genes, relatively little is known regarding their specific functions in T-cell development.33 Because germline transcription is a key step in the early development of T cells, we sought to identify factors that control this process. We report here that reiterated GC-rich motifs regulate Dβ1 transcription through interaction with Krüppel family transcription factors. Among these, KLF534 (also named IKLF, BTEB2) exhibits restricted lymphoid expression in early T cells and transactivates the Dβ1 promoter.
Materials and methods
Electrophoretic mobility shift assays
Single-stranded oligonucleotides were end-labeled with [γ-32P] adenosine triphosphate (ATP) using standard protocols and purified with Centri-sep columns (Princeton Separations, Adelphia, NJ) before duplexing equal molar amounts of each oligonucleotide and its complement in 10 mM Tris pH 7.4, 1 mM EDTA (ethylenediaminetetraacetic acid), 25 mM KCl by boiling for 5 minutes and slowly cooling to room temperature. Nuclear extracts35 containing 20 μg protein were incubated in 20 mM HEPES (N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid) pH 7.9, 1 mM EDTA, 5 mM dithiothreitol (DTT), 5% glycerol, 1% IGEPAL, with 10 μg bovine serum albumin (BSA) and 3 μg double-stranded poly dI-dC before adding labeled, double-stranded oligonucleotides as indicated. After incubating the samples for 20 minutes at room temperature, 3 μL DNA loading buffer was added, and the samples were run on a 6% nondenaturing 0.5 × TBE polyacrylamide gel at 120 V for 3.75 hours. Gels were dried and exposed to film or a phosphorimaging screen at room temperature. Cold competition electrophoretic mobility shift assays (EMSAs) were performed by preincubating the nuclear extracts in 100-fold molar excess of unlabeled probes for 15 minutes before adding the labeled probe, and they were incubated for 20 minutes as described. Supershift assays were performed as described by Martino et al36 using anti-Sp1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-KLF5 antibody37 (generous gift from Dr Christina Teng, National Institute of Environmental Health Sciences, NIH, Research Triangle Park, NC), with goat immunoglobulin G (IgG) as control. Oligonucleotides used in competition experiments were as follows (mutations are shown in italics): p35, GGTAGACCTATGGGAGGGTCCTTTTTTG; p35m, GGTAGACCTATGTCAGGGTCCTTTTTTG; p281, GAGAAGGGCGGATACAAGAGGGAATCCAG; p281m, GAGAAGTCCGGATACAAGAGGGAATCCAG; p314, TCATAGGGTGGTTCCCTTATATGACAAAAATTTGAGAAGG; p314m, TCATAGTCTGGTTCCCTTATATGACAAAAATTTGAGAAGG; BS4, TCAGCTTTTGGGAATGTATTCCCTGTCA.
Cross-linking studies
EMSAs were performed as above, except that the gels were exposed to a short-wave transilluminator 10 cm away for 0 to 60 minutes before exposure to film without drying. Individual complexes were localized by alignment with the autoradiogram, excised from the gel, and extracted by crushing in 100 mM Tris pH 7.4, 10 mM EDTA, and 0.5% sodium dodecyl sulfate (SDS) with incubation for 15 minutes at room temperature. Extracted complexes were diluted 2-fold with 2 × reducing sample buffer (2.5 mM Tris pH 6.8, 10% glycerol, 3.2% SDS, 0.1% bromophenol blue, 0.7 M 2-mercaptoethanol) and were boiled for 5 minutes before electrophoresis on a 10% polyacrylamide gel and autoradiography.
Promoter constructs and transcription reporter assays
The p5424 and p4980 pro–T-cell lines derived from thymic lymphomas of RAG-deficient mice have been described.24 EL-4, A20, and WEHI 231 were from ATCC (Manassas, VA) and were cultured as described.24 Promoter and enhancer constructs were cloned into the HindIII and BamHI sites, respectively, of the pGL3 Basic vector from Promega (Madison, WI). Promoter fragments, numbered relative to the first base coding Dβ1, have been described previously.24 Site-directed mutagenesis was performed using the U.S.E. mutagenesis kit (Pharmacia) using the manufacturer's instructions and was confirmed by DNA sequencing. Mutations of the -35 and -74 sites have been described.24 The -255 putative Ikaros site, AGAGGGAATCC, was changed to AGAGATAATCC; the -276 GATA motif, GGCGGATACAA, was changed to GGCGTCCGCAA; the -281 GC-rich motif, GAAGGGCGGAT, was changed to GAAGTCCGGAT; and the -314 GC-rich motif, ATAGGGTGGTT, was changed to ATAGTCTGGTT. Luciferase reporter assays were performed as before, using cotransfection of the pRL vector to control for transfection efficiency.24 The Dual-Luciferase Reporter System (Promega) was used to assay firefly and Renilla luciferase activity in each sample.
Yeast one-hybrid assays
The mouse Dβ1 promoter (-317 to +6, PDβ1) amplified by PCR and 3 copies of a 26-bp synthesized -35 GC-rich motif (-35 × 3) were cloned into the pHisi-1 and pLacZi vectors (BD Biosciences Clontech, Palo Alto, CA), respectively. Constructs were linearized and stably integrated into the YM4271 yeast genome to generate PDβ1 and -35 × 3 yeast strains containing His3 and LacZ reporters according to the manufacturer's instructions. Reporter strains were transformed using a commercial pACT2 human thymus cDNA library fused to the yeast Gal-4 activation domain (BD Biosciences Clontech) and were selected in Leu-His- media in the presence of 50 mM 3-AT (3-amino-1,2,4-triazole), the concentration that suppresses leaky His3 expression in our yeast strains. Positive colonies were picked and restreaked on His-/3-AT plates, and repeatedly positive clones were screened for β-galactosidase activity using a colony lift assay as described in the user's manual. Positive candidates were rescued using a freeze/thaw-based method and sequenced. The -35 × 3 wild-type and -35 × 3 mutant strains were generated by using pHisi-1 constructs containing 3 copies of the wild-type or mutant form of the -35 motif and were used to test the ability of candidates binding the -35 GC-rich motif.
Reverse transcription–polymerase chain reaction
Total RNAs were extracted from cultured cells or primary tissues as previously described38 and were reverse transcribed using Superscript II reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA) with oligo(dT)15 primer. Reverse transcription (RT) products were serially diluted (1:5:25:125) and amplified using gene-specific primers. The oligonucleotide primer sequence was as follows: Klf3 sense, 5′-AGAGCCTGGAATTGAACCACAG-3′, antisense, 5′-AGCTGGAGACAGGTTTTCAGAC-3′; Klf5 sense, 5′-GCTCAGAGCCTGGAAGTCCC-3′, antisense, 5′-CGTTCGCTCGCTCAGTTCTG-3′; Klf6 sense, 5′-TCTTTTCCAACCCGACATGG-3′, antisense, 5′-CATTAAAGTGGCACCGATG-3′; β-actin sense, 5′-CCTAAGGCCCAACCGTGAAAAG-3′, antisense, 5′-TCTTCATGGTGCTAGGAGCCA-3′. Polymerase chain reaction (PCR) products were electrophoresed on 1.1% agarose gels and visualized using ethidium bromide.
Chromatin immunoprecipitation assays
Both p5424 and A20 cells were fixed in 1% formaldehyde and sonicated. Cross-linked chromatin samples were subjected to immunoprecipitation using a chromatin immunoprecipitation assay kit (Upstate Biotechnology, Lake Placid, NY) following the manufacturer's instructions. Antibodies used included anti-KLF5, anti-Sp1, and goat IgG (as described above). Immunoprecipitated DNA samples were reverse–cross-linked and subjected to nested PCR analysis. PCR primers for amplifying the Dβ1 promoter were: exterior forward, 5′-AGCTAGGCTAGATTGGGGGCTGTTACTTCTTCATAGGGTG-3′; exterior reverse, 5′-TGTCCCCACAATGTTACAGC-3′; interior forward, 5′-GAGAAGGGCGGATACAAGAGGGAATCCAG-3′; interior reverse, 5′-GGTCTACCCCTTGAGGGCTGGAAGAAACCA-3′. IgH DQ52 promoter control primers were: exterior forward, 5′-TCCCCAGGCCATAGTCACC-3′; exterior reverse, 5′-TGACCTCCAGAAAGCTACCTTTCC-3′; interior forward, 5′-ACCAGAGCTCTGGCAGTGACCAC-3′; interior reverse, 5′-AGCACTGTGGTGCTCCGC-3′.
Results
Nuclear factors binding the Dβ1 promoter
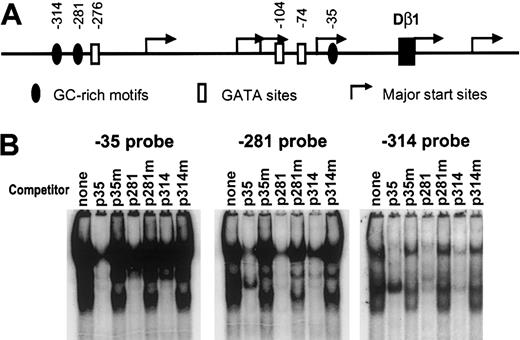
To identify nuclear factors that may regulate TCRβ germline transcription, we performed promoter imaging using EMSA with overlapping oligonucleotide probes spanning the -350 to +150 positions of the Dβ1 promoter. DNA mobility shift patterns were visualized for nuclear extracts derived from the p5424 pro–T-cell line, which exhibits high levels of endogenous germline TCRβ transcripts and has the highest activity of lymphoid lines tested for expression of reporter genes driven by Eβ and Dβ1 promoter.24 These were compared with gel-shift patterns using nuclear extracts from A20 B cells, which transcribe Eβ/Dβ1 promoter reporters poorly (Figure 1). Although a number of DNA-protein complexes were detected in both cell lines, 3 oligonucleotide probes—p35, p281, and p314—exhibited patterns that were distinct in p5424 nuclear extracts. Each of these probes contained a GC-rich region (Table 1), suggesting that they may bind an overlapping set of proteins. To test this, we performed cross-competition experiments with each of the 3 oligonucleotide probes. In each case, the labeled probe was competed by unlabeled versions of all 3 probes (Figure 2). To further investigate the specificity of the binding we made mutations, GG→TC, in the center of each of the GC-rich regions in the 3 probes and tested self- and cross-competition. For all 3 probes, all binding activity was competed by the cold wild-type probes but not the mutant probes, indicating that the GC-rich motifs centered at -314, -281, and -35 in the Dβ1 promoter mediate binding under EMSA conditions to a common set of nuclear proteins.
Identification of Dβ1 promoter sequences binding nuclear proteins in T and B cells. Overlapping double-stranded oligonucleotide probes spanning -350 to +150 were labeled and incubated with either no extract, p5424, or A20 nuclear extracts, as indicated, and were analyzed by EMSA on nondenaturing polyacrylamide gels. Results from probes centered at -35, -281, and -314 relative to Dβ1 are presented.
Identification of Dβ1 promoter sequences binding nuclear proteins in T and B cells. Overlapping double-stranded oligonucleotide probes spanning -350 to +150 were labeled and incubated with either no extract, p5424, or A20 nuclear extracts, as indicated, and were analyzed by EMSA on nondenaturing polyacrylamide gels. Results from probes centered at -35, -281, and -314 relative to Dβ1 are presented.
Alignment and characterization of GC-rich motifs in the Dβ1 promoter
Site . | Sequences . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -35 | C | C | T | A | T | G | G | G | A | G | G | G | T | C | C | T | |||||||||||||||
| -281 | G | A | G | A | A | G | G | G | C | G | G | A | T | A | C | A | |||||||||||||||
| -314 | T | C | A | T | A | G | G | G | T | G | G | T | T | C | C | C | |||||||||||||||
| Consensus | - | - | - | A/T | A/T | G | G | G | N | G | G | N | T | N | C | - | |||||||||||||||
| Binding | - | - | - | C | A/c | a | G | G | t | t | T | C | - | - | - | - | |||||||||||||||
| Nonbinding | - | - | - | - | - | T/C | T | C | - | - | - | - | - | - | - | - | |||||||||||||||
Site . | Sequences . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -35 | C | C | T | A | T | G | G | G | A | G | G | G | T | C | C | T | |||||||||||||||
| -281 | G | A | G | A | A | G | G | G | C | G | G | A | T | A | C | A | |||||||||||||||
| -314 | T | C | A | T | A | G | G | G | T | G | G | T | T | C | C | C | |||||||||||||||
| Consensus | - | - | - | A/T | A/T | G | G | G | N | G | G | N | T | N | C | - | |||||||||||||||
| Binding | - | - | - | C | A/c | a | G | G | t | t | T | C | - | - | - | - | |||||||||||||||
| Nonbinding | - | - | - | - | - | T/C | T | C | - | - | - | - | - | - | - | - | |||||||||||||||
Bases in lowercase have a reduced ability to compete with wild-type sequences. — indicates not applicable.
Three Dβ1 promoter probes bind the similar set of nuclear proteins. (A) Diagram of TCR Dβ1 promoter. GC-rich and GATA motifs in the Dβ1 promoter and major transcription start sites (from Doty et al24 ) are shown. (B) EMSA using nuclear extracts from the p5424 cell line were performed using labeled probes representing the 3 GC-rich motifs with no competitor or with competitors representing sequences with wild-type or mutant GC-rich motifs for the -35 probe (p35 or p35m), the -281 probe (p281 or p281m), or the -314 probe (p314 or p314m).
Three Dβ1 promoter probes bind the similar set of nuclear proteins. (A) Diagram of TCR Dβ1 promoter. GC-rich and GATA motifs in the Dβ1 promoter and major transcription start sites (from Doty et al24 ) are shown. (B) EMSA using nuclear extracts from the p5424 cell line were performed using labeled probes representing the 3 GC-rich motifs with no competitor or with competitors representing sequences with wild-type or mutant GC-rich motifs for the -35 probe (p35 or p35m), the -281 probe (p281 or p281m), or the -314 probe (p314 or p314m).
Sequence specificity for proteins recognizing the GC-rich motif was further investigated using 1 or 2 base substitutions made throughout the length of the -35 probe (Figure 3; Table 1). These mutant oligonucleotides were used in cold competition assays with the wild-type -35 probe. A representative experiment containing mutations throughout the core motif is shown. A complete summary of these experiments, along with alignment of the 3 binding sites in the Dβ1 promoter, is shown in Table 1. These experiments define the boundaries of a GC-rich core region spanning 6 nucleotides, with a consensus GGGNGG as critical for nuclear protein binding. The Ikaros consensus motif GGGAA39 is similar to the defined GC-binding site, but it does not compete for complex binding (Figure 3).
Scanning mutagenesis of the –35 probe to characterize the binding site for nuclear proteins. EMSA assays using p5424 nuclear extracts and the -35 probe, with wild-type or indicated mutant competitors. The wild-type p35 sequence is indicated at the top of the figure, with the respective mutations indicated by the arrows. p35 and p35m are wild-type and mutant -35 probe, respectively; BS4 is a probe with a consensus Ikaros binding site.
Scanning mutagenesis of the –35 probe to characterize the binding site for nuclear proteins. EMSA assays using p5424 nuclear extracts and the -35 probe, with wild-type or indicated mutant competitors. The wild-type p35 sequence is indicated at the top of the figure, with the respective mutations indicated by the arrows. p35 and p35m are wild-type and mutant -35 probe, respectively; BS4 is a probe with a consensus Ikaros binding site.
To further characterize complexes formed with the -35 probe, we performed in-gel ultraviolet (UV) cross-linking following EMSA with p5424 extracts and analyzed individual bands using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 4). The rapidly migrating EMSA complex in p5424 cells (complex 1) yielded a single band with Mr of 90 kDa after cross-linking. Complex 2 showed a single band after cross-linking that migrated with Mr of 120 kDa, whereas complex 3 was composed of at least 4 distinct bands after cross-linking. These studies indicate that the 3 major complexes detected in this assay represent distinct sets of proteins, and they suggest that at least 6 proteins in p5424 extracts can interact with the probe in a manner that is susceptible to UV-induced cross-linking to DNA.
Characterization of –35 binding complexes using in-gel cross-linking. Following EMSA with p5424 nuclear extracts and labeled -35 probe, gel slices were exposed to UV radiation for 0, 15, 30, or 60 minutes, then exposed to film without drying. Individual complexes 1, 2, and 3 were then identified and extracted from the gel in denaturing/reducing loading buffer and were analyzed by SDS-PAGE. Approximate molecular weights are indicated.
Characterization of –35 binding complexes using in-gel cross-linking. Following EMSA with p5424 nuclear extracts and labeled -35 probe, gel slices were exposed to UV radiation for 0, 15, 30, or 60 minutes, then exposed to film without drying. Individual complexes 1, 2, and 3 were then identified and extracted from the gel in denaturing/reducing loading buffer and were analyzed by SDS-PAGE. Approximate molecular weights are indicated.
Role of GC-rich motifs in Dβ1 promoter activity
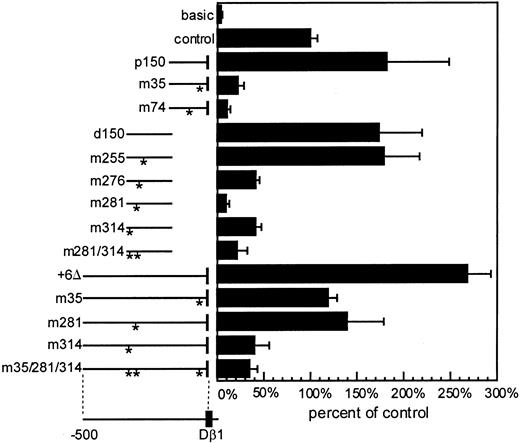
To test the functional relevance of the 3 GC-rich sites in promoter activity, we introduced the mutations that abrogated the binding of EMSA complexes into Dβ1 promoter reporter constructs and determined the effect on transcriptional activity. As previously shown,24 promoter subfragments comprising either the proximal (-163 to +6) or distal (-324 to -146) segments of the promoter were capable of directing transcription of a luciferase reporter gene in the context of Eβ (Figure 5). We confirmed our previous finding that mutation of the -35 GC-rich motif in the p150 promoter subfragment led to markedly reduced promoter activity.24 In addition, the mutation of core GC-rich motifs at the -281 or -314 positions, shown to abrogate binding of nuclear proteins, markedly reduced reporter expression. When these mutations were introduced into the full-length promoter fragment (approximately 500 bp), mutation of either the -35 or the -281 site reduced transcriptional activity by approximately half, consistent with partial redundancy of regulatory sites. Mutation of the -314 GC-rich site had a more severe effect, reducing the activity of the promoter by approximately 85%, which was similar to the combined effect of all 3 mutations. Mutations of an element similar to the GC motif (GGGAA) at position -255, which is a consensus-binding site for the Ikaros family of transcription factors, did not have any effect on transcription activity in the distal 150-bp promoter fragment (Figure 5). GATA consensus sequences at the -74 and -104 positions have previously been shown to affect promoter activity in reporter assays.24,25 In the current studies, we confirmed that mutation in the -74 GATA motif diminished reporter activity and demonstrated in addition that a third GATA motif at -276 also affected reporter activity. In additional experiments, the effect of mutations in the 3 GC-rich motifs was tested in the absence of the enhancer, with similar results, except that the overall activity was approximately 10-fold less (data not shown). These studies demonstrate that reiterated GC-rich motifs and GATA elements play an important role in activating transcription of the Dβ1 promoter.
Role of the GC-rich and GATA elements in Dβ1 promoter activity. Luciferase reporter assays in p5424 cells in vectors containing Eβ. Promoters included full-length Dβ1 (+6Δ) and proximal (p150) or distal (d150) promoter fragments in wild-type form, with individual GC-rich mutations (m35, m281, m314), GATA site mutations (m74 or m276), Ikaros motif mutation (m255), or combinations of the GC-rich sites mutated as indicated by asterisks. The measured luciferase assay was corrected for transfection efficiency based upon Renilla luciferase controls. Average and standard error of 4 to 8 independent transfections for each construct is presented as a percentage of the luciferase activity obtained with the SV40 promoter/enhancer control vector.
Role of the GC-rich and GATA elements in Dβ1 promoter activity. Luciferase reporter assays in p5424 cells in vectors containing Eβ. Promoters included full-length Dβ1 (+6Δ) and proximal (p150) or distal (d150) promoter fragments in wild-type form, with individual GC-rich mutations (m35, m281, m314), GATA site mutations (m74 or m276), Ikaros motif mutation (m255), or combinations of the GC-rich sites mutated as indicated by asterisks. The measured luciferase assay was corrected for transfection efficiency based upon Renilla luciferase controls. Average and standard error of 4 to 8 independent transfections for each construct is presented as a percentage of the luciferase activity obtained with the SV40 promoter/enhancer control vector.
Identification of protein binding to functional GC-rich motifs in the Dβ1 promoter
To identify DNA-binding proteins that may regulate germline Dβ1 transcription, we conducted a yeast one-hybrid screen using either the Dβ1 promoter or 3 copies of the -35 GC-rich motif (-35 × 3) as bait. Both segments were cloned into the pHisi-1 and pLacZi yeast vectors. For each bait, pHisi-1 and pLacZi reporter vectors were stably integrated into the YM4271 yeast strain to generate PDβ1 or -35 × 3 dual-reporter strains. The PDβ1 reporter strain was transformed using a human thymus cDNA library fused to the yeast Gal-4 activation domain and was selected in His- media in the presence of 3-AT. Positive colonies were picked and restreaked on His-/3-AT plates, and repeatedly positive clones were screened for β-galactosidase activity using a colony lift assay. Of 4.6 × 106 primary transformants, 27 were repeatedly positive on the screening assay. The clones were rescued and sequenced. Of these, 21 were deemed artifacts (ribosomal protein genes, other nontranscription factor genes, and nongene short inserts), and 6 independent clones were zinc-finger transcription factor genes. These included KLF family members Klf3,40 Klf5,34 and Klf641 and a novel zinc-finger factor gene (GenBank accession number AY117438) (Table 2). A screen of the same library using the -35 × 3 bait yielded a greater number of artifacts and one example of Klf6 (Table 2).
Transcription factor genes interacting with the Dβ1 promoter in the yeast one-hybrid assay
Bait . | No. clones . |
|---|---|
| PDβ1 | |
| KLF3/BKLF | 1 |
| KLF5/IKLF | 3 |
| KLF6/ZF9 | 1 |
| Novel ZF protein | 1 |
| -35 × 3 | |
| KLF6/ZF9 | 1 |
Bait . | No. clones . |
|---|---|
| PDβ1 | |
| KLF3/BKLF | 1 |
| KLF5/IKLF | 3 |
| KLF6/ZF9 | 1 |
| Novel ZF protein | 1 |
| -35 × 3 | |
| KLF6/ZF9 | 1 |
Because KLF family members bind to GC-rich elements, we surmised that the multiple KLF family members identified in the yeast screen may bind to the GC-rich elements in the Dβ1 promoter. To test this, a pHisi-1 yeast reporter strain was generated containing 3 copies of a 26-bp sequence containing the -35 GC-rich motif (-35 × 3 wild-type). A similar strain was made using the same motif containing a substitution of TC for GG in the center of the consensus GC-rich motif (-35 × 3 mutant), which we had shown led to reduced binding of nuclear proteins and impaired transcriptional promoter activity (Figures 3, 5). Both reporter strains were transformed with purified plasmids representing each of the factors identified in the initial screen, along with a control plasmid identified in the initial screen that contained a small nongene insert. We found that each of the KLF clones and the novel zinc-finger gene conferred preferential growth in His-/3-AT medium for the reporter strain containing the wild-type but not the mutant -35 sequence (Figure 6; Table 3). When transformed into the original -35 × 3 and PDβ1 dual-reporter strains, all 3 KLFs and the novel zinc-finger protein, but not the control, could restore β-gal activity (Table 3). These results suggest that the 3 KLF and the novel zinc-finger protein genes identified in the yeast screen using the entire Dβ1 promoter bound to the -35 GC-rich element in a sequence-specific manner.
Specificity of yeast one-hybrid clones for binding the –35 GC-rich motif. Gal-4 fusion clones representing 3 Krüppel-like factors and a novel zinc-finger (ZF) protein (GenBank accession number AY117438) were transformed into yeast His3 reporter strains driven by 3 copies of wild-type (WT) or mutant (Mu) -35 motif. Strains were plated on Leu-His- plates with or without 3-AT.
Specificity of yeast one-hybrid clones for binding the –35 GC-rich motif. Gal-4 fusion clones representing 3 Krüppel-like factors and a novel zinc-finger (ZF) protein (GenBank accession number AY117438) were transformed into yeast His3 reporter strains driven by 3 copies of wild-type (WT) or mutant (Mu) -35 motif. Strains were plated on Leu-His- plates with or without 3-AT.
Sequence specificity of DNA binding with yeast one-hybrid clones
Candidate . | Rescue His- in -35 × 3 wild-type strain . | Rescue His- in -35 × 3 mutant strain . | Restore LacZ+ in -35 × 3-His3/LacZ strain . | Restore LacZ+ in PDβ1-His3/LacZ strain . |
|---|---|---|---|---|
| KLF3/BKLF | Yes | No | Yes | Yes |
| KLF5/IKLF | Yes | No | Yes | Yes |
| KLF6/ZF9 | Yes | No | Yes | Yes |
| Novel ZF protein | Yes | No | Yes | Yes |
| Control | Yes | Yes | No | No |
Candidate . | Rescue His- in -35 × 3 wild-type strain . | Rescue His- in -35 × 3 mutant strain . | Restore LacZ+ in -35 × 3-His3/LacZ strain . | Restore LacZ+ in PDβ1-His3/LacZ strain . |
|---|---|---|---|---|
| KLF3/BKLF | Yes | No | Yes | Yes |
| KLF5/IKLF | Yes | No | Yes | Yes |
| KLF6/ZF9 | Yes | No | Yes | Yes |
| Novel ZF protein | Yes | No | Yes | Yes |
| Control | Yes | Yes | No | No |
Expression of KLF factors in lymphoid cells
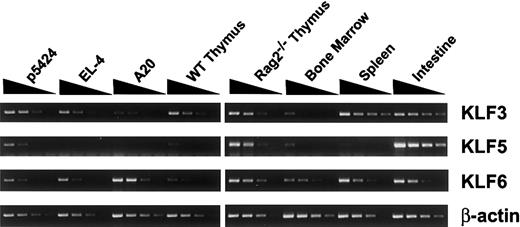
Cell lines and primary tissues were tested for the expression of Klf3, Klf5, and Klf6 mRNAs using RT-PCR (Figure 7). Northern blot analysis was not sufficiently sensitive to detect these mRNAs in lymphoid tissues and cell lines. Consistent with previous studies, Klf3 and Klf6 were widely expressed at various levels in all detected cell lines and tissues.40,42 As previously shown, Klf5 was found at high levels in intestine.37,43 In lymphoid cell lines, Klf5 mRNA was found in the pro–T-cell line p5424 but not in the more mature T cell line EL-4 or the A20 B cell line. In primary tissues, Klf5 was enriched in RAG-2–deficient thymus and was present at lower levels in healthy thymus and bone marrow while being minimally detectable in spleen. These observations suggest that pro-T cell expresses multiple KLF factors and that KLF5, which is known to be expressed at high levels in intestine, exhibits a restricted expression pattern in lymphoid cells that correlates with TCRβ germline transcription.
Klf3, Klf5, and Klf6 mRNA expression in lymphoid cells. Total RNA from p5424 pro-T, EL-4 mature T, A20 B-cell lines, and primary mouse tissues, including wild-type and RAG-2–deficient thymus, spleen, bone marrow, and intestine, were serially diluted (1:5:25:125) and amplified by RT-PCR using specific primer pairs under optimal conditions with β-actin as a control.
Klf3, Klf5, and Klf6 mRNA expression in lymphoid cells. Total RNA from p5424 pro-T, EL-4 mature T, A20 B-cell lines, and primary mouse tissues, including wild-type and RAG-2–deficient thymus, spleen, bone marrow, and intestine, were serially diluted (1:5:25:125) and amplified by RT-PCR using specific primer pairs under optimal conditions with β-actin as a control.
We next asked whether KLF5 was among the several proteins from p5424 cells that formed complexes with the Dβ1 promoter -35 GC-rich motif. The ubiquitous transcription factor Sp1 was previously shown to be among nuclear proteins binding to the (-281) GC-rich motif.25 Antibody-interference EMSA was performed using anti-Sp1 and anti-KLF5 antibodies (Figure 8A). The result indicates that KLF5 and Sp1 are among nuclear proteins binding to the -35 GC-rich motif. To examine the interaction of KLF5 binding the Dβ1 promoter in vivo, chromatin immunoprecipitation assay was carried out using the same anti-KLF5 and anti-Sp1 antibodies (Figure 8B). Anti-KLF5 efficiently precipitated the Dβ1 promoter in p5424 cells, but a faint signal was detectable with anti-Sp1. The anti-KLF5 signal appeared to be specific inasmuch as parallel immunoprecipitations with A20 cells (where KLF5 mRNA is not detectable in our assays) did not yield the Dβ1 promoter. These data indicate that KLF5 associates with the Dβ1 promoter in vivo.
KLF5 binds Dβ1 promoter in vitro and in vivo. (A) KLF5 and Sp1 are among nuclear proteins in pro-T cells binding the -35 motif in the Dβ1 promoter. Nuclear extracts from p5424 cells were incubated with no antibody, anti-Sp1 antiserum, anti-KLF5 antiserum, or control antiserum at room temperature for 20 minutes before EMSA. (B) KLF5 binds the Dβ1 promoter in p5424 cells in vivo. p5424 and A20 cells were cross-linked by 1% formaldehyde, sonicated, and subjected to chromatin immunoprecipitation assay with anti-KLF5 antiserum, anti-Sp1 antiserum, control antiserum, or without antibody. Immunoprecipitates were assayed for Dβ1 or IgH DQ52 promoter sequences using nested PCR.
KLF5 binds Dβ1 promoter in vitro and in vivo. (A) KLF5 and Sp1 are among nuclear proteins in pro-T cells binding the -35 motif in the Dβ1 promoter. Nuclear extracts from p5424 cells were incubated with no antibody, anti-Sp1 antiserum, anti-KLF5 antiserum, or control antiserum at room temperature for 20 minutes before EMSA. (B) KLF5 binds the Dβ1 promoter in p5424 cells in vivo. p5424 and A20 cells were cross-linked by 1% formaldehyde, sonicated, and subjected to chromatin immunoprecipitation assay with anti-KLF5 antiserum, anti-Sp1 antiserum, control antiserum, or without antibody. Immunoprecipitates were assayed for Dβ1 or IgH DQ52 promoter sequences using nested PCR.
KLF5 activates Dβ1 promoter activity
To test whether the binding of KLF5 to GC-rich motifs in the Dβ1 promoter correlates with transcriptional activity, we performed cotransfection experiments with expression constructs containing either full-length KLF5 or a truncated KLF5 containing only the DNA-binding domain, which we predicted, based on previous studies with Sp3, would act as a dominant-negative mutant43 (Figure 9). In NIH/3T3 cells, the activity of Dβ1 promoter reporter constructs was enhanced 3- to 4-fold in the presence of the KLF5 expression construct (Figure 9B). This transactivation was dependent on the GC-rich motifs but was not affected by the presence of the Eβ enhancer, suggesting the effect was specific for GC-rich motifs in the promoter. With the dominant-negative mutant of KLF5, luciferase expression was reduced (Figure 9C). These results suggest that KLF5 may act as a positive regulator of Dβ1 transcriptional activity. In contrast, a KLF6 expression construct did not substantially enhance Dβ1 promoter activity, either in the absence or the presence of Eβ (Figure 9D).
KLF5 but not KLF6 transactivates Dβ1 promoter expression. (A) Expression constructs representing full-length KLF5, containing putative activating domain (AD), nuclear localization signal (N), and the zinc-finger (ZF) domain. A dominant-negative KLF5 mutant construct (KLF5DN) lacks the N-terminal 358 amino acids within the activating domain. (B) Effect of vector or KLF5 on reporter gene activity in NIH/3T3 cells. Reporters included the Dβ1 promoter (+6Δ) or a construct containing mutations within the -35, -281, and -314 GC-rich motifs (asterisks). Each was tested in the presence or absence of the Eβ enhancer. (C) Effect of wild-type or KLF5DN expression constructs on Dβ1 promoter constructs without or with the Eβ enhancer. (D) Effect of KLF6 expression on Dβ1 promoter constructs in the absence or presence of Eβ enhancer. Luciferase activity in these assays is expressed relative to the vector-only control cotransfected with the wild-type Dβ1 promoter construct. Average and standard error for 3 independent transfections are presented.
KLF5 but not KLF6 transactivates Dβ1 promoter expression. (A) Expression constructs representing full-length KLF5, containing putative activating domain (AD), nuclear localization signal (N), and the zinc-finger (ZF) domain. A dominant-negative KLF5 mutant construct (KLF5DN) lacks the N-terminal 358 amino acids within the activating domain. (B) Effect of vector or KLF5 on reporter gene activity in NIH/3T3 cells. Reporters included the Dβ1 promoter (+6Δ) or a construct containing mutations within the -35, -281, and -314 GC-rich motifs (asterisks). Each was tested in the presence or absence of the Eβ enhancer. (C) Effect of wild-type or KLF5DN expression constructs on Dβ1 promoter constructs without or with the Eβ enhancer. (D) Effect of KLF6 expression on Dβ1 promoter constructs in the absence or presence of Eβ enhancer. Luciferase activity in these assays is expressed relative to the vector-only control cotransfected with the wild-type Dβ1 promoter construct. Average and standard error for 3 independent transfections are presented.
Discussion
Specification of the major T-cell lineage in developing multipotential hematopoietic cells includes events that lead to rearrangement of the TCRβ locus, including the activation of TCRβ germline transcription.22,44 Germline transcription of the reiterated DJC complex is regulated by the Eβ enhancer, the Dβ1 promoter, and likely a second promoter element near Dβ2.24, 25, 26 In transcriptional reporter assays, the Dβ1 promoter, in conjunction with Eβ, is sufficient to recapitulate preferential activity in cell lines with characteristics of pro-T cells and is less active in T-cell lines representing later stages of T-cell development or in B-cell lines.24 Recent studies using transgenic mice expressing green fluorescence protein (GFP) under control of the Dβ1 promoter and Eβ enhancer also suggest that these elements are sufficient to direct a restricted pattern of expression at early stages of T-cell development in thymus and bone marrow (R.T.D., D.M.W., unpublished observation, 2002). Although the involvement of these cis elements is clear, the regulatory factors that bind these elements in pro-T cells and the signals that lead to transcriptional activation are unknown.
To identify potentially significant cis elements in the Dβ1 promoter, we used EMSA to screen for regions that exhibited differential binding to nuclear extracts from p5424 cells, which have high-level TCRβ germline transcription and Eβ/Dβ1 promoter activity, compared with extracts from the A20 B-cell line (Figure 1). The most prominent differences were found with probes containing 1 of 3 GC-rich motifs. Characterization of binding to these elements using cold competition and mutation studies suggests that these motifs bind a similar set of nuclear proteins and may, therefore, act as reiterated transcriptional regulatory motifs. Each of these motifs was functional in transcriptional reporter assays, in which mutation of the central residues (GG→TC) abrogated the binding of nuclear proteins in EMSA assays (Figure 2) and also led to a substantial reduction in transcriptional activity in reporter experiments (Figure 5). These findings contribute to explaining the ability of subfragments of the Dβ1 promoter to efficiently direct transcription because each fragment with promoter activity contained at least one of these GC-rich motifs (Figure 5). This redundancy of independent sites for transcriptional activation correlates with heterogeneous initiation of Dβ1 germline transcripts observed in pro–T-cell lines and RAG-2-/- thymocytes and might contribute to achieving the necessary configuration for recombinase accessibility.24
Oligonucleotide probes containing each of the 3 GC-rich elements bind multiple nuclear proteins, most or all of which are dependent on the central GGGNGG residues in each of the motifs. Cross-linking studies suggest that as many as 6 proteins in the p5424 pro–T-cell extract bind to or are otherwise in proximity to DNA (Figure 4). Rather than indicating a large multiprotein complex, these observations suggest that a number of individual proteins in the cell are capable of binding the core motif. This motif is consistent with GC-rich elements present in a wide variety of transcriptional regulatory regions and they bind to transcription factors of the Sp/KLF family. These proteins represent a large group of the C2H2-type, zinc-finger, DNA-binding proteins consisting of at least 20 members.45, 46, 47, 48 Overlapping binding by multiple family members present in a given cell type is a characteristic feature and may contribute to complex patterns of tissue-specific gene regulation.46, 47, 48 In yeast one-hybrid screens, we identified 6 clones that bound to the GC-rich motifs in a sequence-specific manner, including KLF3, KLF5, KLF6, and a novel zinc-finger protein. Of these, KLF5 was identified most frequently and was confirmed as a member of the group of nuclear proteins from pro–T-cell extracts binding GC-rich motifs in the Dβ1 promoter.
KLF5 was previously identified as a bifunctional factor binding GC boxes, and it is expressed at high levels in intestine, where it localizes to crypts of Lieberkühn.37,43 KLF5 transactivates the SM22α gene promoter in smooth muscle cells through the TCE GC-rich element (GAGTGGGGCG) and the β-globin promoter in NIH/3T3 fibroblast cells through 2 CACCC motifs.43,49 KLF5 was reported to exhibit repressor activity for the lactoferrin promoter in human endometrial carcinoma RL95-2 cells.37 Targeted inactivation of the KLF5 locus in mice leads to early embryonic lethality in homozygotes. In heterozygous mice, there is a reduction in cardiac hypertrophy and angiogenesis in response to injury, indicating KLF5 plays a role in the regulation of vascular tissue remodeling.50 We found that Klf5 is also expressed in lymphoid cells, most prominently in pro-T lines and RAG-2-/- thymocytes, whereas expression in healthy thymus and bone marrow is much lower (Figure 7). This suggests that in addition to its function in the intestine, KLF5 may also play a role in early thymocyte development.
One potential mechanism for lineage-specific gene regulation is the selective expression of transcriptional regulatory factors. Among the proteins that can interact with GC-rich motifs in the Dβ1 promoter, Sp1, KLF3, and KLF6 appear to be broadly expressed in lymphoid cells, though the expression of KLF5 appears more restricted. Which of these interactions is relevant for regulation of the Dβ1 promoter in vivo? The demonstration of KLF5 binding to the Dβ1 promoter in p5424 cells by chromatin immunoprecipitation (Figure 8B) indicates that this binding interaction occurs in vivo. This experiment also suggested that Sp1 interacts with the Dβ1 promoter to some extent. The potential for KLF5 to function as a transcriptional activator of the Dβ1 promoter is supported by its transactivating activity in reporter assays, which were specific for the GC-rich motifs (Figure 9B). Moreover, a truncated KLF5, lacking the putative N-terminal activation domain (aa 1-358), suppressed basal activity of the Dβ1 promoter in NIH/3T3 cells (Figure 9C). In contrast, KLF6 exhibited no transactivating activity in this assay, whereas KLF3 behaves as a transcriptional repressor by recruitment of CtBP2, a general corepressor protein interacting with a Pro-X-(Asp/Asn)-Leu-(Ser/Thr) motif located in the KLF3 repression domain.46,48 Taken together, these observations suggest that KLF5 contributes to the regulation of TCRβ germline transcription. Further genetic studies are required to confirm this hypothesis in vivo.
Our data do not necessarily indicate that KLF5 is an exclusive regulator of the GC-rich motifs in the Dβ1 promoter, and it is possible that other GC-binding proteins may also participate in the regulation of this locus. Limited sequence specificity is characteristic of Sp/KLF family proteins, and many cell types express multiple family members. This has important, though poorly understood, implications for lineage-specific transcriptional regulation, where competition for binding sites among different family members may hinge on subtle changes in expression levels, protein–protein interactions, or posttranslational modifications. Inasmuch as Sp/KLF family members may participate in transcriptional activation or repression (or, in some cases, both), these complex interactions may yield highly regulated outcomes with respect to gene expression. Examples include the GC-rich motifs in the β-globin promoter, which interact with Sp1, EKLF, and BKLF/KLF3 to regulate developmental expression of the β-globin gene.40 In smooth muscle cells, the differentiation marker SM22α is regulated through interaction of a GC box (TCE) with GKLF/KLF4 and BTEB2/KLF5.49 Finally, both KLF4 and KLF6 have been implicated in controlling the differentiation-linked human keratin 4 promoter in esophagus and cornea and the pregnancy-specific glycoprotein genes during human placenta development.51,52
Lineage-specific regulation by KLF family members is also suggested by gene deletion studies. EKLF is expressed throughout development during erythropoiesis; however, EKLF deficiency leads to a specific hemoglobin deficiency at the time of the fetal to adult (γ to β) hemoglobin switch.53,54 LKLF is broadly expressed, is required for healthy blood vessel morphology and stability,55 is required for maintaining mature T-cell quiescence and survival, and is rapidly degraded on T-cell activation.56 Osterix is a newly identified member of this family that is required for bone formation.45 The fact that each of these KLF family members is absolutely required for at least one developmental function, in spite of other members with similar binding specificity being coexpressed, supports the hypothesis that coexpressed Sp/KLF family members may be able to fine-tune gene expression through interaction (cooperation or competition), but they cannot replace or adequately compensate for a key member in a specific developmental process.
Our data suggest that TCRβ germline transcription is under complex regulatory control by reiterated GC-rich elements that bind multiple Sp/KLF family transcription factors expressed in early T cells. Of these, differentially expressed KLF5 may contribute to regulating lineage- and stage-specific germline TCRβ expression. Further efforts to understand the regulation of germline transcription through GC-rich motifs and the factors that bind them appear warranted.
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2002-08-2579.
Supported by National Institutes of Health grants AI41051 and CA88075 (D.M.W.) and by National Research Service Award fellowship AI10600 (R.T.D).
X.O.Y. and R.T.D. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Christina Teng for providing anti-KLF5 antiserum and KLF5 expression construct and Dr Anthony Martino for technical advice on antibody interference studies.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal