Abstract
Fanconi anemia complementation group C (Fancc)–deficient murine bone marrow progenitors demonstrate increased sensitivity to growth inhibition by interferon γ (IFNγ), tumor necrosis factor α (TNFα), and macrophage inflammatory protein 1α (MIP-1α). This property has been proposed as a possible pathogenic factor in the marrow failure seen in Fanconi anemia. Supporting our hypothesis that nitric oxide (NO) production might be a common effector in this sensitivity, we found that cytokine-mediated growth inhibition ofFancc−/− bone marrow cells was prevented by inhibiting NO synthase activity. Interestingly,Fancc−/− hematopoietic cells also exhibited increased growth inhibition on exposure to 2 distinct NO-generating agents, S-nitroso-N-acetyl-D, L-penicillamine (SNAP) and diethylenetriamine nitric oxide adduct (DETA/NO). In keeping with the sensitivity of Fancc−/− cells to IFNγ, inducible nitric oxide synthase (iNOS) levels and nitrite release were both increased following stimulation ofFancc−/− macrophages with this cytokine, either alone or in combination with bacterial lipopolysaccharide. Suggesting a plausible mechanism for the increased expression of iNOS, IFNγ-stimulated Fancc−/− macrophages generated higher levels of phospho-Stat1, a positive regulator ofinos (nos2) gene expression. These observations, while confined to C57BL/6 Fancc−/−hematopoietic cells, raise the possibility that nitric oxide has a role in the pathogenesis of Fanconi anemia.
Introduction
Fanconi anemia (FA) is an autosomal recessive disorder characterized by progressive bone marrow failure, hypersensitivity to DNA cross-linking agents, chromosomal instability, and a predisposition to acute myeloid leukemia.1,2Through complementation analysis, 6 FA genes (FANCA, FANCC, FANCD2, FANCE, FANCF, FANCG) have been identified whose gene products interact in a specific sequence within the cytoplasm with subsequent nuclear translocation.3-5 In the nucleus, additional FA proteins are then recruited to form a complex that is required for the eventual ubiquitination of FANCD2, a molecule that is part of the BRCA1-containing recognition/repair complex in response to DNA damage.6 As well as supporting the assembly of a multimeric FA protein complex, FANCC may have additional intrinsic roles. For example, the correction of mitomycin C toxicity requires FANCC to be cytoplasmic.7 In addition, via yeast 2-hybrid and other approaches, FANCC has been shown to interact with a variety of cytoplasmic and nuclear molecules, including the chaperone glucose-regulated protein 94 (GRP94),8 nicotinamide adenine dinucleotide phosphate (NADPH) cytochrome P450 reductase,9 the zinc finger–containing protein Fanconi anemia zinc finger (FAZF),10 and the phase II detoxification enzyme glutathione S-transferase P1-1 (GSTP1).11
Although FA genes are ubiquitously expressed in humans and mice, the principal pathological manifestation of FA mutations is progressive bone marrow (BM) failure. In keeping with this, a specific role for FANCC in the survival and/or proliferation of hematopoietic progenitor cells (HPCs) has been suggested.12 Interestingly,Fancc−/− HPCs were shown to be hypersensitive to the growth-inhibiting effects of 3 unrelated cytokines: interferon γ (IFNγ), tumor necrosis factor α (TNFα), and macrophage inflammatory protein 1α (MIP-1α).13-15 In keeping with these results, obtained inFancc−/− murine cells, HPCs fromFANCC-deficient individuals were shown to up-regulatefas and interferon response factor 1 (IRF-1) expression at significantly lower doses of IFNγ than those required for control cells.16 The apoptotic responses in these cells were mediated via the caspase 8–dependent activation of caspase 3.17 Paradoxically, although FANCC- andFancc-deficient cells demonstrated increased sensitivity to the growth-inhibitory effects of IFNγ, depending on the specificFANCC mutations involved,18 activation of signal transducer and activator of transcription 1 (STAT1) in response to IFNγ in both FANCC-deficient lymphoblastoid cells andFancc−/− embryonic fibroblasts was shown to be impaired.19 20
We hypothesized that a common mechanism might account for the inhibition of Fancc−/− hematopoietic cell growth by IFNγ, TNFα, and MIP-1α. Several lines of evidence pointed toward nitric oxide (NO) as a candidate for mediating the inhibitory effects of these 3 unrelated cytokines. NO, a free radical enzymatically generated from L-arginine, is capable of reacting with oxygen to yield various noxious species, ranging from stable anions to highly reactive peroxides.21 NO is involved in a wide variety of biological processes. For example, NO generated by endothelial NOS (eNOS) and neuronal NOS (nNOS) has been implicated in neuronal function, innate immune responses, tumor killing, control of vascular tone, and chemotaxis,22 while NO produced by iNOS in response to cell stimulation by a variety of factors (eg, proinflammatory cytokines, Fas-L, and bacterial cell wall components) plays a direct role in inflammatory responses.22 Both IFNγ and TNFα, known inhibitors of hematopoiesis,23,24 are capable of inducing iNOS expression, and hence NO production, from various cell types.25,26 Similarly, the chemokine MIP-1α, which was shown to suppress hematopoiesis,27,28 is able to trigger NO release from human peripheral blood mononuclear cells.29 Finally, there is evidence that NO itself is capable of suppressing hematopoiesis in vitro.30 Given this background, we set out to determine whether NO was involved in mediating the inhibitory effects of IFNγ, MIP-1α, and TNFα on Fancc−/−bone marrow cells, and also whether Fancc deficiency might be associated with an abnormal sensitivity to this agent.
Materials and methods
Mice and cell isolations
Fancc-deficient mice were generously provided by M. Buchwald (Hospital for Sick Children, Toronto, ON, Canada).31Fancc−/− mice on a C57BL/6 background (N = 7) and Fancc+/+ littermate controls were killed at 8 weeks of age by CO2 asphyxiation for BM harvesting or macrophage isolation. Locus-specific polymerase chain reaction (PCR) was used to genotype the mutant mice. Viral antibody–free mice were housed in the Animal Resources Center barrier facility according to protocols approved by the Animal Care Committee of the University of Calgary.
Unfractionated BM samples were collected by flushing both femurs from each mouse with α Modified Eagle Medium (αMEM) with 5% fetal calf serum (FCS). Cell viability was more than 90% in all samples, as determined by trypan blue exclusion. HPC isolations were performed as previously described.32 Briefly, lineage-depleted (Lin−) samples were collected by resuspending unfractionated BM cells at 5.0 × 107nucleated cells per milliliter in phosphate-buffered saline (PBS) with 2% fetal bovine serum (FBS) plus 5% rat serum for 15 minutes at 4°C. Samples were incubated with an antibody cocktail (CD5, CD11b, CD45R, GR1, 7-4, and TER-119) and subsequently with an antibiotin tetrameric antibody complex (antibodies from StemCell Technologies, Vancouver, BC, Canada); each incubation was for 15 minutes at 4°C. A magnetic colloid was added for cell separation as recommended (StemCell Technologies). The BM samples were applied to a primed, 0.3-inch magnetic column and washed 3 times with PBS containing 2% FBS. HPC (Lin−) cells were grown in Iscove Modified Dulbecco Medium (IMDM) with 15% FBS, 50 ng/mL stem cell factor (SCF), 10 ng/mL interleukin 3 (IL-3), and 10 ng/mL IL-6 in a 37°C incubator with 5% CO2 in air, humidity 95% or higher. Viability was measured by trypan blue exclusion. Cells were plated at a density of 2.0 × 105 in 1 mL media, in triplicate in a 24-well plate (n = 4 mice for each genotype). IFNγ and NG-monomethyl-L-arginine (L-NMMA) were diluted directly into IMDM (with growth factors) at the indicated concentrations.
For bone marrow–derived macrophage (BMDM) cultures, BM samples were centrifuged at 1200 rpm and resuspended at a density of 107cells/mL in a 10-cm2 dish in Dulbecco Modified Eagle Medium (DMEM) with 10% FCS and 5% colony-stimulating factor 1 (CSF-1)–conditioned (cell-free) media. The next day all suspension cells were removed to sterile 50-mL Falcon tubes and the adherent population discarded. Cells were centrifuged and resuspended in twice the original volume of DMEM with 10% FCS and 5% CSF-1–conditioned medium, then plated at a density of 8.5 × 106 cells per well of a 6-well tissue-culture dish and allowed to grow in a humidified 5% CO2 incubator for 8 to 10 days or until cultures became confluent. Fresh media was added to the cultures every third day.
For peritoneal macrophage isolation, 1 mL 3% thioglycollate (suspended in PBS and autoclaved) was injected intraperitoneally. On day 5 after injection, the mice were killed and 10 mL DMEM with 10% FCS was injected into the peritoneal cavity. The cell suspension was then collected using a syringe and 18-gauge needle. After centrifugation at 1200 rpm for 5 minutes, the cell pellet was resuspended at 0.75 × 106 cells/mL media and incubated for 5 hours. Adherent cells were washed twice with warm PBS to remove suspension cells and DMEM with 10% FCS was added. The following day recombinant murine IFNγ (10 ng/mL; R&D Systems), with or without lipopolysaccharide (LPS; 100 ng/mL), was added to the macrophage cultures. Supernatants were collected for nitrite assay and cell protein lysates were also prepared for immunoblotting experiments.
Clonogenic assays for committed hematopoietic progenitor cells
Methylcellulose assays for committed progenitors were performed as previously described.32 Briefly, unfractionated BM cells were plated in 1.1 mL 1% methylcellulose media supplemented with erythropoietin, IL-3, IL-6, and SCF (StemCell Technologies). Cells were cultured at a density of 8.5 × 103 cells per 35-mm dish (each sample done in duplicate) and the dishes incubated for 10 days at 37°C, 5% CO2, humidity 95% or higher. Methylcellulose plates with fewer than 12 colonies were not included in the data set. Colonies (> 20 cells) were counted (and scored morphologically) using a gridded stage on an inverted microscope.
Immunoblotting and densitometry
Macrophages were lysed in phosphorylation solubilization buffer (PSB; 50 mM HEPES [N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid], 100 mM NaF, 10 mM Na4P2O7, 2 mM Na3VO4, 2 mM EDTA [ethylenediaminetetraacetic acid], 2 mM NaMoO4, 1% Triton X freshly added; pH 7.35) in the presence of protease inhibitors (leupeptin [1:1000], aprotinin [1:1000], phenylmethylsulfonyl fluoride [PMSF; 1:1000]; Roche Diagnostics, Mannheim, Germany). Whole cell lysates were centrifuged for 5 minutes at 12 000 rpm to remove cellular debris and supernatants were collected in tubes and stored at −20°C. Protein concentrations were determined by Bradford method–based assay. Lysate volumes corresponding to 40 and 80 μg total protein (for the iNOS and Stat1 immunoblots, respectively) were diluted 6:1 with Laemmli sample buffer and then boiled for 5 minutes prior to electrophoresis. Total cell lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) at 150 V and transferred to polyvinylidine difluoride (PVDF) membranes by electroblotting using a semidry transfer method at 25 V for 45 minutes at room temperature in a solution containing semidry transfer buffer (192 mM glycine, 25 mM Tris [tris(hydroxymethyl)aminomethane], 10% SDS, and 20% methanol). Filters were blocked for 1 hour at room temperature in TBST (10 mM Tris, pH 8.0; 150 mM NaCl; and 0.05% Tween-20) containing 5% bovine serum albumin (BSA). Filters were incubated overnight at 4°C in TBST with 1% BSA with one of the following antibodies (working dilutions shown): anti-iNOS (1:1000; Upstate Biotechnology, Lake Placid, NY); anti-Stat1 (1:1000) or anti–P-Stat1 (1:1000; Cell Signaling Technology, Beverly, MA); or anti–α-tubulin (1:500; Sigma, St Louis, MO). After 3 TBST washes, filters were incubated for 1 hour at room temperature with a horseradish peroxidase–conjugated secondary antibody (Jackson ImmunoResearch Labs, West Grove, PA). Proteins were detected by chemiluminescence (Amersham, Arlington Heights, IL), using a Fluor-S Multi Imager equipped with densitometry software (Bio-Rad Laboratories, Mississauga, ON, Canada).
Nitrite assay
Macrophage culture supernatants were collected and stored at −20°C. For assays, 10 μL 30% (wt/vol) ZnSO4 was added to microfuge tubes containing 250-μL samples of each supernatant, mixed using a vortex, and incubated at room temperature for 15 minutes. Samples were centrifuged at 4000 rpm for 5 minutes to collect the pellets and the cleared supernatants were transferred to a microfuge tube containing 0.5-g cadmium beads. Samples were nutated overnight at room temperature, then transferred to a clean tube, where the cadmium beads were removed and the supernatants cleared by centrifugation at 10 000 rpm for 5 minutes. Then 100 μL nitrite standards and 100 μL of each sample were loaded in duplicate onto a 96-well ImmunoSorp ELISA plate (NUNC, Rochester, NY). Color Reagent 1 (50 μL) was added to each well and the samples were briefly mixed; then 50 μL Color Reagent 2 was added to each well and the whole plate was incubated at room temperature for 15 minutes. Color reagents and nitrite standards were from Oxford Biomedical Research (Oxford, MI). Absorbance was measured at 540 nm in a Multiskan Ascent Microtiter Plate reader (Dynex Labsystems, Chantilly, VA). Data were collected as micromols of nitrite based on a standard curve done for each individual plate and normalized to total protein (Bradford method).
Flow cytometry
For flow cytometry, 1 × 106 cells were resuspended in 500 μL PBS with 2% FCS (fluorescence-activated cell sorter [FACS] buffer) and blocked on ice with 1 μg of anti-FcγRIIb (2.4G2; Pharmingen, Mississauga, ON, Canada) for 30 minutes. Cells were washed once in FACS buffer and then stained with one of the following antibodies for 1 hour at 4°C; 0.5 μg anti-CD11b–fluorescein isothiocyanate (FITC), 0.5 μg anti-CD14-FITC or 0.5 μg anti-CD119-FITC (Pharmingen, Mississauga, ON, Canada). Cells were washed 3 times with FACS buffer and resuspended in 500 μL buffer before analysis on a FACSCalibur (Becton Dickinson, Mountain View, CA) flow cytometer equipped with CellQuest software (Becton Dickinson). Peritoneal macrophages were analyzed using antibodies against cell surface markers: CD11b, CD14, and CD119 (IFNγR α chain). There was no difference in the percentage of cells staining with any of these antibodies betweenFancc−/− and wild-type macrophage samples (n = 3 per genotype; data not shown).
Chemicals
Diethylenetriamine nitric oxide adduct (DETA/NO) was purchased from Sigma–RBI (St Louis, MO). S-nitroso-N-acetyl-D, L-penicillamine (SNAP) and NG-monomethyl-L-arginine (L-NMMA) were purchased from Calbiochem (San Diego, CA). Recombinant murine IFN-γ, TNF-α, and MIP-1α were purchased from R&D Systems (Minneapolis, MN). All chemicals were diluted in αMEM.
Statistical methods
The Student t test (Microsoft Excel) was used when analyzing the results. P < .05 was considered significant.
Results
Cytokine inhibition of Fancc−/−BM colony growth and response to L-NMMA
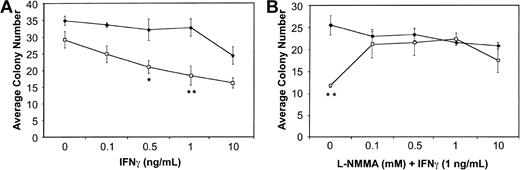
BM cells from wild-type and Fancc−/− mice were plated in methylcellulose in the presence of increasing doses of IFNγ. Consistent with previous reports,13 14Fancc−/− BM cells exhibited a dose-dependent inhibition of colony number in response to IFNγ. Figure1A represents the average total colony number, including both myeloid and erythroid colonies, with inhibition of total colony number from Fancc−/− mice being maximal at 1 ng/mL (P = .03). We did observe a modest difference in IFNγ sensitivity based on colony type, withFancc−/− myeloid and erythroid colonies being maximally inhibited by 1 ng/mL, and 0.5 ng/mL IFNγ (P = .04 and .05, respectively). Both myeloid and erythroid colonies were significantly different from untreated (no-IFNγ) Fancc−/− controls at all doses tested.
Cytokine-mediated inhibition of
Fancc−/− BM colony formation is blocked by the addition of L-NMMA. (A) Hematopoietic colony formation by Fancc−/− (■) and littermate wild-type (♦) BM progenitor cells plated in methylcellulose in the presence of increasing concentrations of IFNγ. (B) Colony formation by BM cells from wild-type (♦) and Fancc−/−(■) mice grown in the presence of IFNγ (1 ng/mL) and increasing concentrations of L-NMMA. Data points represent the average number of colonies counted. n = 3 mice per genotype. *P < .05; **P < .005.
Cytokine-mediated inhibition of
Fancc−/− BM colony formation is blocked by the addition of L-NMMA. (A) Hematopoietic colony formation by Fancc−/− (■) and littermate wild-type (♦) BM progenitor cells plated in methylcellulose in the presence of increasing concentrations of IFNγ. (B) Colony formation by BM cells from wild-type (♦) and Fancc−/−(■) mice grown in the presence of IFNγ (1 ng/mL) and increasing concentrations of L-NMMA. Data points represent the average number of colonies counted. n = 3 mice per genotype. *P < .05; **P < .005.
BM cells were then plated in the presence of 1 ng/mL IFNγ and increasing concentrations of the iNOS inhibitor NG-monomethyl-L-arginine (L-NMMA). As shown in Figure1B, at 0.1, 0.5, and 1.0 mM L-NMMA there was complete rescue of Fancc−/− total colony formation in the methylcellulose cultures. The average totalFancc−/− colony numbers at these 3 L-NMMA doses were not significantly different from those generated by the wild-type controls. In contrast, they were significantly different (P = .01, .02, and .03 for L-NMMA 0.1, 0.5, and 1.0 mM, respectively) from Fancc−/− colony numbers when the latter were grown in the presence of 1 ng/mL IFNγ without L-NMMA. Interestingly, while myeloid colonies generated fromFancc−/− progenitors were rescued at all doses of L-NMMA, compared with wild-type controls, they were significantly different from Fancc−/− progenitors grown in 1 ng/mL IFNγ alone only when grown in the presence of 0.5 mM L-NMMA (data not shown). Erythroid colony numbers generated fromFancc−/− progenitors were not significantly different from those of wild-type mice at all doses of L-NMMA tested; however, they were significantly different fromFancc−/− colonies grown in 1 ng/mL IFNγ alone (data not shown).
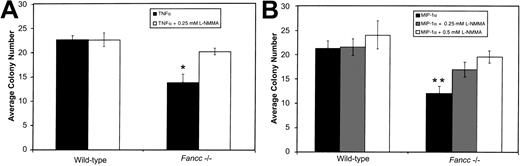
Given the above results, and the ability of both TNFα and MIP-1α to induce NO production by hematopoietic cells, we determined whether inhibition of colony formation by these factors was also preventable by L-NMMA. BM cells were plated in methylcellulose in the presence of either 0.5 ng/mL TNFα or 1 ng/mL MIP-1α, with or without L-NMMA (Figure 2A and 2B, respectively). As Figure 2A shows, the growth of Fancc−/−progenitors treated with TNFα alone was significantly suppressed as compared with wild-type TNFα-treated cultures (P = .007). As seen in the case of the IFNγ-treated BM progenitor cultures, TNFα-mediated inhibition ofFancc−/− total colony growth was blocked by the addition of 0.25 mM L-NMMA. Growth of TNFα-treatedFancc−/− cultures was significantly different from that of Fancc−/− cultures grown in the presence of TNFα plus L-NMMA (P = .01).
L-NMMA prevents TNFα- and MIP-1α–mediated inhibition of
Fancc−/− BM colony growth.(A) In the presence of 0.5 ng/mL TNFα, colony formation byFancc−/− BM cells was reduced, as compared with wild-type littermate controls. This was prevented by addition of 0.25 mM L-NMMA. (B) Reduction in Fancc−/− BM cell colony numbers in the presence of MIP-1α (1 ng/mL) was inhibited by the addition of 0.5 mM L-NMMA. Data points represent the average number of colonies counted. n = 4 mice per genotype. *P < .05; **P < .005.
L-NMMA prevents TNFα- and MIP-1α–mediated inhibition of
Fancc−/− BM colony growth.(A) In the presence of 0.5 ng/mL TNFα, colony formation byFancc−/− BM cells was reduced, as compared with wild-type littermate controls. This was prevented by addition of 0.25 mM L-NMMA. (B) Reduction in Fancc−/− BM cell colony numbers in the presence of MIP-1α (1 ng/mL) was inhibited by the addition of 0.5 mM L-NMMA. Data points represent the average number of colonies counted. n = 4 mice per genotype. *P < .05; **P < .005.
As Figure 2B shows, Fancc−/− progenitors treated with MIP-1α were significantly inhibited as compared with MIP-1α–treated wild-type cultures (P = .003). In contrast, growth of Fancc−/− BM cells in the presence of MIP-1α plus 0.25 mM L-NMMA was no longer significantly different from that of wild-type cells maintained under the same conditions; also, the average total colony number was not significantly different from Fancc−/− MIP-1α–treated cultures. Increasing L-NMMA to 0.5 mM, however, restoredFancc−/− progenitor growth to wild-type levels under the same conditions, and this was significantly different from cultures lacking L-NMMA (P = .02). As rescue of MIP-1α–treated progenitors occurred at a somewhat higher dose of L-NMMA, it raised the possibility that at the concentration tested, MIP-1α might be generating higher levels of NO than either of the other 2 cytokines. Erythroid colonies grown in either TNFα or MIP-1α were rescued by L-NMMA and were significantly different fromFancc−/− colonies grown in cytokine alone, while myeloid colonies from L-NMMA cultures were not different fromFancc−/− colonies grown in cytokine alone (data not shown). Together, these results implicate NO generation as a common mechanism through which these 3 cytokines bring about inhibition of Fancc−/− colony formation.
Sensitivity of Fancc−/−BM cells to NO donors
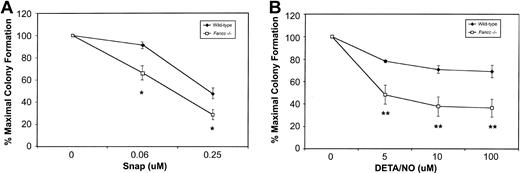
Based on their sensitivity to 3 different NO-generating cytokines, we hypothesized that Fancc−/− BM cells might be hypersensitive to NO. To test this, colony formation was carried out in the presence of 2 mechanistically distinct NO donors. First, BM cells from wild-type and Fancc−/− mice were plated in increasing concentrations of S-nitroso-N-acetyl-D, L-penicillamine (SNAP). As depicted in Figure3A, both wild-type andFancc−/− progenitors exhibited dose-dependent inhibition of colony numbers in the presence of this compound, withFancc−/− progenitors generating fewer colonies at 0.06 and 0.25 μM SNAP as compared with wild-type controls (P = .04 and .05, respectively). Since SNAP produces NO over a wide concentration range and generates additional reactive nitrogen and oxygen species, in addition to sulfhydryls,21 any response of the cells to this chemical would be difficult to attribute solely to NO.33 Thus, wild-type and Fancc−/− colony formation was also assessed in the presence of diethylene triamine nitric oxide adduct (DETA/NO). This member of the NONOate class of NO donors, with a half-life of approximately 20 hours in cell culture, has less potential for generating unwanted reactive species.33 As shown in Figure 3B, there was a significant reduction inFancc−/− BM total colony formation, commencing at 5 μM DETA/NO (P = .004), with progressive reductions being observed up to the highest concentration tested, 100 μM (P = .0009). Furthermore, we observed a strong trend for erythroid colony formation to be preferentially inhibited (data not shown). As seen in Figure 3B, the effect of DETA/NO on wild-type colony formation was minimal at all concentrations tested. These results suggested that committed hematopoietic progenitors ofFancc−/− mice were more sensitive than control cells to the growth-inhibitory effects of 2 distinct NO donors.
Fancc−/− BM progenitors demonstrate increased sensitivity to NO-generating compounds.
(A) Committed progenitor cell growth in the presence of increasing concentrations of the NO donor SNAP; 100% represents 89.5 total colonies (control) and 70.5 total colonies (Fancc−/−). (B) Growth of committed progenitor cells in the presence of increasing concentrations of DETA/NO; 100% represents 104.5 total colonies (control) and 82.5 total colonies (Fancc−/−). Percentage maximal colony formation was determined by dividing the number of colonies scored at a given concentration of NO donor by the number of colonies scored in the absence of the NO donor. n = 3 or 4 mice per genotype. *P ≤ .05; **P < .005.
Fancc−/− BM progenitors demonstrate increased sensitivity to NO-generating compounds.
(A) Committed progenitor cell growth in the presence of increasing concentrations of the NO donor SNAP; 100% represents 89.5 total colonies (control) and 70.5 total colonies (Fancc−/−). (B) Growth of committed progenitor cells in the presence of increasing concentrations of DETA/NO; 100% represents 104.5 total colonies (control) and 82.5 total colonies (Fancc−/−). Percentage maximal colony formation was determined by dividing the number of colonies scored at a given concentration of NO donor by the number of colonies scored in the absence of the NO donor. n = 3 or 4 mice per genotype. *P ≤ .05; **P < .005.
Effect of L-NMMA on IFNγ-treatedFancc−/−HPCs
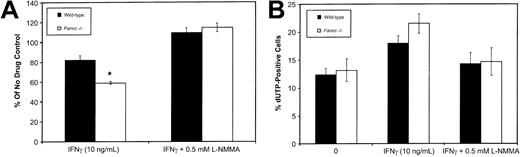
To establish whether IFNγ-induced NO production would inhibit the growth of HPC-enriched populations, we isolated Lin−cells (obtained using Lin+ cell depletion as described in “Materials and methods”) and maintained these in the presence of 50 ng/mL SCF, 10 ng/mL IL-3, and 10 ng/mL IL-6. Flow cytometry of Lin− cells using monoclonal antibodies against CD34, Sca1, and c-kit revealed no significant differences between the wild-type andFancc−/− mice. Immediately following isolation, column-purified Lin− HPCs were cultured in the presence of IFNγ (10 ng/mL), either with or without 0.5 mM L-NMMA. After 3 days in culture, cell counts were used to ascertain the effects of these growth conditions (Figure 4A). IFNγ inhibited the growth of both wild-type andFancc−/− HPCs (to 82% and 58% of the untreated controls, respectively; P = .04 forFancc−/− HPCs only). However, when HPCs were cultured in the presence of IFNγ plus 0.5 mM L-NMMA, HPC growth was fully restored in both cultures. As a 3-day culture period was unlikely to allow the differentiation of early progenitors into macrophages and granulocytes, this suggested that IFNγ was exerting a direct effect on progenitors. However, we are unable to exclude the possibility that an indirect effect on HPCs is mediated by IFNγ activation of small contaminating populations of mature cells, such as macrophages.
Inhibition of
Fancc−/− HPC growth by IFNγ was prevented by L-NMMA. (A) Fancc−/−(■) and control (▪) HPCs grown in the presence of IFNγ (10 ng/mL) with and without 0.5 mM L-NMMA. Bars indicate percentage of control (no cytokine added) for each of the 2 genotypes. (B) Flow cytometry TUNEL analysis showing percentage of cell nuclei that were dUTP positive in untreated cultures (0), cultures treated with IFNγ (10 ng/mL), and cultures treated with IFNγ plus 0.5 mM L-NMMA. Flow data were based on 10 000 events. n = 4 mice per genotype. *P < .05.
Inhibition of
Fancc−/− HPC growth by IFNγ was prevented by L-NMMA. (A) Fancc−/−(■) and control (▪) HPCs grown in the presence of IFNγ (10 ng/mL) with and without 0.5 mM L-NMMA. Bars indicate percentage of control (no cytokine added) for each of the 2 genotypes. (B) Flow cytometry TUNEL analysis showing percentage of cell nuclei that were dUTP positive in untreated cultures (0), cultures treated with IFNγ (10 ng/mL), and cultures treated with IFNγ plus 0.5 mM L-NMMA. Flow data were based on 10 000 events. n = 4 mice per genotype. *P < .05.
To investigate the potential contribution of apoptosis to the effect of IFNγ, day 6 cultures were examined for the presence of apoptotic nuclei using a flow cytometry–based terminal deoxynucleotidyl transferase–mediated dUTP nick-end labeling (TUNEL) assay (Figure 4B). Day 6 was selected to allow for sufficient growth of cells (106 cells required) to enable flow cytometry. This analysis revealed that IFNγ treatment ofFancc−/− cultures was accompanied by a trend toward increased levels of apoptosis. This trend was reversed by the addition of 0.5 mM L-NMMA to the IFNγ-containing cultures of both wild-type and Fancc−/− HPCs (Figure 4B). While these results could be reflective of a direct effect of IFNγ on progenitor populations, it is important to note that by day 6 the cultures would undoubtedly contain significant numbers of differentiating myeloid cells. The latter, likely having a greater potential than HPCs for releasing large amounts of NO in response to IFNγ, might account for the results of the TUNEL assay.
Expression of iNOS following activation ofFancc−/−macrophages
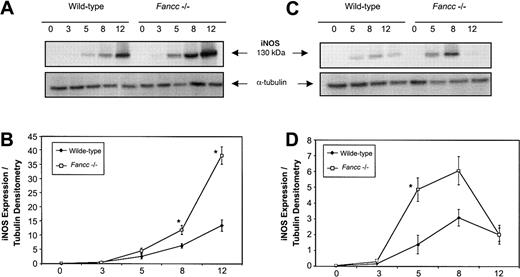
Given the sensitivity of Fancc−/− BM cells to NO-generating cytokines, and the ability of L-NMMA to blunt the negative effects of these cytokines, we hypothesized that altered regulation of iNOS might be present in Fancc-deficient cells. Since progenitor cells that give rise to colonies in methylcellulose experiments would be difficult to purify in sufficient numbers to enable signal transduction analyses, an alternate BM-derived cell source was selected to facilitate a study of the response of iNOS in a primary BM cell population. We first investigated the response of thioglycollate-elicited primary peritoneal macrophages to the combined effects of IFNγ plus bacterial lipopolysaccharide (LPS), a potent iNOS-inducing stimulus. Figure 5A (top panel) shows a representative immunoblot of iNOS expression in peritoneal macrophages from wild-type andFancc−/− mice following stimulation with the combination of IFNγ (10 ng/mL) and LPS (100 ng/mL). Expression of iNOS protein was increased in the Fancc-deficient cells and reached a higher level at the 12-hour time point than in controls. Figure 5B represents the average densitometry ratio from 5 independent experiments; it can be seen that iNOS expression was on average significantly higher in Fancc−/− macrophages than in controls at 8 and 12 hours after stimulation (P = .02, .04, respectively). This was consistent with altered regulation of iNOS in Fancc−/−thioglycollate-elicited peritoneal macrophages exposed to the potent inductive stimulus of IFNγ plus LPS.
Elevated iNOS expression in
Fancc−/− macrophages stimulated with IFNγ and LPS. Peritoneal or bone marrow–derived macrophages were stimulated for 0 to 12 hours with IFNγ (10 ng/mL) with or without LPS (100 ng/mL), and whole cell lysates were assayed for iNOS expression by immunoblotting. (A) Representative filter showing iNOS protein expression in Fancc−/−and control peritoneal macrophages following IFNγ plus LPS stimulation (top panel), with α-tubulin (bottom panel) as the loading control. (B) Densitometric representation of 5 independent experiments showing a significant difference in iNOS expression betweenFancc−/− (■) and wild-type (♦) peritoneal macrophages at 8 and 12 hours after stimulation (P = .02, .04, respectively). (C) Representative filter showing iNOS expression in BMDMs following IFNγ stimulation (top panel), with α-tubulin (bottom panel) as the loading control. (D) Densitometric representation of 4 independent experiments showing a significant increase in iNOS expression within Fancc−/− BMDMs at 5 hours after stimulation, with expression of iNOS reaching a maximum at 8 hours. *P < .05.
Elevated iNOS expression in
Fancc−/− macrophages stimulated with IFNγ and LPS. Peritoneal or bone marrow–derived macrophages were stimulated for 0 to 12 hours with IFNγ (10 ng/mL) with or without LPS (100 ng/mL), and whole cell lysates were assayed for iNOS expression by immunoblotting. (A) Representative filter showing iNOS protein expression in Fancc−/−and control peritoneal macrophages following IFNγ plus LPS stimulation (top panel), with α-tubulin (bottom panel) as the loading control. (B) Densitometric representation of 5 independent experiments showing a significant difference in iNOS expression betweenFancc−/− (■) and wild-type (♦) peritoneal macrophages at 8 and 12 hours after stimulation (P = .02, .04, respectively). (C) Representative filter showing iNOS expression in BMDMs following IFNγ stimulation (top panel), with α-tubulin (bottom panel) as the loading control. (D) Densitometric representation of 4 independent experiments showing a significant increase in iNOS expression within Fancc−/− BMDMs at 5 hours after stimulation, with expression of iNOS reaching a maximum at 8 hours. *P < .05.
iNOS expression in peritoneal macrophages from wild-type andFancc−/− mice, stimulated with IFNγ alone, did not consistently demonstrate increased iNOS expression inFancc−/− cells, as this was seen in only 3 of 5 independent experiments. It should be noted, however, that the intraperitoneal injection of thioglycollate broth constitutes a proinflammatory stimulus that may well affect the basal activation state of macrophages, potentially leading to animal-to-animal variability when responses to IFNγ alone are assessed. For this reason we also evaluated IFNγ induction of iNOS in BM-derived macrophages (BMDMs) cultured from total marrow cells for 7 days in the absence of any proinflammatory stimulus prior to IFNγ challenge. As shown in Figure 5C (upper panel), Fancc−/−BMDMs stimulated with 10 ng/mL IFNγ expressed higher levels of iNOS than controls. Densitometry analysis (Figure 5D) of 5 independent experiments showed that iNOS expression was maximal in IFNγ-treatedFancc−/− BMDMs at 8 hours after stimulation, while the greatest difference between the IFNγ-treatedFancc−/− cells and control BMDMs was at 5 hours (P = .04). These results indicated that Fancc-deficient BM-derived monocytic cells were able to generate higher levels of iNOS after IFNγ stimulation than control cells, providing a plausible explanation for the increased sensitivity of these cells to the growth-inhibitory effects of this cytokine.
NO (as nitrite) production by activatedFancc−/−macrophages
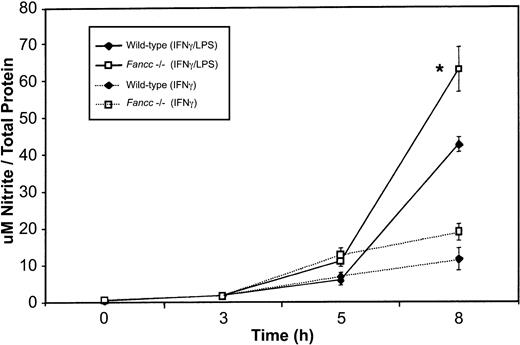
To determine whether the increased expression of iNOS in Fancc-deficient macrophages was accompanied by increased release of NO, we measured levels of this species (as nitrite) in culture supernatants. As shown in Figure 6, we found a significant increase in nitrite production byFancc−/− macrophages when these were stimulated with IFNγ plus LPS. This increase was statistically different from that of wild-type samples at the 8-hour time point (P = .04). Fancc−/−macrophages stimulated with IFNγ also revealed an increase in NO production, compared with wild-type samples, at 5 and 8 hours; however, this increase was not significant. Thus, there was a correlation between the levels of iNOS and in vitro NO production by the macrophage populations.
Increased NO (as nitrite) production by activated
Fancc−/− macrophages.Supernatants from the peritoneal macrophages used in iNOS expression studies (Figure 5) were harvested for nitrite quantitation. There was a significant increase in nitrite levels in the supernatants ofFancc−/− cells (at 8 hours) when macrophages were stimulated with IFNγ plus LPS or with IFNγ alone (although the latter did not reach significance). *P < .05.
Increased NO (as nitrite) production by activated
Fancc−/− macrophages.Supernatants from the peritoneal macrophages used in iNOS expression studies (Figure 5) were harvested for nitrite quantitation. There was a significant increase in nitrite levels in the supernatants ofFancc−/− cells (at 8 hours) when macrophages were stimulated with IFNγ plus LPS or with IFNγ alone (although the latter did not reach significance). *P < .05.
IFNγ-stimulated Stat1 phosphorylation inFancc−/−macrophages
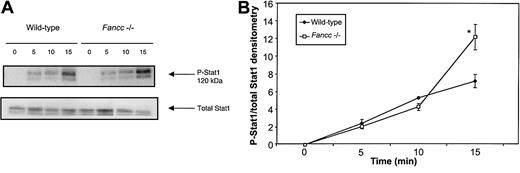
Several transcription factors, including Stat1, regulate expression of the inos gene in response to IFNγ.21 Given our results, which revealed elevated iNOS levels in Fancc−/− cells, we were interested in determining the phosphorylation status of Stat1 following exposure to IFNγ. Peritoneal macrophages from control andFancc−/− mice were stimulated with IFNγ, and phospho-Stat1 (P-Stat1) levels were assessed. Figure7A shows a representative experiment showing P-Stat1 levels in wild-type andFancc−/− peritoneal macrophages following stimulation with IFNγ (top panel) and normalized for loading using a Stat1 antibody (lower panel). Densitometry results from 4 independent experiments (Figure 7B) revealed that Fancc−/−macrophages generated higher levels of P-Stat1 at 15 minutes after stimulation (P = .04) than did wild-type controls. The possibility that increased expression of IFNγ receptors in Fancc-deficient cells might account for the increased levels of phospho-Stat1 was excluded by flow cytometry using anti-CD119 antibody staining (data not shown). As Stat1 is a positive regulator of iNOS expression,34 these results provided a possible explanation for the increased levels of iNOS observed in the IFNγ-stimulated Fancc-deficient BM cells.
Stat1 phosphorylation is augmented in IFNγ-stimulated
Fancc−/− macrophages.Peritoneal macrophages from control andFancc−/− mice were stimulated with IFNγ and their cell lysates subjected to Stat1 immunoblotting analysis. (A) Representative filter showing phospho-Stat1 (top panel) and total Stat1 protein (bottom panel). (B) Densitometry analyses of 4 independent experiments demonstrated a significant increase in the phospho-Stat1 signal within Fancc−/− macrophages at 15 minutes after stimulation with IFNγ. *P < .05.
Stat1 phosphorylation is augmented in IFNγ-stimulated
Fancc−/− macrophages.Peritoneal macrophages from control andFancc−/− mice were stimulated with IFNγ and their cell lysates subjected to Stat1 immunoblotting analysis. (A) Representative filter showing phospho-Stat1 (top panel) and total Stat1 protein (bottom panel). (B) Densitometry analyses of 4 independent experiments demonstrated a significant increase in the phospho-Stat1 signal within Fancc−/− macrophages at 15 minutes after stimulation with IFNγ. *P < .05.
Discussion
It has been proposed that IFNγ, TNFα, and MIP-1α, whether released constitutively or as a result of intercurrent illnesses,35 may play a role in human FA.16Although murine FA models generated to date lack spontaneous marrow aplasia, increased sensitivity of Fancc−/− BM cells to these 3 cytokines has been demonstrated.13,14Given that NO is suppressive to normal hematopoiesis,30and the fact that all 3 cytokines can up-regulate iNOS levels in target cells, it was of interest to evaluate the effects of the broad-spectrum NOS inhibitor L-NMMA on cytokine-inhibitedFancc−/− colony formation. Our results support the hypothesis that cytokine-inhibitedFancc−/− progenitor growth in vitro is mediated primarily through NO generation, although, as noted above, an indirect effect involving cytokine-mediated NO release from contaminating mature cells is a possibility. The effects of L-NMMA on IFNγ-mediated inhibition of hematopoietic cells was not confined to the committed progenitor methylcellulose colony-forming assays, since L-NMMA also rescued 3-day column-purified Lin− HPC culture growth from the inhibitory effects of IFNγ.
The finding that NO donors were inhibitory at lower concentrations inFancc−/− hematopoietic cells than controls was of particular interest, as it suggests that C57BL/6Fancc-deficient cells were more sensitive to the toxic effects of this radical. In keeping with this notion, we found increased numbers of TUNEL-positive cells in IFNγ-treated HPC cultures, an effect that was reversed by the addition of L-NMMA. The sensitivity of Fancc-deficient BM cells to NO might conceivably be related to the sensitivity some FA cells show toward oxygen,36 37 given the potential for reactive nitrogen and oxygen species to combine and generate highly toxic molecules (see next paragraph). To our knowledge, this study provides the first indication that reactive nitrogen species, specifically NO, may play a role in the pathogenesis of the FA defect.
Among its myriad effects, NO has the potential to induce DNA damage, necrosis, and apoptosis.22 NO and its derivatives (N2O3, NO, and NO+) are genotoxic, leading to DNA damage both directly and indirectly.38 Of particular interest, NO is able to combine with superoxide, which may be elevated in FA hematopoietic cells,39 leading to the formation of the highly reactive species peroxynitrite (ONOO−).22 ONOO− is capable of causing single- and double-strand DNA breaks, oxidative lesions (such as 8-oxoguanine), induction of polyADP ribose polymerase, Fe2+ release, glutathione depletion, and cell death.38,40-42 NO can also act as an indirect genotoxin via inhibition of specific DNA repair enzymes.43 44 In light of these effects, the increased sensitivity ofFancc−/− cells to NO-generating compounds and NO-inducing cytokines might be due to the impairment of DNA repair present in these cells. If NO were found to be involved in human FA, many of the above-mentioned DNA lesions would have the potential to induce chromosomal aberrations and other types of mutations that might predispose to leukemic transformation.
To gain some insight into signal transduction events that might provide a mechanistic explanation for the increased sensitivity ofFancc−/− BM-derived cells to IFNγ, a population of primary cells that could be obtained in sufficient numbers for biochemical analyses was required. Thus, peritoneal and BM-derived macrophages were selected for study. Macrophages may be appropriate to the study of FA, given that cytokines and other agents released from these cells may possibly play a role in this condition. Induction of iNOS expression and NO production is readily achieved with IFNγ and LPS costimulation. Using this stimulus, we found thatFancc−/− peritoneal macrophages generated elevated levels of iNOS and NO, as compared with controls. When IFNγ alone was used to stimulate Fancc−/−peritoneal macrophages, iNOS expression levels were variable, possibly owing to differences in the ratios and activation states of macrophages and neutrophils recruited into the peritoneal cavity after thioglycollate broth administration. This nonspecific stimulus results in the variable activation of the incoming cells, in part owing to chemokine and cytokine production by resident peritoneal macrophages that occurs following thioglycollate injection.45 As it was also important to assess the iNOS response to IFNγ in nonactivated cells, BMDMs were evaluated. Similar to the peritoneal cells, BMDMs from Fancc−/− mice generated higher levels of iNOS than controls after IFNγ stimulation. These results provide a plausible explanation for the increased sensitivity of Fancc-deficient hematopoietic cells to IFNγ, namely, that the increased levels of NO generated by these cells result in growth inhibition greater than that in Fancc-proficient cells.
iNOS expression has been detected in CD34+ progenitor cells following exposure to IFNγ, and in this system NO was found to be antiproliferative.30 Although the effect of IFNγ on day 3 Lin− cell cultures (Figure 4A) provided evidence that this cytokine was exerting a direct effect on early progenitors, an indirect effect, mediated via mature macrophages, was also a possibility, as cells of the monocyte-macrophage lineage are able to respond to a variety of stimuli with strong inductions of iNOS and NO.21 Thus, it would be of interest to determine whether mature bone marrow–derived macrophages represent the primary source of NO within the bone marrow microenvironment, and also whether NO produced by small numbers of macrophages within the in vitro assays was ultimately responsible for IFNγ-mediated inhibition ofFancc−/− progenitors. Interestingly, constitutive expression of iNOS was found in BM macrophages of patients with myelodysplastic syndromes as well as in idiopathic aplastic anemia samples.46 47 Clearly, many factors could potentially be involved in promoting marrow aplasia in FA. These factors include the uniqueness of marrow cell populations, in which progenitors coexist with cells having the ability to elaborate not only high levels of reactive nitrogen and oxygen species, but also NO-inducing cytokines in response to various stimuli; the sinusoidal nature of the marrow vascular system, which may favor accumulation of reactive species, chemokines, and cytokines; and the potential myelosuppressive effects of these factors.
Regulation of the inos gene promoter is complex, involving a variety of transcription factors, including nuclear factor (NF)–κB, STAT1, and hypoxia-inducible factor (HIF)1α.21 As iNOS expression was higher in IFNγ-stimulated Fancc-deficient cells, it was important to begin to define signal transduction pathways that might be responsible for this differential effect. In this respect, we found increased P-Stat1 levels in Fancc-deficient cells after IFNγ stimulation, a finding that contrasts with the diminished levels of IFNγ-induced Stat1 activation observed in FANCC-deficient lymphoblastoid cells andFancc−/− embryonic fibroblasts.18-20 There are several potential reasons for this difference. For example, we analyzed Stat1 responses to IFNγ in primary murine macrophages rather than in Epstein-Barr virus (EBV)–transformed lymphoblasts or murine embryonic fibroblasts. Second, the lack of Stat1 activation seems to be cell type–specific, as FANCC-deficient fibroblasts failed to show this defect.20 Thus, the differences in phosphorylation status of Stat1 are likely attributable to differences in the cell types analyzed (for example, lymphoblasts vs macrophages) or even the differentiation state of the cells. Finally, our observations may be characteristic of the predominantly C57BL/6 genetic background of ourFancc−/− colony. Indeed, since our results have been confined to Fancc-deficient murine BM cells, it will be critical to establish whether dysregulation of NO production and/or increased sensitivity to NO-generating compounds are attributes shared by human FA bone marrow cells.
We are indebted to Dr M. Buchwald for providing us withFancc+/− mice and to Dr G. C. Bagby for his advice. Our thanks also to S. Lines and J. Gorday for maintaining the animal colony and performing the intraperitoneal injections, and to C. Huang for his assistance.
Prepublished online as Blood First Edition Paper, January 9, 2003; DOI 10.1182/blood-2002-10-3147.
Supported by the National Cancer Institute of Canada with funds from The Canadian Cancer Society and by an Establishment Grant from Alberta Heritage Foundation for Medical Research (AHFMR). S.H. held a Natural Sciences and Engineering Research Council (NSERC) Scholarship award, and F.R.J. was the recipient of AHFMR Scientist and Canada Research Chair awards.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Frank R. Jirik, Department of Biochemistry and Molecular Biology, University of Calgary, 3330 Hospital Dr NW, Calgary, AB, Canada T2N 4N1; e-mail:jirik@ucalgary.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal