Abstract
Hermansky-Pudlak syndrome (HPS) is a disorder of organelle biogenesis affecting 3 related organelles—melanosomes, platelet dense bodies, and lysosomes. Four genes causing HPS in humans (HPS1-HPS4) are known, and at least 15 nonallelic mutations cause HPS in the mouse. Where their functions are known, the HPS-associated proteins are involved in some aspect of intracellular vesicular trafficking, that is, protein sorting and vesicle docking and fusion. Biochemical and genetic evidence indicates that the HPS-associated genes encode components of at least 3 distinct protein complexes: the adaptor complex AP-3; the HPS1/HPS4 complex; and BLOC-1 (biogenesis of lysosome-related organelles complex-1), consisting of the proteins encoded at 2 mouse HPS loci, pallid (pa) and muted (mu), and at least 3 other unidentified proteins. Here, we report the cloning of the mouse HPS mutation cappuccino (cno). We show that the wild-type cno gene encodes a novel, ubiquitously expressed cytoplasmic protein that coassembles with pallidin and the muted protein in the BLOC-1 complex. Further, we identify a frameshift mutation in mutant cno/cno mice. The C-terminal 81 amino acids are replaced with 72 different amino acids in the mutant CNO protein, and its ability to interact in BLOC-1 is abolished. We performed mutation screening of patients with HPS and failed to identify any CNO defects. Notably, although defects in components of the HPS1/HPS4 and the AP-3 complexes are associated with HPS in humans, no defects in the known components of BLOC-1 have been identified in 142 patients with HPS screened to date, suggesting that BLOC-1 function may be critical in humans.
Introduction
In the autosomal recessive Hermansky-Pudlak syndrome (HPS) defects in lysosome-related organelles (melanosomes, platelet dense bodies, lysosomes) cause albinism, prolonged bleeding, and lysosomal storage disease.1,2 The worldwide incidence of HPS is unknown, but it occurs in all ethnic groups and its frequency is very high in some populations due to founder effects (eg, Northwest Puerto Rico, 1/1800).3 HPS is associated with considerable morbidity and mortality. Melanosome defects lead to oculocutaneous albinism resulting in nystagmus, decreased visual acuity, and skin damage on exposure to sunlight.4 Platelet dense body defects result in prolonged bleeding, which can be severe.4,5 Pulmonary fibrotic disease, possibly the result of ceroid lipofuscin accumulation in lysosomes, is progressive and is known to occur in patients with HPS-1 and HPS-4. In such individuals, death typically occurs in the fourth to fifth decades.5, 6, 7
In mice, at least 15 nonallelic mutations cause HPS.8 Eight are cloned, including the orthologues of the human genes HPS1-HPS4 (pale ear [ep], pearl [pe], cocoa [coa], light ear [le]), and mocha (mh), pallid (pa), ashen (ash), and muted (mu).9, 10, 11, 12, 13, 14, 15, 16, 17 The known or suspected functions of the HPS proteins implicate them in some aspect of intracellular vesicular trafficking in the biogenesis of lysosome-related organelles. The pearl and mocha mutations involve defects in the β3A and δ subunits, respectively, of the AP-3 complex, which functions in endosomal-lysosomal protein trafficking.12,15,18 In ashen (ash), Rab27a, a protein involved in targeting and fusing transport vesicles to their acceptor membrane, is defective.16 Pallid is a defect in a novel protein, pallidin, which binds to syntaxin-13, a t-snare protein that mediates vesicle docking and fusion.14 Although the function of the muted protein is unknown, it was recently shown to interact with pallidin and at least 3 other unidentified proteins in a complex designated BLOC-1 (biogenesis of lysosome-related organelles complex 1).19,20 Here, we report the positional cloning of the mouse HPS mutation, cappuccino (cno; GenBank accession no. AY186603), and we show that the wild-type CNO protein is a component of BLOC-1 and interacts with pallidin and muted. In cno/cno mice, an 11-bp deletion causes a frameshift that alters the C-terminal one third of the protein sequence. As a result, the mutant protein is unable to coassemble with pallidin. In addition, we performed mutation screening of patients with HPS for CNO defects. None were found. Notably, the pa, mu, and cno mutations are the most phenotypically severe of the mouse HPS models, and no human HPS patients with orthologous defects are known, suggesting that BLOC-1 function may be essential in humans.
Materials and methods
Mice
All mice were raised at The Jackson Laboratory (Bar Harbor, ME). They were housed in humidity- and temperature-controlled rooms with a 12-hour light cycle and given free access to acidified water and chow. All protocols were approved by The Jackson Laboratory Animal Care and Use Committee. The Jackson Laboratory is fully accredited by the American Association for Accreditation of Laboratory Animal Care (AAALAC).
Molecular characterization of cappuccino
Total RNA was prepared using TRIzol reagent (Invitrogen, Carlsbad, CA). Polyadenylated RNA was purified on oligo-dT columns from Stratagene (La Jolla, CA). DNA was prepared from tail tissue as previously described.21 Northern and Southern blotting were performed using standard techniques.22 Human and mouse premade multiple tissue Northern blots were purchased from Clontech (Palo Alto, CA) and Origene Technologies (Rockland, MD), respectively, and were hybridized and washed as recommended by the manufacturers. Polymerase chain reaction (PCR) and reverse transcription-PCR (RT-PCR) amplifications were performed using standard techniques as previously described.23 Specific primer sequences for each reaction are given in the appropriate figure legend. Primers were obtained from Ransom Hill Bioscience (Ramona, CA). Sequencing was performed using the automated dye termination technique (ABI PRISM Model 3700 Genetic Analyzer, Applied Biosystems, Foster City, CA). We searched for candidate transcripts within the cno interval on chromosome 5 using the Celera Discovery System and Celera Genomics associated databases (www.celeradiscoverysystem.com/glance/home.cfm). We searched for sequence homologies using BLAST analysis through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), The Institute for Genomic Research (http://tigrblast.tigr.org/tgi/), and the Wisconsin GCG Package (Version 10.3). Protein sequences were analyzed using ExPASy Proteomics Tools (http://us.expasy.org/tools/; PSORT, Motif-Scan, TMpred, Jpred, Coils) and Network Protein Sequence Analysis tools (Pôle BioInformatique Lyonnais; http://npsa-pbil.ibcp.fr/).
Quantitative PCR
RNA was reversed transcribed using oligo-dT. The primers for cno amplification and detection were as follows: forward, 5′-GTGACAGACCGCAGCTCTGA-3′; reverse, 5′-CACTTGGGACAAGAGTCTCGCT-3′; detection probe, 5′-FAM TGTGCGCCTCCACCGCATGA-3′-TAMRA. 18S RNA was amplified with the forward probe 5′-CCGCAGCTAGGAATAATGGAAT-3′, reverse probe 5′-CGAACCTCCGACTTTCGTTCT-3′, and detection probe 5′-VIC ACCGGCGCAAGACGGACCAGA-3′-TAMRA. Detection probes were obtained from Applied Biosystems. The PCR reaction mix (25 μL) contained 1 μL cDNA, 250 nM detection primer, 900 nM cno or 18s primers, and 12.5 μL 2 × Taq Man Universal PCR Master Mix (Applied Biosystems). The reactions were performed using an Applied Biosystems PRISM 7700 Sequence Detection System. All reactions were carried out in triplicate, and the experiment was performed a total of 3 times.
Cells and antibodies
The mouse fibroblast cell lines and the culture conditions have been described previously.20 Megakaryocytes were cultured from 13.5 days after coitus (dpc) fetal livers and purified as described.24 Platelets were isolated from whole blood, and Western blots were prepared from megakaryocyte and platelet lysates as described.23 Peptide antibodies to the N-terminus of CNO (amino acids 36-55) and the mutant C-terminus (amino acids 140-159) were generated by Research Genetics (Huntsville, AL). All other antibodies have been previously described.20
Cell transfection and immunofluorescence
The complete coding sequences of both normal and mutant cno were subcloned in-frame into the pCMV-Tag4 FLAG vector (Stratagene). Normal C57BL/6 fibroblasts were plated (5 × 106 cells) onto 24-well tissue culture plates and allowed to grow to 60% to 80% confluence. Cells were transfected with 0.2 μg DNA using LIPOFECTAMINE PLUS reagent (Invitrogen) according to the manufacturer's protocol and cultured for 2 weeks in the presence of G418. Stable transfectants were grown on coverslips overnight, washed in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 5 minutes, and washed 3 times with methanol. The cells were then fixed in methanol for an additional 10 minutes. All washes and fixation steps were carried out at 4°C. All subsequent steps were carried out at room temperature. Nonspecific antibody binding was blocked by incubating the cells in 5% normal goat serum (NGS; Jackson Immunoresearch, West Grove, PA) in PBS for 15 minutes. The coverslips were washed and incubated with mouse monoclonal anti-FLAG antibody (Stratagene) diluted to 10 ng/μL in 5% NGS/PBS or 5% NGS/PBS alone (negative control) for 1 hour at room temperature, washed, and incubated with 5 ng/μL goat antimouse IgG (Molecular Probes, Eugene, OR) for 30 minutes. The coverslips were washed and mounted on glass slides using SlowFade Antifade (Molecular Probes). Confocal images were collected with a Leica TCS NT confocal microscope (Leica Microsystems, Exton, PA) using either a 63 × numerical aperture (NA) 1.2 water immersion or 100 × NA 1.4 oil immersion lens. Single 0.5- to 0.6-mm optical sections were collected at the level of the nucleus. Filter selection and image collection parameters were optimized to provide images with a full range of 256 Gy values and minimal fluorescent bleed-through, as assayed with the appropriate single-labeled controls. Multiple color fluorescent and differential interference contrast microscopy (DIC) images were collected simultaneously.
Immunoprecipitation-recapture assays
Sedimentation velocity on sucrose gradients
H4 cells were metabolically labeled with 35S-methionine, extracted with 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; pH 7.4), 0.15 M NaCl, 0.05% (wt/vol) Triton X-100, and sedimented on 2% to 15% (wt/vol) sucrose gradients as previously described.27 Sixteen fractions were collected for immunoprecipitation-recapture experiments.
Screening of patients with HPS
PCR amplification products from fibroblast DNA were sequenced using a Beckman CEQ 2000 and the CEQ Dye Terminator Cycle Sequencing kit according to the manufacturer's instructions.
Results
Positional cloning of the cappuccino gene
The cappuccino (cno) mutation arose spontaneously on the C3H/HeJ inbred strain and is a classic mouse model of severe HPS (Figure 1A).21,28 The skin and eye melanosomes are immature and dramatically decreased in number.21,29 Platelet dense body contents (serotonin, adenosine triphosphate) are profoundly reduced resulting in abnormal platelet aggregation and prolongation of the bleeding time, and lysosomal enzyme levels are elevated in the kidney and liver, reflecting aberrant secretion.21
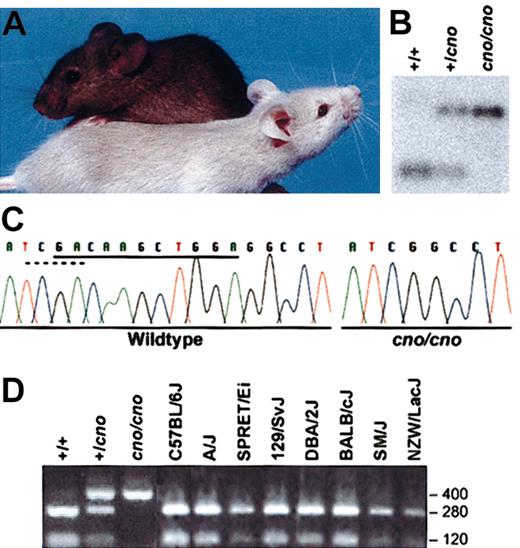
Cappuccino is a defect in a novel protein. (A) A C3H/HeJ-cno/cno mouse (foreground) showing its characteristic severe albinism with a normal littermate (background). (B) Southern blot showing a TaqI polymorphism in cno. gDNA was cut with TaqI, blotted, and probed with a 403-bp fragment (bp 314-716) of BC019169 amplified from normal gDNA with forward primer 5′-AGTTCGTGGGCATGCTGGACATGAT-3′ and reverse primer ACAAGAGTCTCGCTGGAGTCCTCAT-3′. In normal (+/+) DNA, the probe detects a 0.4-kb fragment. In cno/cno DNA, deletion of a TaqI site leaves a larger, 0.8-kb fragment. (C) Direct sequencing reveals an 11-bp deletion (underlined bases) in the cno/cno gene that eliminates a TaqI restriction site (dashed line). (D) Amplification with the above primers and TaqI digestion of gDNA from wild-type littermates (C3H/HeJ-+/+) produces the expected fragments (280 and 120 bp), whereas the cno/cno fragment is not cut. Note the presence of both the wild-type and mutant fragments in heterozygous +/cno DNA. Notably, all other inbred strains tested show the wild-type pattern.
Cappuccino is a defect in a novel protein. (A) A C3H/HeJ-cno/cno mouse (foreground) showing its characteristic severe albinism with a normal littermate (background). (B) Southern blot showing a TaqI polymorphism in cno. gDNA was cut with TaqI, blotted, and probed with a 403-bp fragment (bp 314-716) of BC019169 amplified from normal gDNA with forward primer 5′-AGTTCGTGGGCATGCTGGACATGAT-3′ and reverse primer ACAAGAGTCTCGCTGGAGTCCTCAT-3′. In normal (+/+) DNA, the probe detects a 0.4-kb fragment. In cno/cno DNA, deletion of a TaqI site leaves a larger, 0.8-kb fragment. (C) Direct sequencing reveals an 11-bp deletion (underlined bases) in the cno/cno gene that eliminates a TaqI restriction site (dashed line). (D) Amplification with the above primers and TaqI digestion of gDNA from wild-type littermates (C3H/HeJ-+/+) produces the expected fragments (280 and 120 bp), whereas the cno/cno fragment is not cut. Note the presence of both the wild-type and mutant fragments in heterozygous +/cno DNA. Notably, all other inbred strains tested show the wild-type pattern.
Previously we localized cno by linkage analysis to a 0.9-cM interval between D5Ertd521e and D5Mit230 on chromosome 5, a region conserved with human 4p15-16.21 Using the Celera Discovery System and Celera Genomics–associated databases, 10 transcripts were identified within this 387-kb interval. The gene encoding epididymis-specific α-D-mannosidase (Man2b2; GenBank accession no. U29947),30 an unlikely candidate, was the only known gene within the interval. We screened the remaining candidates by a combination of Northern blot analysis, Southern blot analysis of gDNA digested with multiple restriction enzymes, and direct sequencing. One transcript within the interval, BC019169, showed a TaqI restriction fragment length polymorphism on Southern blots (Figure 1B). Sequencing revealed an 11-bp deletion (nucleotides 427-437) in cno mice. The deletion causes a frameshift and removes a TaqI site (Figure 1C). PCR amplification of a 403-bp fragment flanking the deletion using BC019169-specific primers followed by digestion with TaqI produced the expected fragments from +/+, +/cno, and cno/cno gDNA (Figure 1D), and demonstrated that the mutation segregates with the cno phenotype. Similar analysis of 8 other inbred strains revealed the presence of the TaqI site (Figure 1D), and sequencing confirmed that the 11-bp deletion was unique to the cno mutation.
Expression of the cappuccino gene
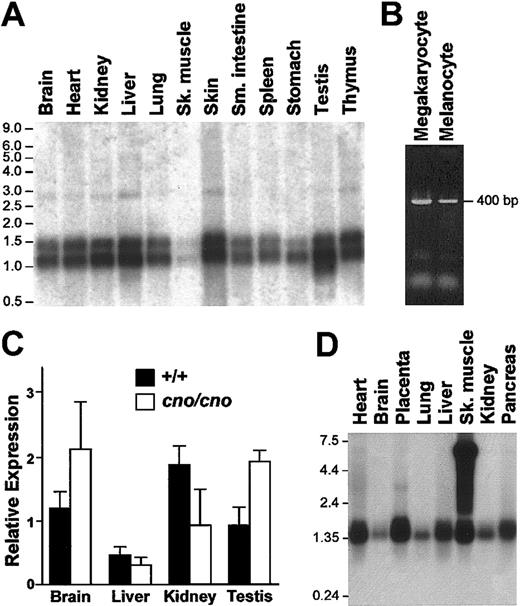
The BC019169 (hereafter cno) cDNA contains 1044 bp and includes a consensus polyadenylation signal and a short poly A tail. As with all HPS-causing genes, cno appears to be ubiquitously expressed. In normal mice, the cno gene is expressed in all tissues examined including isolated megakaryocytes and cultured melanocytes (Figure 2A-B). In mutant cno/cno tissues, expression appeared normal by Northern blot analysis (data not shown). Quantitative PCR analyses confirmed that the mutant allele was expressed at levels comparable to the wild-type allele (Figure 2C).
Widespread expression ofcnoin normal tissues and cells. (A) OriGene Technologies Northern blot showing expression of cno in mouse tissues. The blot was hybridized with a 738-bp probe amplified from gDNA that included the entire open reading frame of mouse cno (forward 5′-TAACGCAATCGCTCAGCACCGGAAG-3′, reverse 5′-ACAAGAGTCTCGCTGGAGTCCTCAT-3′). (B) By RT-PCR, cno is detected in RNA isolated from megakaryocytes and RNA from cultured C57BL/6J melanocytes. Total RNA was reverse transcribed using oligo-dT. The PCR primer sequences were as described in Figure 1B. (C) Quantitative real-time PCR analysis of wild-type and mutant tissues. Expression of cno in each tissue is presented relative to 18S RNA expression. The data are expressed as the ×±SD of 3 separate experiments. No significant differences in expression were seen in cno/cno tissues (□) versus +/+ tissues (▪). (D) Clontech human multiple tissue Northern blot hybridized with a human CNO probe (bp 396-917 of AK002092) amplified from fibroblast gDNA using forward primer 5′-GGCATGCTGGACATGCTTCGCGG-3′ and reverse primer 5′-CTATACATTCTACACTGAATCATG-3′. The origin of the high-molecular-weight band in skeletal muscle is unknown. The migration of RNA standards is shown to the left of panels A and D.
Widespread expression ofcnoin normal tissues and cells. (A) OriGene Technologies Northern blot showing expression of cno in mouse tissues. The blot was hybridized with a 738-bp probe amplified from gDNA that included the entire open reading frame of mouse cno (forward 5′-TAACGCAATCGCTCAGCACCGGAAG-3′, reverse 5′-ACAAGAGTCTCGCTGGAGTCCTCAT-3′). (B) By RT-PCR, cno is detected in RNA isolated from megakaryocytes and RNA from cultured C57BL/6J melanocytes. Total RNA was reverse transcribed using oligo-dT. The PCR primer sequences were as described in Figure 1B. (C) Quantitative real-time PCR analysis of wild-type and mutant tissues. Expression of cno in each tissue is presented relative to 18S RNA expression. The data are expressed as the ×±SD of 3 separate experiments. No significant differences in expression were seen in cno/cno tissues (□) versus +/+ tissues (▪). (D) Clontech human multiple tissue Northern blot hybridized with a human CNO probe (bp 396-917 of AK002092) amplified from fibroblast gDNA using forward primer 5′-GGCATGCTGGACATGCTTCGCGG-3′ and reverse primer 5′-CTATACATTCTACACTGAATCATG-3′. The origin of the high-molecular-weight band in skeletal muscle is unknown. The migration of RNA standards is shown to the left of panels A and D.
Two mRNAs of approximately 1.1 and 1.4 kb are present on Northern blots (Figure 2A). A BLAST search of the TIGR Unique Gene Indices database revealed a second 1443-bp clone (gene identifier TC242970) that is identical BC019169 but has a longer 3′ untranslated sequence. The corresponding human cDNA (AK002092) contains 1403 bp and also is widely expressed (Figure 2D). As in the mouse, a search of dbEST revealed 2 mRNAs, a 1250-bp mRNA containing a poly A tail, and the longer 1403-bp spliceoform. (These species are not resolved on the Northern blot shown in Figure 2D.) Comparison with the Celera consensus sequence indicates that the entire cDNA sequence is contained within a single exon in both mouse and human. Because a strong consensus Kozak sequence surrounds the ATG initiation codon in each case,31 both mRNA species likely arise through utilization of different polyadenylation signals.
Structure of the normal and mutant cappuccino proteins
The normal mouse CNO protein consists of 215 amino acids with a predicted molecular mass of 23.1 kDa. It shares 77% identity with the human orthologue (Figure 3A). There are no predicted transmembrane domains, suggesting the protein is cytosolic. No known motifs are present that might give clues to its function. Searches of BLASTn, BLASTp, and Swiss-Prot using the normal mouse CNO sequence suggest that the gene is unique to metazoans. An unnamed human protein product (AK000502) derived from a breast carcinoma cell line exhibited low homology (38% identity/56% similarity). The region, extending from amino acids 94-213, is also weakly homologous to a Drosophila melanogaster gene product, AY071122 (23%/44%). In addition, a short region (amino acids 127-205) is weakly homologous to the Caenorhodditis elegans protein AY071122 (31%/44%).
Amino acid sequence of the CNO protein. (A) Comparison of human and mouse CNO amino acid sequence reveals an overall identity of 77% (shaded residues). The predicted coiled-coil domains are boxed. In the mutant protein, the 81 amino acids from position 135 (aspartic acid, double underline) are replaced with those shown in panel B.
Amino acid sequence of the CNO protein. (A) Comparison of human and mouse CNO amino acid sequence reveals an overall identity of 77% (shaded residues). The predicted coiled-coil domains are boxed. In the mutant protein, the 81 amino acids from position 135 (aspartic acid, double underline) are replaced with those shown in panel B.
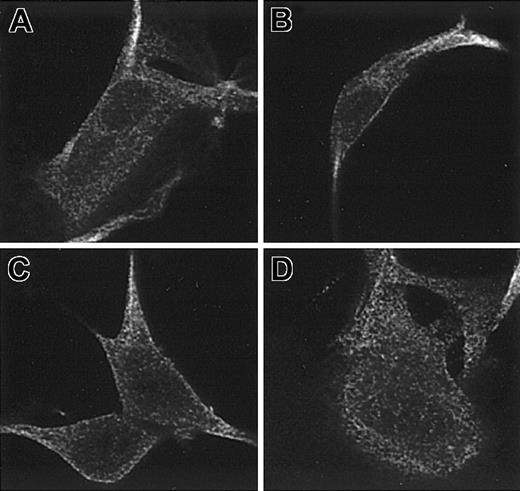
In mutant cno/cno mice, the 11-bp deletion leads to a frameshift that replaces the C-terminal 81 amino acids with 72 new amino acids (Figure 3B). Examination of the predicted secondary structure of CNO revealed that the wild-type protein has an unstructured amino terminus of approximately 55 amino acids followed by a domain with high α-helical content (45%) containing 2 coiled-coil domains. These structures mediate protein-protein interactions.32 The first coiled coil spans amino acids 80-107 and the second 141-165. The mutant protein has reduced α-helical content (33%) and lacks the second coiled coil. To assess whether the dramatic change in primary and secondary structure resulted in mislocalization of the mutant protein within cells, we transfected fibroblasts with either a full-length normal or mutant FLAG-tagged cDNA. By immunofluorescence staining, the distribution of the mutant CNO protein appears essentially normal, indicating that the altered C-terminal amino acid sequence does not affect localization of the protein (Figure 4 A-D).
Distribution of the normal and mutant CNO proteins in stably transfected fibroblasts. C57Bl/6J fibroblasts were transfected with pCMV-Tag4 FLAG expressing the normal (A-B) and mutant (C-D) CNO proteins and stained with anti-FLAG antibody. No disruption of the overall distribution of the mutant CNO protein is discernable. Original magnification × 1000.
Distribution of the normal and mutant CNO proteins in stably transfected fibroblasts. C57Bl/6J fibroblasts were transfected with pCMV-Tag4 FLAG expressing the normal (A-B) and mutant (C-D) CNO proteins and stained with anti-FLAG antibody. No disruption of the overall distribution of the mutant CNO protein is discernable. Original magnification × 1000.
Wild-type but not mutant cappuccino coassembles with components of BLOC-1
Both pallidin and the muted protein show predicted secondary structures that are strikingly similar to CNO.20 Each contains an unstructured amino terminal domain followed by a domain of high α-helical content containing 2 coiled coils. Pallidin and muted interact in the BLOC-1 complex with at least 3 other proteins of approximately 32 kDa, 20 kDa, and 15 kDa.20 To examine whether CNO could be part of this complex, we produced antipeptide antibodies to the N-terminus of CNO and to the C-terminus of the mutant protein. In cultured normal (C57BL/6J) fibroblasts, the wild-type CNO, pallidin and muted proteins can be detected by immunoprecipitation-recapture assays (Figure 5A). As expected, the mutant CNO protein (CNO*) was not detected in these cells. In pallid fibroblasts, no pallidin is detected, but levels of CNO and muted are also significantly reduced compared with their levels in C57BL/6J fibroblasts (Figure 5B). Likewise, in cappuccino fibroblasts, reduced levels of pallidin and muted are seen, and the mutant CNO protein (CNO*) is now present (Figure 5C). These data suggest that CNO, pallidin, and muted may interact and that the absence of one protein destabilizes the others.
CNO, pallidin, and muted interact. (A-C) Fibroblasts from C57BL/6 (A), pallid (B), and cappuccino (C) fibroblasts were metabolically labeled with 35S-methionine and subjected to immunoprecipitation-recapture assays using preimmune serum (PI) or the combination of antibodies indicated at the top of each panel. In this technique, tissue extracts are subjected to immunoprecipitation under nondenaturing conditions. Antibody-protein complexes are recovered using protein A Sepharose, denatured, and immunoprecipitated again using the same or a different antibody. Relative to C57BL/6 fibroblasts, CNO and muted (MU) are decreased in pallid fibroblasts, as are pallidin (PA) and muted (MU) in cappuccino fibroblasts, suggesting that these proteins interact. The mutant form of CNO (CNO*) is only detectable in cappuccino fibroblasts, as expected. (D) Immunoprecipitation-recapture assays in H4 cells reveal that pretreatment with CNO antibodies precipitates PA and pretreatment with PA antibodies precipitates CNO, indicting an interaction between CNO and PA. The doublet seen with anti-PA is characteristic of H4 cells.20 Its origin is unknown, but may represent proteolysis during handling. (E) Pretreatment with CNO antibodies precipitates MU in H4 cells, indicating an interaction between and CNO and MU. (F) The mutant form of CNO (CNO*) is unable to associate with pallidin in fibroblasts, indicating that failure to produce an intact BLOC-1 complex underlies the HPS phenotype in cno/cno mice. (G) CNO does not interact with the σ3 subunit of the AP3 complex in H4 cells. The migration of molecular weight markers (kDa) is shown to the left of each panel.
CNO, pallidin, and muted interact. (A-C) Fibroblasts from C57BL/6 (A), pallid (B), and cappuccino (C) fibroblasts were metabolically labeled with 35S-methionine and subjected to immunoprecipitation-recapture assays using preimmune serum (PI) or the combination of antibodies indicated at the top of each panel. In this technique, tissue extracts are subjected to immunoprecipitation under nondenaturing conditions. Antibody-protein complexes are recovered using protein A Sepharose, denatured, and immunoprecipitated again using the same or a different antibody. Relative to C57BL/6 fibroblasts, CNO and muted (MU) are decreased in pallid fibroblasts, as are pallidin (PA) and muted (MU) in cappuccino fibroblasts, suggesting that these proteins interact. The mutant form of CNO (CNO*) is only detectable in cappuccino fibroblasts, as expected. (D) Immunoprecipitation-recapture assays in H4 cells reveal that pretreatment with CNO antibodies precipitates PA and pretreatment with PA antibodies precipitates CNO, indicting an interaction between CNO and PA. The doublet seen with anti-PA is characteristic of H4 cells.20 Its origin is unknown, but may represent proteolysis during handling. (E) Pretreatment with CNO antibodies precipitates MU in H4 cells, indicating an interaction between and CNO and MU. (F) The mutant form of CNO (CNO*) is unable to associate with pallidin in fibroblasts, indicating that failure to produce an intact BLOC-1 complex underlies the HPS phenotype in cno/cno mice. (G) CNO does not interact with the σ3 subunit of the AP3 complex in H4 cells. The migration of molecular weight markers (kDa) is shown to the left of each panel.
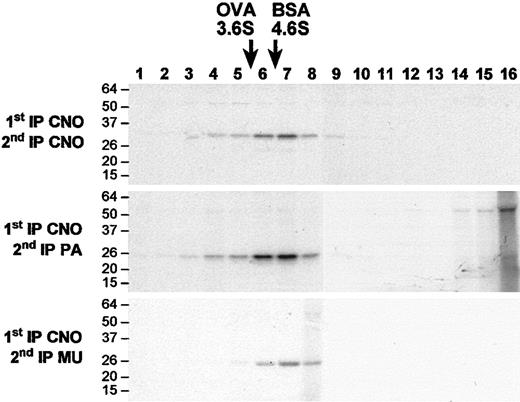
In immunoprecipitation-recapture assays in H4 human neuro-glioma cells, which express high levels of pallidin,20 wild-type CNO coimmunoprecipitates with both pallidin and muted (Figure 5D-E). Notably, the mutant CNO protein is unable to coassemble with pallidin in cultured fibroblasts (Figure 5F). Analysis of H4 extracts by sedimentation velocity on sucrose gradients followed by immunoprecipitation-recapture reveals that the native CNO, pallidin, and muted proteins sediment as a complex with a sedimentation coefficient of about 5.1 Svedberg units (Figure 6), confirming that CNO is a component of BLOC-1. Previous studies19,20 showed that neither pallidin nor muted interact with the components of the AP-3 complex. Likewise, CNO does not interact with the AP-3 complex; the β3A subunit of AP-3 appears at normal levels on Western blots of platelets and megakaryocytes from cno/cno mice (not shown), and the σ3 subunit of AP-3 does not coimmunoprecipitate with CNO in H4 cells (Figure 5G).
Sedimentation velocity on sucrose gradients.35S-methionine–labeled H4 cells were extracted and sedimented on a linear sucrose gradient. Sixteen fractions were collected for immunoprecipitation-recapture experiments using the combination of antibodies shown to the left of each panel. CNO, pallidin and muted all peak in fractions 6 and 7, and pallidin (middle panel) and muted (bottom panel) coprecipitate with CNO. These data indicate that native CNO is part of a complex that includes pallidin and muted. The position of standards, ovalbumin (OVA, 3.6 Svedberg units) and bovine serum albumin (BSA, 4.6 Svedberg units) are indicated at the top. The migration of molecular weight markers (kDa) are shown to the left of each panel.
Sedimentation velocity on sucrose gradients.35S-methionine–labeled H4 cells were extracted and sedimented on a linear sucrose gradient. Sixteen fractions were collected for immunoprecipitation-recapture experiments using the combination of antibodies shown to the left of each panel. CNO, pallidin and muted all peak in fractions 6 and 7, and pallidin (middle panel) and muted (bottom panel) coprecipitate with CNO. These data indicate that native CNO is part of a complex that includes pallidin and muted. The position of standards, ovalbumin (OVA, 3.6 Svedberg units) and bovine serum albumin (BSA, 4.6 Svedberg units) are indicated at the top. The migration of molecular weight markers (kDa) are shown to the left of each panel.
Human mutation screening
We screened 18 HPS patients with no defects in HPS1, HPS2 (APB31), HPS3, or HPS4 for CNO mutations by amplification of gDNA from cultured fibroblasts and sequencing. No defects were observed.
Discussion
Using a classical positional cloning approach, we have identified the gene defect in cappuccino, a mouse model of HPS. The normal cno gene is ubiquitously expressed and encodes a novel 215-amino acid protein that is a subunit of BLOC-1. In mutant mice, a deletion leads to frameshift. As a result, 72 new amino acids are encoded that replace the wild-type C-terminal 82 amino acids and significantly alter the predicted secondary structure by eliminating a coiled-coil domain. Interestingly, the expression and subcellular localization of the mutant protein are not affected by the drastically altered C-terminal sequence. However, the ability of the protein to interact with components of the BLOC-1 complex is abolished, indicating that failure to produce an intact BLOC-1 complex underlies the HPS phenotype in cno/cno mice as well as pa and mu homozygotes. Three unidentified proteins of about 32, 20, and 15 kDa are known to interact with pallidin and muted in BLOC-1.20 Results of coprecipitation and cosedimentation indicate that CNO is the previously unidentified 32-kDa component of BLOC-1. Thus, CNO shares with pallidin and the muted protein the property of migrating at a higher than predicted apparent molecular mass (Figures 5 and 6).
The wild-type CNO protein shows no significant homologies to any protein except its human counterpart. It is noteworthy that pallidin, CNO, and the muted protein share strikingly similar predicted secondary structures and appear to be confined to metazoans.20 Similarly, no yeast orthologues exist for the pale ear mutation (ep, HPS1) or the light ear mutation (le, HPS4). In contrast, components of the AP-3 complex are found in all eukaryotes. These observations suggest that both the BLOC-1 and HPS1/HPS4 complexes evolved to serve specialized functions in the biogenesis of those lysosome-related organelles (platelet dense bodies, melanosomes) that are specific to higher eukaryotes.
In humans, defects in HPS1 and HPS4 result in the most severe forms of HPS, emphasizing the critical function of the recently evolved HPS1/HPS4 complex in the biogenesis of lysosome-related organelles in man. Notably, the pa, mu, and cno mouse mutations are the most severe of all, even more so than the HPS1 and HPS4 orthologues. Interestingly, of the 9 published (including cno) mouse HPS genes, only the 3 known components of the BLOC-1 complex and mocha (mh), which encodes another component (δ) of the AP-3 complex, have not been associated with human disease. Pale ear, pearl, cocoa, and light ear are orthologous to HPS1-4, respectively, and ashen encodes RAB27a, which has been associated with Griscelli syndrome in humans.33 Moreover, while this manuscript was in press, the mouse ruby-eye-2 (ru2) and ruby-eye (ru) mouse HPS mutations were cloned and shown to be orthologous to human HPS-5 and HPS-6, respectively.39 In a total of 142 HPS patients screened to date,9,34, 35, 36, 37, 38 no defects in the known BLOC-1 components have been detected, suggesting that BLOC-1 function is critical in humans or, alternatively, that BLOC-1 defects in humans are exceedingly rare.
Although BLOC-1 clearly functions in the development of lysosome-related organelles, its precise mechanism of action remains unknown. Some clue to the function of BLOC-1 may come from pallidin, which binds syntaxin-13, a SNARE protein involved in vesicle fusion and docking.14 This suggests that the BLOC-1 complex might be involved in vesicle tethering and fusion processes. Elucidation of the remaining components and the precise function of BLOC-1 will lead to a better understanding of the biogenesis of platelet dense bodies and melanosomes in higher eukaryotes and the pathophysiology of HPS.
Note: The mouse cappuccino gene name and symbol (cno) have been approved by the Mouse Genome Nomenclature Committee (http://www.informatics.jax.org/mgihome/nomen/index.shtml). The mouse gene is not related to the Drosophila cappuccino (capu) gene.40
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2003-01-0020.
Supported by National Institutes of Health grant HL55321 (L.L.P.) and RR01183 (The Jackson Laboratory Mouse Mutant Resource) and the National Cancer Institute CA34196 (The Jackson Laboratory). Presented in abstract form at the 44th Annual Meeting of the American Society of Hematology, Philadelphia, PA, December 6-10, 2002.28
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Eva Eicher and Sue Ackerman for critical review of the manuscript, Richard Swank for the generous gift of polyclonal antiserum to the muted protein, and Jennifer Smith for computer graphics.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal