Abstract
Activated protein C resistance (APCR) is the most prevalent risk factor for thrombosis, accounting for 20% to 60% of familial thrombophilia. A mutation in the F5 gene, factor V Leiden (FVL), is a major determinant of pathological APCR in some populations. However, APCR predicts risk for thrombosis independently of FVL. This suggests that other genetic factors may influence risk of thrombosis through quantitative variation in APCR. To search for these unknown loci, we conducted a genome-wide linkage screen for genes affecting normal variation in APCR in the 21 Spanish families from the Genetic Analysis of Idiopathic Thrombophilia (GAIT) project. Conditional on FVL, the strongest linkage signal for APCR was found on chromosome 18 near D18S53. Bivariate linkage analyses with a genetically correlated trait, levels of clotting factor VIII, strengthened evidence for the chromosome 18 quantitative trait locus (QTL; logarithm of the odds [LOD], 4.5; P = 3.08 × 10−5). However, the region on chromosome 1 that contains the F5structural gene showed little evidence of linkage to APCR (LOD, < 1). This indicates that apart from the FVL, the F5 locus itself plays a relatively minor role in normal variation in APCR, including the HR2 haplotype polymorphisms. A second bivariate analysis of APCR with thrombosis liability suggested that this QTL also influences the risk of thrombosis (P = .0016). These results indicate that a locus on chromosome 18 pleiotropically influences normal variation in the APCR phenotype and factor VIII (FVIII) levels as well as susceptibility to thrombosis. Importantly, there are no known thrombosis-related candidate genes in this region, implying that this QTL represents a completely novel thrombosis risk factor.
Introduction
Venous and arterial thrombosis may be life-threatening events and are of great importance in public health. Very little is known about the relative importance of genetic factors in thrombosis risk in the general population.1 Recently, as part of the GAIT (Genetic Analysis of Idiopathic Thrombophila) project, we have quantified the genetic contribution to susceptibility to thrombosis and related phenotypes in the Spanish population.2,3 Of the quantitative risk factors studied, activated protein C resistance (APCR) had the highest heritability (0.71), and it was genetically correlated with thrombosis (ςg = − 0.65;P = 1 × 10−6),2 3 indicating that some of the genes that influence quantitative variation in this phenotype also influence susceptibility to thrombosis.
APCR is the most prevalent risk factor for thrombosis,4accounting for 20% to 60% of familial thrombophilia.5 A mutation in the F5 gene (factor V Leiden [FVL]) produces a coding change from Arg506 to Gln at the first cleavage site, where APC acts to inactivate FV. As a consequence, this mutation produces a protein that is intrinsically resistant to APC, causing the APCR pathological phenotype.6 Prevalence of FVL in different European countries ranges between 2% and 6%.7Moreover, APCR is a genetic risk factor for thrombosis independent of FVL.8,9 Therefore, because of its implication in thrombotic disease, there has been a growing interest in studying other genetic factors that are likely to affect thrombosis risk through quantitative variation in this important intermediate phenotype. The APCR phenotype could theoretically result from a variety of other mutations of critical sites in the F5 or F8genes. However, no mutations of F8 have yet been identified in patients with the APCR phenotype,10,11 whereas 2 point mutations in F5 (Arg306Gly and Arg306Thr) affecting the Arg306 APC cleavage site have been described in, respectively, 2 patients and 1 patient with venous thromboembolism (VTE).12 13 Although these mutations cause pathological values of APCR, they are too rare to constitute the primary genetic influences on APCR variability in the normal population.
Moreover, in recent years, many studies have focused onF5 polymorphisms to explain APC sensitivity, particularly the His1299Arg polymorphic variant as part of the HR2 haplotype. This haplotype has been associated with a mild APCR phenotype14 and with variability in plasma FV levels.15 It also has been associated with an increased risk of thrombosis.16,17 However, there are studies18 19 that fail to detect these associations.
In addition, high factor VIII (FVIII) levels, a common risk factor for thrombotic disease,20 have been associated with reduced sensitivity for APC in the absence of FVL.21 Although the precise role of high FVIII levels in affecting risk is still unknown, it is possible that FVIII levels influence thrombosis susceptibility via an effect on the APC sensitivity ratio.
However, it seems to be clear that with the exception of the mutations in the APC cleavage sites of the F5 gene, very little information is available on the genetic factors influencing APCR in the general population. To investigate this question, we performed a genome scan to identify specific genes that affect APCR values. To our knowledge, our study represents the first genome-wide scan designed to identify genes that influence variation in susceptibility to thrombosis and its intermediate phenotypes.
Patients and methods
Subjects and phenotypes
The recruitment, sampling, and phenotyping methods used in the GAIT project have been extensively described in previous publications.3,22 Briefly, our sample included 398 individuals belonging to 3- to 5-generation–extended pedigrees. The participants ranged in age from younger than 1 year to 88 years and included approximately equal numbers of males and females. Twelve families were selected through a proband with idiopathic thrombophilia, and 9 families were randomly selected without regard to phenotype. APCR and FVIII levels were measured on fresh plasma samples by automated coagulometry as previously described.3
All procedures were reviewed and approved by the institutional review board of the Hospital de la Santa Creu i Sant Pau (Barcelona, Spain). Adult subjects gave informed consent for themselves and for their minor children.
Genotypes
DNA was extracted by means of a standard protocol.23 The genome scan used 363 highly informative microsatellite DNA markers, spaced at a density of 9.5 centimorgans (cMs). Microsatellites consisted primarily of the ABI-Prism (Applied Biosystems, Foster City, CA) genotyping set MD-10. Linkage mapping was undertaken by means of the PE LMS II fluorescent marker set (ABI Prism, Foster City, CA) with multiplex polymerase chain reaction (PCR) as described.24 In a few instances, nearby Genethon (Centre National de Genotypage, Evry, France) markers were substituted for LMS II markers to improve robustness of the scan (http://www.cng.fr/). The PCR products were analyzed on the PE 310, PE 377, and PE 3700 automated sequencers, and they were genotyped by means of the PE Genotyper software (ABI Prism). Information on microsatellite markers can be found in the publicly accessible Genomic DataBase (http://www.gdb.org). The average heterozygosity of these markers was 0.79.
Markers in or near several hemostasis-related candidate genes were used to augment the genome scan. The FVL mutation and F5polymorphisms (Table 1), including the HR2 haplotype polymorphisms, were genotyped as previously described.6,14,25 26
Results from the linkage analysis: APCR with F5 DNA variants, and APCR conditional on the FVL mutation
| Locus . | DNA variant . | Amino acid change . | Allelic frequencies in the GAIT sample . | LOD score . | |
|---|---|---|---|---|---|
| APCR . | APCR-FVL . | ||||
| FV | — | Ala485Leu | 0.996/0.004 | 0.24 | 0.26 |
| FV | FVL | Arg506Gln | 0.993/0.007 | 2.78 | 0.00 |
| FV | IVS11 | None | 0.222/0.572/0.083/0.122 | 0.00 | 0.00 |
| FV | 2298T > C | None | 0.757/0.243 | 0.00 | 0.00 |
| FV | 2325C > T | None | 0.810/0.189 | 0.00 | 0.00 |
| FV | 2391G > A | None | 0.750/0.250 | 0.00 | 0.00 |
| FV | 2833A > T | None | 0.962/0.038 | 0.00 | 0.00 |
| FV | 4070A > G | His1299Arg | 0.920/0.079 | 0.00 | 0.00 |
| FV | 5380A > G | None | 0.656/0.344 | 0.00 | 0.00 |
| FV | IVS16 | None | 0.049/0.951 | 3.05 | 0.00 |
| Locus . | DNA variant . | Amino acid change . | Allelic frequencies in the GAIT sample . | LOD score . | |
|---|---|---|---|---|---|
| APCR . | APCR-FVL . | ||||
| FV | — | Ala485Leu | 0.996/0.004 | 0.24 | 0.26 |
| FV | FVL | Arg506Gln | 0.993/0.007 | 2.78 | 0.00 |
| FV | IVS11 | None | 0.222/0.572/0.083/0.122 | 0.00 | 0.00 |
| FV | 2298T > C | None | 0.757/0.243 | 0.00 | 0.00 |
| FV | 2325C > T | None | 0.810/0.189 | 0.00 | 0.00 |
| FV | 2391G > A | None | 0.750/0.250 | 0.00 | 0.00 |
| FV | 2833A > T | None | 0.962/0.038 | 0.00 | 0.00 |
| FV | 4070A > G | His1299Arg | 0.920/0.079 | 0.00 | 0.00 |
| FV | 5380A > G | None | 0.656/0.344 | 0.00 | 0.00 |
| FV | IVS16 | None | 0.049/0.951 | 3.05 | 0.00 |
IVS indicates intervening sequence.
The genotypic data were entered into a database and were analyzed for discrepancies (ie, violations of Mendelian inheritance) by means of the program INFER (PEDSYS, San Antonio, TX).27Discrepancies were checked for mistyping, and were either corrected or excluded from the analysis.
Linkage analysis
Standard multipoint variance component linkage methods, as implemented in the computer program Sequential Oligogenic Linkage Analysis Routines (SOLAR) (NIH, Bethesda, MD), were used for the genome scan.28 Previous studies suggested that these methods may be vulnerable to deviations from multivariate normality and particularly to high levels of kurtosis in the distribution of the trait.29 Levels of factor VIII and APCR in the GAIT individuals exhibited a kurtosis of 0.63 and −0.17, respectively. Recent statistical genetic theory has demonstrated that this level of kurtosis does not affect the distribution of logarithm of the odds (LOD) scores and that the standard nominalP values for LOD scores are appropriate for the APCR and FVIII linkage screens.30 Allelic frequencies were estimated from the GAIT sample, and marker maps for multipoint analyses were obtained from ABI-Prism (http://www.appliedbiosystems.com/molecularbiology/) and from the Marshfield Clinic (Marshfield, WI) (http://research.marshfieldclinic.org/genetics/). As 12 of the families in the GAIT project were ascertained through thrombophilic probands, all analyses included an ascertainment correction achieved by conditioning the likelihood of these pedigrees on the likelihoods of their respective probands.31 Genome-wide Pvalues were calculated by means of the method of Feingold et al.32 Bivariate linkage analyses using the mixed discrete/continuous trait multivariate model were conducted with a modified version of SOLAR.33
Combined linkage/association analysis
Quantitative trait association analyses were performed with the use of the measured genotype approach34 by testing for genotype-specific differences in the means of traits while allowing for the nonindependence among family members. These analyses were performed by means of SOLAR.28 To assess linkage and association simultaneously,35 36 an extension of the variance component–based linkage test was performed by simultaneously incorporating the genotype-specific means of the measured genotype test. If a variant is functional and there are no other functional variants in the candidate gene under investigation, then a linkage analysis conditional on the measured genotypes (ie, a linkage test in which the measured genotypes are controlled for) should yield no residual evidence for linkage. This is because all of the genetic variance due to the quantitative trait locus (QTL) will be removed when the QTL is itself used as a covariate. Alternatively, if a variant is merely in linkage disequilibrium with a functional site, linkage analyses will have additional predictive power over the measured genotype test and will yield evidence for linkage.
Results
The subjects in our study were genotyped for an autosomal genome-wide scan that included 363 microsatellite DNA markers spaced at approximately 9.5-cM intervals. The average heterozygosity of the single tandem repeat (STR) markers was 0.79. We also genotyped several F5 DNA variants associated with APCR phenotype, including the FVL mutation and the HR2 haplotype polymorphisms14 (Table 1). Although none of the individuals in the GAIT project had pathological APCR values, 9 of them were heterozygous for the FVL mutation. Five of these individuals belonged to a randomly ascertained family, and 4 individuals were from a thrombophilic family. In this thrombophilic family, only one FVL carrier individual suffered thrombosis. However, the mutation came from her father, who is not related to the original thrombophilic proband. These 9 carriers exhibited a markedly lower mean APCR ratio than the noncarriers (2.26 versus 3.19), which represented a significant difference of −0.93 (P = 6 × 10−7).
Multipoint variance-component methods were used to assess linkage between autosomal markers and quantitative values of APCR. Age, sex, FVL mutation, and the ABO blood group (in the case of FVIII levels) were used as covariates in all of the analyses. Their effects were estimated simultaneously with the genetic effects.
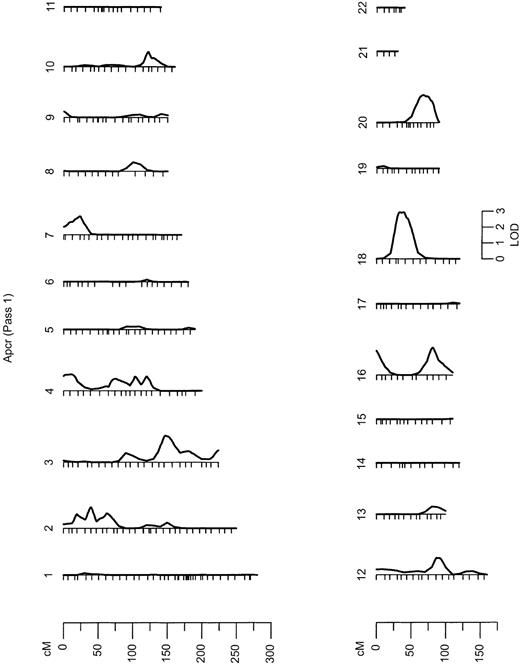
Strong evidence of linkage with a QTL for APCR was obtained in a linkage analysis with FVL (LOD, 2.84;P = 1.84 × 10−4) and with the IVS16 microsatellite in F5 located in intron 16 (LOD, 3.05). Evidence of linkage with both F5 markers completely disappeared when we simultaneously allowed for an association between the FVL mutation and APCR value (Table 1). This is consistent with a direct functional effect of this mutation. However, under the assumption that FVL functionally affects the APCR phenotype, we calculated that this mutation accounts for less than 6% of the variance in the APCR ratio in the GAIT sample. The other F5polymorphisms did not show evidence of linkage with APCR (Table 1). It is worth noting that in the multipoint genetic analysis, conditional on FVL, the region on chromosome 1q23-24 that contains the F5structural gene showed little evidence of linkage to APCR (LOD, less than 1) (Figure 1). This indicates that there is little support for an F5 QTL apart from the influence of FVL on normal variation in the APCR phenotype, including the HR2 haplotype polymorphisms, such as His1299Arg.
Results from the autosomal multipoint genome scan.
LOD scales are shown for all chromosomes on which the maximum LOD score exceeds 1. Hatch marks along the length of the chromosomes indicate the positions of genotyped markers. APCR indicates analyzed phenotypes.
Results from the autosomal multipoint genome scan.
LOD scales are shown for all chromosomes on which the maximum LOD score exceeds 1. Hatch marks along the length of the chromosomes indicate the positions of genotyped markers. APCR indicates analyzed phenotypes.
In the initial marginal multipoint genome scan (Figure 1), 7 regions exhibited LOD scores higher than 1 (Table2), with the strongest linkage signal on chromosome 18 (LOD, 2.93; P = .000 12) (Figure2). We then evaluated all possible multilocus models. In the second linkage pass, conditional on the chromosome 18 QTL detected in the initial screen, evidence for QTLs on chromosome 4 and 10 remained (LOD, greater than 1), whereas LOD scores in other regions previously showing suggestive linkage declined slightly. Conditional on the chromosome 18 and 4 QTLs, little evidence for additional QTLs remained in a third linkage screen (all LOD scores, below 1.0).
Results from the initial and conditional genome-wide linkage screens of APCR
| Chromosome location . | 1st pass . | 2nd pass . | 3rd pass . |
|---|---|---|---|
| 18 (42 cM) | 2.93 | — | — |
| 4 (81 cM) | 1.31 | 1.77 | — |
| 10 (125 cM) | 1.27 | 1.62 | 0.91 |
| 2 (42 cM) | 1.28 | 0.92 | 0.35 |
| 3 (153 cM) | 1.61 | 0.91 | 0.45 |
| 16 (0 cM) | 1.18 | 0.92 | 0.75 |
| 16 (79 cM) | 1.35 | 0.84 | 0.18 |
| Chromosome location . | 1st pass . | 2nd pass . | 3rd pass . |
|---|---|---|---|
| 18 (42 cM) | 2.93 | — | — |
| 4 (81 cM) | 1.31 | 1.77 | — |
| 10 (125 cM) | 1.27 | 1.62 | 0.91 |
| 2 (42 cM) | 1.28 | 0.92 | 0.35 |
| 3 (153 cM) | 1.61 | 0.91 | 0.45 |
| 16 (0 cM) | 1.18 | 0.92 | 0.75 |
| 16 (79 cM) | 1.35 | 0.84 | 0.18 |
All loci with LOD scores over 1.0 in the initial pass are shown. The second linkage pass was conditional on a locus on chromosome 18, and the third pass was conditional on loci on chromosomes 18 and 4.
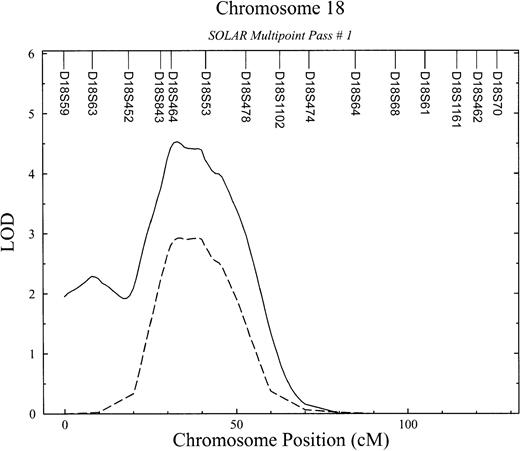
Detailed linkage results for chromosome 18.
APCR phenotype (dashed line) and bivariate APCR-FVIII phenotype (solid line).
Detailed linkage results for chromosome 18.
APCR phenotype (dashed line) and bivariate APCR-FVIII phenotype (solid line).
Because our previous studies have suggested that APCR values are correlated with FVIII levels (ςg = −0.53;P = 2.5 × 10−4) and that this relationship is due in part to genes that jointly influence both traits,2 we performed a bivariate linkage analysis. Bivariate analyses with related phenotypes have been shown to increase power to detect linkage.22 This analysis significantly improved the linkage signal for the APCR phenotype (LOD score, 4.5;P = 3.08 × 10−5; genome-wide, P= .011) (Figure 2) and provided strong evidence that this QTL has a pleiotropic effect on FVIII levels. In other words, this chromosome 18 QTL simultaneously influences the APCR phenotype and plasma levels of FVIII. The peak LOD score occurred near D18S53 in the interval flanked by markers D18S843 and D18S54 in a region that maps to 18p13 (Figure2). The 95% confidence interval surrounding this LOD ranges in chromosomal location from 28 to 51 cM from pter.
Since low APCR values and high FVIII levels predispose individuals to thrombosis,9,20 we tested whether this QTL on chromosome 18 might also contribute to the genetic susceptibility to thrombosis. This was accomplished through bivariate linkage analysis using a mixed discrete/continuous trait multivariate model.37 From this combined analysis, we obtained strong evidence that the chromosome 18 QTL also has a pleiotropic effect on the risk of thrombosis (P = .0016).
Discussion
Despite growing insight regarding the pathogenesis of thrombophilia, the specific cause of many thrombotic episodes remains unknown. Recently, new laboratory phenotypes that are associated with an increased risk of venous thrombosis have been reported.20 The most prevalent among them is APCR. Our results represent the first genome-wide scan undertaken to identify regions containing genes that influence variation in susceptibility to thrombosis disease and their intermediate phenotypes, such as APCR. Initial and subsequent conditional passes of variance-component linkage analyses revealed one region on chromosomes 18 showing strong evidence of linkage with quantitative variation of APCR (LOD, 2.93; P = .000 12), an important intermediate phenotype correlated with thrombosis.3 Given the strong genetic correlation between APCR and FVIII levels (ςg = −0.53;P = 2.5 × 10−4), a bivariate analysis significantly improved the linkage signal for the APCR phenotype (LOD score, 4.5;P = 3.08 × 10−5) and provided strong evidence that this QTL has a pleiotropic effect on FVIII levels. In other words, this chromosome 18 QTL simultaneously influences the APCR phenotype and plasma levels of FVIII.
High FVIII levels in plasma predispose individuals to venous thrombosis,20 coronary heart disease,38,39and stroke40; the genetic contribution to its normal variation is largely unknown. The complexity of FVIII levels is exemplified by its association with von Willebrand factor (VWF),41 whose levels are determined mainly by the ABO blood group.42 We have reported previously that genetic factors appear to be the most important determinants of quantitative variation in FVIII levels, with an additive genetic heritability of 0.4 ± 0.088.3 However, ABO blood group accounts for only a small portion of the total variation in FVIII,42 demonstrating that other genetic factor might be involved in the quantitative variation of this important phenotype. The results that we have described here represent the first QTL that directly influences FVIII levels.
It is important to note that this study confirms and extends our previous observation that variability of APCR values is correlated with the risk of thrombosis and that this relationship is due in part to genes that jointly influence both traits.3This was accomplished through bivariate linkage analysis of APCR and thrombosis.33 From this combined analysis, we obtained strong evidence that the chromosome 18 QTL also has a pleiotropic effect on the risk of thrombosis (P = .0016).
In the region of the linkage signal on chromosome 18, there are no obvious candidate hemostasis-related genes that might influence the APCR ratio or FVIII levels. Further investigations, including fine mapping and gene-identification studies in this region, may enhance our understanding of the factors influencing thrombosis, especially variability in APCR and FVIII levels.
In addition to the chromosome 18 genetic signal, strong evidence of linkage with a QTL for APCR was obtained in a linkage analysis with FVL and the IVS16 microsatellite in F5 intron 16 (LOD, 2.84 and 3.05, respectively). Evidence of linkage with both F5markers completely disappeared when we simultaneously allowed for an association between the FVL mutation and the APCR value (Table 1). This is consistent with a direct functional effect of FVL mutation, since the FVL mutation completely accounts for the genetic linkage signal we observed. However, the FVL variant accounted for less than 6% of the variation in APCR in the GAIT sample. This indicates that there are other QTLs that influence APCR values, as suggested by the highly significant linkage signal on chromosome 18.
The other F5 polymorphisms did not show evidence of linkage with APCR. It is worth noting that in the multipoint genetic analysis, conditional on FVL, the region on chromosome 1 that contains theF5 structural gene showed little evidence of linkage to APCR (LOD, lower than 1) (Figure 1). This indicates that there is little support for an F5 QTL apart from the influence of FVL on normal variation in the APCR phenotype, including the HR2 haplotype polymorphisms, such as His1299Arg. Thus, although individuals with the FVL mutation show a pathological APCR value with a dramatically increased incidence of thromboembolic disorder, theF5 locus itself plays a relatively minor role in normal variation in APCR. These results are important because they emphasize the limitation of classical association studies in assessing the importance of specific variants or genes at the population level.43 Our results are consistant with a naturally occurring mutation that also provided evidence for a nonfunctional role of the His1299Arg polymorphism in the phenotypic variability of APCR values.44
In summary, these results represent the first direct evidence that APCR and FVIII levels, which are major risk factors underlying liability to thrombosis, are jointly influenced by a QTL on chromosome 18. Our results also support the conclusion that this QTL is an important modulator of an individual's susceptibility to thrombosis. In addition, the F5 locus itself plays a relatively minor role in normal variation in APCR.
The identification of this chromosome 18 QTL may help to elucidate the mechanisms underlying the risk of thrombosis, and ultimately may lead to preventive strategies that will reduce morbidity and mortality of thrombosis-related diseases.
We are grateful to a number of doctors who assisted in the ascertainment and recruitment of thrombophilic pedigrees: Dr Javier Rodrı́guez Martorell from Hospital Universitario Puerta del Mar, Dr Carmen Araguás from Hospital Arnau de Vilanova, Dr Francisco Velasco from Hospital Reina Sofia, Dr Montserrat Maicas from Hospital General de Albacete, and Dr Dilia Brito from Hospital Carlos Haya. Finally, we are deeply grateful to all of the families who have participated in the GAIT study.
Prepublished online as Blood First Edition Paper, August 15, 2002; DOI 10.1182/blood-2002-06-1792.
Supported by Fondo de Investigacion Sanitaria (FIS) grants 97/2032 and 00/290; by National Institutes of Health (NIH) grant MH59490 (for statistical genetic analyses); and by FIS 99/3048 (J.M.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
José Manuel Soria, Unitat d'Hemostàsia i Trombosi, Hospital de la Santa Creu i Sant Pau, C/ Sant Antoni M. Claret 167, 08025 Barcelona, Spain; e-mail:jsoria@hsp.santpau.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal