Abstract
Monoclonal gammopathy of undetermined significance (MGUS) can transform to multiple myeloma (MM). In myeloma, mutated VHgenes with sequence homogeneity reveal a postfollicular origin. Previously, some MGUS cases showed mutated VH genes with intraclonal variation, indicating an earlier stage of arrest. We investigated progression from 2 of 2 MGUS to MM, in which VH genes confirmed clonal evolution. In one MGUS case, intraclonal heterogeneity was evident, and transformation to myeloma occurred rapidly with apparent homogeneity in the emergent clone. However, residual MGUS-derived sequences were detectable at this time. Heterogeneity in MGUS does not associate with benign disease, but it indicates an origin from a tumorigenic cell, most likely surface immunoglobulin+, undergoing somatic mutation. The remaining case displayed intraclonal homogeneity at the MGUS stage, conceivably resulting from a self-cloning outgrowth from MGUS with heterogeneity. Transformation can occur at either MGUS stage, but it involves a single cell in which somatic mutation is then silent.
Introduction
Monoclonal gammopathy of undetermined significance (MGUS) is defined by key criteria: a plasma cell infiltration of less than 10% of bone marrow mononuclear cells, serum monoclonal M-protein of equal to or less than 30 g/L, absence of lytic lesions, related anemia, hypercalcemia, and renal insufficiency.1 MGUS varies in disease outcome, in some cases transforming to multiple myeloma (MM). Little is known concerning the underlying pathways of this progression, or indeed if the same clone is always involved. In part, these questions are hampered by the paucity of suitable tumor material for longitudinal analysis. More recently, evolution from MGUS to MM has been shown to exhibit a chromosome 13q abnormality.2 3
Analysis of immunoglobulin variable (V) region genes now provides a means of identifying salient features of B-cell tumor biology. Tumor V genes retain an imprint of the clonal history of the cell of origin.4 They can reveal the impact of somatic mutation, most likely to have occurred in the germinal center (GC).5This is initiated in vitro by ligation of surface immunoglobulin (sIg) in normal and neoplastic B cells.6-8 Tumors residing in the GC display intraclonal variation in V gene sequence patterns, a product of ongoing mutational activity at this site.4 9
Extensive data have characterized a postfollicular origin for MM, with more than 100 VH gene sequences displaying homogeneity of clonal tumor-derived sequences.4 In contrast, 3 of 7 MGUS revealed significant intraclonal variation in VH sequence in tumor clones, suggesting at least in a subset of MGUS a maintenance of the mutational mechanism.10 We now report 2 cases, evaluated during progression from MGUS to myeloma by VH gene analysis. Our findings reveal that the same tumor clone evolves to malignant status, and, importantly, analysis of somatic mutations suggests different pathways of disease progression.
Study design
Patients
Case 1 presented with advanced lung cancer with pulmonary involvement and a lesion in TH10. Immunofixation identified a trace IgAλ band. Bone-marrow (BM) biopsy (sample 1/1) revealed normal composition, with plasma cells (PCs) lower than 5%. Magnetic resonance imaging revealed infiltration of most vertebra 4 months later. Serum M-component was demonstrated (total IgA, 64 g/L; M level, not determined [ND]), with 80% PCs in BM (sample 1/2). A retrospective classification of TH10 could not rule out an isolated plasmacytoma at first biopsy; however, this had excluded any clinical marrow involvement. Further, postmortem examination revealed disseminated myeloma and confirmed concomitant squamous-cell cancer with multiple pulmonary lesions, suggesting a possible metastatic TH10 spread.
Case 2 initially had an M-gradient in her serum electrophoresis (total IgG, 15.7 g/L; M level, ND; PCs, 5% of nucleated BM cells; sample 2/1) that progressed over 67 months (IgG, 26.6 g/L; BM PCs, 30%; sample 2/2).
Cell preparation and VH gene analysis
BM aspirates were taken for magnetic activated cell sorting of CD138+ cells.11 Total RNA isolation from CD138+ cells (1 × 104-2.6 × 106 cells) and cDNA preparation using oligo-dT or constant region (CH) primer were as described.12 cDNA was amplified using VH1-7 leader primers with 3′ IgG or IgA primer. Cloning, sequence analysis, and VH gene alignments were as reported.12 For VH genes, between 16 to 30 clones/sample were sequenced from 2 separate polymerase chain reactions (PCRs).
For each MGUS and MM sample, 246 to 258 bp of the tumor CHwere amplified and cloned to establish the Taqerror rate using single 5′ complementary determining region 3 (CDR3) and Cγ or Cα primers (916-3690 bp analyzed/sample).Taq error depends primarily upon the initial amount of template available. We used the same cDNA preparation and identical amounts of template input for VH and CHamplification. This corrected for variations in amount of RNA isolated and efficiency of cDNA synthesis, reflected in the range ofTaq error observed in CH(3.4 × 10−4 to 1.6 × 10−3bp−1). Replicate analysis confirmed Taqerror.
Results and discussion
Case 1 utilized V3-11 and case 2 V4-30.2in sequential samples. These showed evidence for extensive somatic mutations (nucleotide sequences have been deposited in the European Molecular Biology Laboratory database; accession numbersAJ536045-AJ536051).
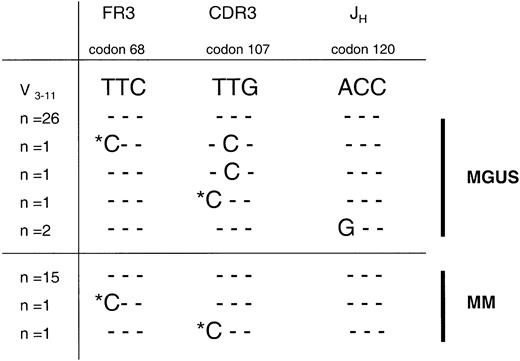
We observed intraclonal variation in sequence at the MGUS stage in case 1. Mutations in codons 107T>C and 120A>G were identified in 2 respective clones obtained from separate PCRs (Figure1). Additional single nucleotide changes were also observed (not shown), with a mutational frequency (1.6 × 10−3 bp−1) exceeding Taqerrors in CH (3.4 × 10−4 bp−1; 11 clones from 2 PCRs). This confirms heterogeneity in MGUS.10 It implicates a cell that is undergoing continual somatic mutation following tumorigenic arrest. This may be occurring in the GC environment, suggesting a less differentiated but sIg+ cell.6-8 This cell has acquired a proliferative potential, perhaps due to a chromosomal event 1 (Figure2). It allows progeny with heterogeneous sequences to survive and migrate to the marrow to differentiate further. This heterogeneity also exists in clonally derived Cμ transcripts, demonstrated in only one MGUS case to date,10and implicating the stage of isotype switch in event 1.
Intraclonal heterogeneity observed in tumor VH gene sequences at the MGUS stage in case 1 is identified as residual, persisting clones at the myeloma stage of disease.
The mutational frequency of variations in clonal sequences exceededTaq error in sample 1/1 from the MGUS stage, with repeated nucleotide changes seen in 2 clones from separate PCRs (codons 107 and 120) confirming intraclonal heterogeneity. In sample 1/2 from the MM stage, some of these mutations were again identified (codons 68 and 107), clearly excluding Taq error. The mutational frequency of intraclonal variation, however, was comparable with the backgroundTaq misincorporation rate in this sample, suggesting a cessation of somatic mutation at the MM stage. Most likely, variant clones here represent residual MGUS cells, as disease evolution was remarkably rapid. Only informative codon sequences at the nucleotide level are shown, with homology to the donor germ line gene V3-11 depicted as dashes. Mutations repeatedly observed in MGUS and MM samples are marked with an asterisk. Numbers of clones with each sequence varied (n = 1-26). FR3 indicates framework region 3; and JH, joining (H) gene.
Intraclonal heterogeneity observed in tumor VH gene sequences at the MGUS stage in case 1 is identified as residual, persisting clones at the myeloma stage of disease.
The mutational frequency of variations in clonal sequences exceededTaq error in sample 1/1 from the MGUS stage, with repeated nucleotide changes seen in 2 clones from separate PCRs (codons 107 and 120) confirming intraclonal heterogeneity. In sample 1/2 from the MM stage, some of these mutations were again identified (codons 68 and 107), clearly excluding Taq error. The mutational frequency of intraclonal variation, however, was comparable with the backgroundTaq misincorporation rate in this sample, suggesting a cessation of somatic mutation at the MM stage. Most likely, variant clones here represent residual MGUS cells, as disease evolution was remarkably rapid. Only informative codon sequences at the nucleotide level are shown, with homology to the donor germ line gene V3-11 depicted as dashes. Mutations repeatedly observed in MGUS and MM samples are marked with an asterisk. Numbers of clones with each sequence varied (n = 1-26). FR3 indicates framework region 3; and JH, joining (H) gene.
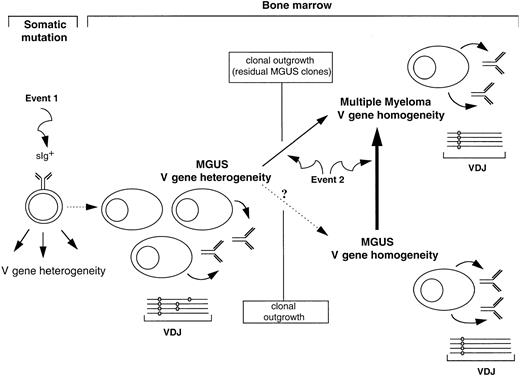
Pathways of clonal progression from MGUS to multiple myeloma.
Mutational patterns in VH genes define diversity in MGUS, with some cases showing intraclonal heterogeneity. Plasma cells in these MGUS cases most likely derive by maturation of a less differentiated sIg+ cell that is undergoing somatic mutation and has acquired a chromosomal event 1 to gain a proliferative potential. Transformation to myeloma can clearly occur in MGUS displaying heterogeneity, but does so in a single cell in which somatic mutation is silent and that acquires neoplastic event 2. This transformation may be rapid, and residual MGUS clones can still be identified at this time but will eventually be lost by clonal outgrowth as myeloma is homogeneous. A self-cloning process may also lead to the MGUS cases identified by intraclonal stability of sequence, which are more common. These cases can then transform to myeloma as a result of event 2. VDJ indicates functionally rearranged variable-diversity–joining gene.
Pathways of clonal progression from MGUS to multiple myeloma.
Mutational patterns in VH genes define diversity in MGUS, with some cases showing intraclonal heterogeneity. Plasma cells in these MGUS cases most likely derive by maturation of a less differentiated sIg+ cell that is undergoing somatic mutation and has acquired a chromosomal event 1 to gain a proliferative potential. Transformation to myeloma can clearly occur in MGUS displaying heterogeneity, but does so in a single cell in which somatic mutation is silent and that acquires neoplastic event 2. This transformation may be rapid, and residual MGUS clones can still be identified at this time but will eventually be lost by clonal outgrowth as myeloma is homogeneous. A self-cloning process may also lead to the MGUS cases identified by intraclonal stability of sequence, which are more common. These cases can then transform to myeloma as a result of event 2. VDJ indicates functionally rearranged variable-diversity–joining gene.
Following transformation to myeloma in this patient (sample 1/2; Figure1), the observed ongoing mutational activity appeared to be silenced, as the frequency of single nonclonal mutations (1.5 × 10−3 bp−1) was similar toTaq error (1.6 × 10−3 bp−1). However, we observed 2 mutational changes at codons 68T>C and 107T>C in single clones at the MM stage that were identical to mutations at the early stage of disease (Figure 1). These 2 mutations were detected in samples taken at 2 different disease stages, negating random PCR error. Most likely, they represent residual MGUS clones as transformation was rapid. In the absence of paired samples, a very low level of nonshared mutations in some MM clones would have been assigned as Taq error in V gene analysis. Reports of stability of myeloma V gene sequence indicate the rarity of identifying such persisting residual clones by current technology.4However, there have been sporadic reports that hint at this possibilty. In 1 of 4 MM cases with more than 40% tumor cells, 2 clonally-related subclones with heterogeneity were observed,13 possibly reflecting a narrowing of a wider variation that existed previously. In a further MM case, significant intraclonal diversity was noted in the IgA variant transcripts but not the tumor isotype, suggesting emergence of a dominant clone but with residual variants.14 Given that MM is clonally homogeneous even in Cμ tumor-derived sequences,15-17 this suggests that the final transformation event (event 2, Figure 2) occurs in one cell of the population, at a site where somatic mutation is silent, possibly the bone marrow.
In case 2, we observed intraclonal homogeneity at both the MGUS and MM stage, confirming previous observations in 4 of 7 MGUS cases.10 A self-cloning phenomenon may underlie this observed sequence homogeneity in MGUS, being analyzed at a stage where hypermutation has ceased and heterogeneity has been lost because of clonal outgrowth. Transformation to myeloma then occurs at the invariant stage, involving event 2 (Figure 2).
VH gene analysis has revealed diversity in MGUS and has suggested that transformation may involve different pathways. MGUS may derive from an tumorigenic sIg+ cell undergoing somatic mutation, which differentiates into Ig-secreting plasma cells. This population may persist and possibly self clone to homogeneity. However, it is susceptible to further transforming events that can occur in one cell at the heterogeneous or homogeneous stage, giving rise to homogeneous MM that is independent of the sIg+ population. Progression to myeloma then involves factors intrinsic to this cell.
We wish to thank Dr Judith Schuster and Irene Assmann for assistance in cell sorting.
Prepublished online as Blood First Edition Paper, January 16, 2003; DOI 10.1182/ blood-2002-09-2825.
Supported by a Novartis-UICC Translational Cancer Research Fellowship (funded by Novartis AG, Switzerland), the European Society for Medical Oncology (ESMO), the International Myeloma Foundation (United Kingdom) and The Leukaemia Research Fund (United Kingdom).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Surinder S. Sahota, Molecular Immunology Group, Tenovus Laboratory, Cancer Sciences Division, Southampton University Hospital Trust, Southampton, SO16 6YD, United Kingdom; e-mail: s.s.sahota@soton.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal