Abstract
Familial multiple coagulation factor deficiency (FMFD) of factors II, VII, IX, X, protein C, and protein S is a very rare bleeding disorder with autosomal recessive inheritance. The phenotypic presentation is variable with respect to the residual activities of the affected proteins, its response to oral administration of vitamin K, and to the involvement of skeletal abnormalities. The disease may result either from a defective resorption/transport of vitamin K to the liver, or from a mutation in one of the genes encoding γ-carboxylase or other proteins of the vitamin K cycle. We have recently presented clinical details of a Lebanese family and a German family with 10 and 4 individuals, respectively, where we proposed autosomal recessive inheritance of the FMFD phenotype. Biochemical investigations of vitamin K components in patients' serum showed a significantly increased level of vitamin K epoxide, thus suggesting a defect in one of the subunits of the vitamin K 2,3-epoxide reductase (VKOR) complex. We now have performed a genome-wide linkage analysis and found significant linkage of FMFD to chromosome 16. A total maximum 2-point LOD score of 3.4 at θ = 0 was obtained in the interval between markers D16S3131 on 16p12 and D16S419 on 16q21. In both families, patients were autozygous for 26 and 28 markers, respectively, in an interval of 3 centimorgans (cM). Assuming that FMFD and warfarin resistance are allelic, conserved synteny between human and mouse linkage groups would restrict the candidate gene interval to the centromeric region of the short arm of chromosome 16.
Introduction
Familial multiple coagulation factor deficiency (FMFD, MIM #277450) is a very rare bleeding disorder, with 14 cases described as yet.1-13 Clinical symptoms of the disease are episodes of intracerebral hemorrhage in the first weeks of life sometimes leading to a fatal outcome. Deficiency of all vitamin K–dependent clotting factors leads to a bleeding tendency which is usually completely reversed with oral administration of vitamin K. The disease can be caused by intestinal malabsorption of vitamin K,14 mutations in the γ-glutamyl carboxylase (reported in 2 kindreds11,12), or functional deficiency of the multiprotein complex vitamin K 2,3-epoxide reductase (VKOR) (reported in 3 kindreds7,13). The pathomechanism of FMFD is based on a reduced hepatic γ-carboxylation of glutamic acid residues of all vitamin K–dependent blood coagulation factors. A number of proteins, such as the procoagulant factors II, VII, IX, and X, the anticoagulant factors protein C, protein S, and nonhemostatic proteins such as matrix gla protein and osteocalcin are subjected to γ-carboxylation.15 This posttranslational modification enables the calcium-dependent attachment of the modified proteins to the phospholipid bilayer of membranes,16,17 which is an essential prerequisite for the mechanism of blood coagulation. Vitamin K1 acts as a cofactor for the vitamin K–dependent carboxylase (γ-glutamyl carboxylase) in liver microsomes.18,19Gamma-carboxylation of Glu-residues is coupled to the stoichiometric formation of vitamin K 2,3-epoxide.20 The regeneration of the active cofactor vitamin K1 hydroquinone (vitamin K1H2) is mediated in the vitamin K cycle by either one of 2 pathways. The flavoprotein DT-diaphorase (NMOR1, EC 1.6.99.2) reduces only the quinone form21 and is of minor importance under physiologic conditions, whereas the multiprotein complex VKOR is capable of reducing the quinone as well as the epoxide form of vitamin K1.22 Cain et al20 presented evidence that a yet unknown member of the glutathione S-transferase (GST) gene family associates with microsomal epoxide hydrolase (EPHX1, EC3.3.2.3) and γ-carboxylase in the endoplasmic reticulum membrane to form the VKOR complex. This complex is the target of warfarin, which is used as an important anticoagulant to prevent thrombosis23 and in rodent pest control.24Resistance to warfarin has been reported in mice,25rats,26 and humans.27-29 Resistance seems to be caused by an autosomal dominant mutant allele of vitamin K reductase which either displays a decreased affinity for the coumarin drugs or an increased affinity for vitamin K.
We have recently described 2 pedigrees showing an autosomal recessive transmission of FMFD.13 The presence of mutations in γ-glutamyl carboxylase, microsomal epoxide hydrolase, and DT-diaphorase was excluded by sequencing the coding regions of the corresponding genes. Biochemical investigations clearly showed an elevated ratio of vitamin K epoxide/vitamin K hydrochinone in affected individuals, which implicates an impaired recycling of the cofactor of γ-glutamyl carboxylation.
In the present study, we performed a genome-wide linkage analysis for the FMFD locus in 2 affected pedigrees. As consanguinity was known to occur in one of the families, homozygosity mapping could be used to narrow down the candidate gene region.
Patients, materials, and methods
Patients
Clinical aspects of the 2 nonrelated families suffering from congenital deficiency of all vitamin K–dependent coagulation factors have been described elsewhere.13 Figure 2A depicts an abridged pedigree of family A, which is a consanguineous kindred of Lebanese origin with 8 children alive. There were 2 further children (not shown) who died, one on day 3 after birth from intracerebral hemorrhage, and one during the first year of life. There are 4 children (II:3, II:4, II:6, II:8) who experienced severe bleedings in the past. To the best of our knowledge, no consanguinity was present in family B (Figure 2B), which is of German origin. However, distant consanguinity cannot be ruled out since both maternal and paternal grandparents originate from neighboring villages. Hydrocephalus internus caused by intracranial bleeding was diagnosed in the first child (II:1).
The coagulation factor status of family members was determined by measuring the activity of vitamin K–dependent clotting factors II, VII, IX, and X and determination of vitamin K1H2 and vitamin K 2,3-epoxide.13 All affected individuals from both families showed a mild deficiency of the vitamin K–dependent coagulation factors ranging from 20% to 60%. Vitamin K 2,3-epoxide levels of 19.5 ng/mL to 66.2 ng/mL after oral administration of 10 mg vitamin K clearly indicate a defect in one of the proteins of the VKOR complex. Normal levels of vitamin K–dependent clotting factors and vitamin KO were found in all unaffected members of both families, including the parents, who are expected to be heterozygous carriers of the putative disease alleles.
Sequence analysis
Mutation screening was performed by direct sequencing of the candidate genes. In both families, we examined 8 exons and their flanking regions of microsomal epoxide hydrolase1 as well as the promoter sequence, and 6 exons and their flanking regions of NAD(P)H:menadione oxidoreductase 1(NMOR1, also known as DT-diaphorase orcytochrome b5 reductase). Mutations in theγ-glutamyl carboxylase gene had already been excluded in a previous report.13 Both strands of the polymerase chain reaction (PCR) templates were sequenced by the dideoxynucleotide chain termination method and the thermo sequenase radiolabeled terminator cycle sequencing kit (USB Corporation).
Genotype analysis
Blood samples were collected after obtaining informed consent and genomic DNA was extracted by standard procedures. A genome-wide scan, based on 374 microsatellite markers with an average spacing of 11 cM, was performed on family A at the Max Delbrück Center, Berlin, Germany. Markers were amplified by PCR in a final reaction volume of 10 μL containing 10 mM Tris, 1.5 mM MgCl2, 100 μM each dNTP, 0.4 U DNA polymerase (Invitek, Berlin, Germany), 7 pmol of each primer, and 20 ng genomic DNA. One of the primers was end-labeled with fluorescent dye. Pooled products were electrophoresed on ABI PRISM 377 automated DNA sequencers (Applied Biosystems, Foster City, CA). Further fine mapping with an additional 54 microsatellites from the pericentromeric region of chromosome 16 was performed in both families. PCR was performed in 25 μL reactions containing 100 ng genomic DNA, 20 pmol of each primer, 1.5 mM MgCl2, 0% to 4% formamide, 0% to 20% betaine, 1.25 UTaq DNA polymerase ([Gibco] Invitrogen, Carlsbad, CA), 200 μM dNTPs, and 1.0 μCi (0.037 MBq) α[32P]-dCTP. Alleles were resolved on 6% denaturing sequencing gels. After drying, the gels were exposed to Retina x-ray films overnight at room temperature (Retina, Berlin, Germany).
Linkage analysis
Microsatellite data from the whole-genome scan were analyzed using the computer programs GENESCAN v3.0 and GENOTYPER v2.5 (Applied Biosystems). For the calculation of multipoint logarithmic odds (LOD) scores GENEHUNTER 2.030 was used, while 2-point LOD scores were calculated using the MLINK program of the LINKAGE package 5.2031 in the FASTLINK 4.0P implementation.32 Inheritance of the disease was assumed to be autosomal recessive with complete penetrance. According to the low incidence of the disease, the frequency of the disease allele was set to 0.0001. For simplicity, marker allele frequencies were assumed to be equal. Markers in Figure1 were arranged according to the Genethon microsatellite map.33 Marker orders in Figure2 correspond to the current order in the gmap section of the Genetic Location Database (last updated March 2001).
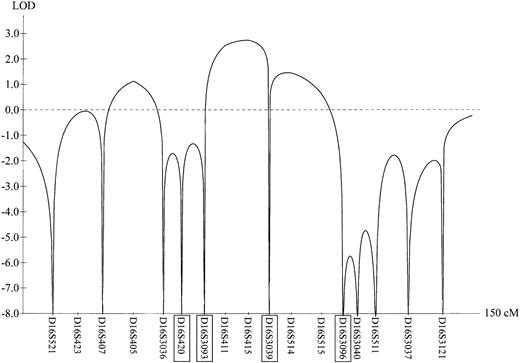
Results of the initial genome scan for chromosome 16.
Multipoint LOD scores of the FMFD phenotype in family A (see Figure 2A) were calculated with GENEHUNTER 3.0 and plotted against the Genethon microsatellite map. Markers highlighted with boxes represent backbone markers, which are also shown in Figure 2.
Results of the initial genome scan for chromosome 16.
Multipoint LOD scores of the FMFD phenotype in family A (see Figure 2A) were calculated with GENEHUNTER 3.0 and plotted against the Genethon microsatellite map. Markers highlighted with boxes represent backbone markers, which are also shown in Figure 2.
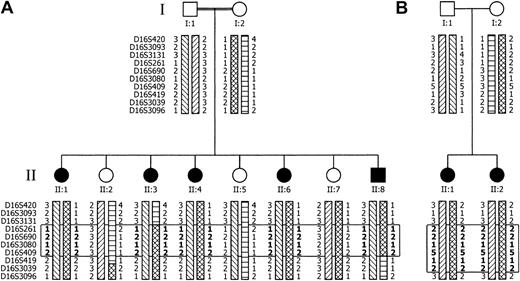
Pedigrees of FMFD families with informative haplotypes from selected markers.
For clarity, only a subset of additional markers between the backbone markers of Figure 1 is shown. The open rectangle demarcates the candidate gene region, which comprises only homozygous markers. Markers in the rectangle, presumably inherited identical by descent (IBD), are denoted in bold. The 4 parenteral haplotypes are indicated by the various box patterns. Combination of 2 box patterns in one haplotype indicates a recombination event. Family A (panel A) is of Lebanese origin; the parents are first-degree cousins. Haplotype analysis implies a heterozygous carrier state for individuals II:5 and II:7. However, this cannot be proven since the biochemical assay does not differentiate between wild-type and heterozygous carrier. The nuclear family B (panel B) originates in Germany and parents claim to be not related, although their ancestors lived in neighboring villages.
Pedigrees of FMFD families with informative haplotypes from selected markers.
For clarity, only a subset of additional markers between the backbone markers of Figure 1 is shown. The open rectangle demarcates the candidate gene region, which comprises only homozygous markers. Markers in the rectangle, presumably inherited identical by descent (IBD), are denoted in bold. The 4 parenteral haplotypes are indicated by the various box patterns. Combination of 2 box patterns in one haplotype indicates a recombination event. Family A (panel A) is of Lebanese origin; the parents are first-degree cousins. Haplotype analysis implies a heterozygous carrier state for individuals II:5 and II:7. However, this cannot be proven since the biochemical assay does not differentiate between wild-type and heterozygous carrier. The nuclear family B (panel B) originates in Germany and parents claim to be not related, although their ancestors lived in neighboring villages.
Results
Before starting a genome-wide scan for linkage in the Lebanese family (family A, Figure 2A), we scanned potential candidate genes for FMFD for mutations by direct sequencing. Potential candidate loci for this disease are (1) microsomal epoxide hydrolase 1 on chromsome 1q42.1, (2) γ-glutamyl carboxylase on chromosome 2p1.2, and (3)NAD(P)H:menadione oxidoreductase 1(NMOR1, aka DT-diaphorase orcytochrome b5 reductase) located on chromosome 16q22.1. Sequence analysis of the promotor and coding regions of these genes revealed no mutations in both families.
A whole-genome linkage analysis of the FMFD locus in family A yielded a multipoint LOD score of 2.78 at θ = 0 for marker D16S415 (Figure 1) indicating location of the FMFD locus in the centromeric region of chromosome 16. The interval with the highest likelihood of harboring the FMFD gene was confined by the markers D16S3131 and D16S419. Fine mapping studies using additional markers (Figure 2A-B) confirmed linkage to this interval in both families, with family B adding another 0.61 LOD scores. In both kindreds all affected individuals share a common region of homozygosity in the pericentromeric region of chromosome 16. Haplotyping allowed the reconstruction of recombination events which define the FMFD candidate gene region. The upper boundary of the critical region is determined by a recombination event in patient A II:3 between D16S3131 and D16S261 while the lower boundary is delimited by a recombination event between D16S409 and D16S419 in patient A II:8 (Figure 2A). These recombination events demarcate a region of homozygosity of approximately 3 centimorgans (cM) containing 26 homozygous markers (for clarity only some of them are shown in Figure 2). No recombination was observed in family B but an extended overlapping area of homozygosity is apparent from the haplotypes (Figure 2B). Combined 2-point LOD scores for both families are shown in Table 1; a maximum LOD of 3.4 was reached with marker D16S261 at θ = 0, implying significant linkage.
Values for 2-point LOD score
| Order . | Comp . | McM . | FcM . | Logarithm of the odds (LOD score) . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| θ = 0 . | θ = .01 . | θ = .05 . | θ = .1 . | θ = .2 . | θ = .3 . | θ = .4 . | ||||
| D16S420 | 31 130 | 51.65 | 43.44 | -infini | 1.32 | 1.75 | 1.71 | 1.31 | 0.77 | 0.25 |
| D16S3093 | 36 625 | 52.51 | 52.12 | -infini | −0.74 | −0.11 | 0.09 | 0.18 | 0.13 | 0.04 |
| D16S3131 | 37 256 | 52.61 | 53.17 | -infini | −0.15 | 0.40 | 0.52 | 0.45 | 0.26 | 0.08 |
| D16S261 | 40 460 | 53.69 | 64.17 | 3.39* | 3.32* | 3.03* | 2.67 | 1.91 | 1.13 | 0.38 |
| D16S690 | 52 906 | 53.69 | 65.97 | 3.39* | 3.32* | 3.03* | 2.67 | 1.91 | 1.13 | 0.38 |
| D16S3080 | 55 810 | 53.69 | 66.39 | 1.88 | 1.84 | 1.68 | 1.48 | 1.05 | 0.61 | 0.20 |
| D16S409 | 62 310 | 53.69 | 67.33 | 3.08* | 3.02* | 2.77 | 2.45 | 1.78 | 1.06 | 0.36 |
| D16S419 | 63 668 | 56.83 | 78.35 | -infini | 1.32 | 1.75 | 1.71 | 1.31 | 0.77 | 0.25 |
| D16S3039 | 65 124 | 56.83 | 92.29 | -infini | 1.32 | 1.73 | 1.68 | 1.26 | 0.72 | 0.23 |
| D16S3096 | 91 611 | 69.66 | 120.05 | -infini | −1.39 | −0.17 | 0.20 | 0.34 | 0.24 | 0.08 |
| Order . | Comp . | McM . | FcM . | Logarithm of the odds (LOD score) . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| θ = 0 . | θ = .01 . | θ = .05 . | θ = .1 . | θ = .2 . | θ = .3 . | θ = .4 . | ||||
| D16S420 | 31 130 | 51.65 | 43.44 | -infini | 1.32 | 1.75 | 1.71 | 1.31 | 0.77 | 0.25 |
| D16S3093 | 36 625 | 52.51 | 52.12 | -infini | −0.74 | −0.11 | 0.09 | 0.18 | 0.13 | 0.04 |
| D16S3131 | 37 256 | 52.61 | 53.17 | -infini | −0.15 | 0.40 | 0.52 | 0.45 | 0.26 | 0.08 |
| D16S261 | 40 460 | 53.69 | 64.17 | 3.39* | 3.32* | 3.03* | 2.67 | 1.91 | 1.13 | 0.38 |
| D16S690 | 52 906 | 53.69 | 65.97 | 3.39* | 3.32* | 3.03* | 2.67 | 1.91 | 1.13 | 0.38 |
| D16S3080 | 55 810 | 53.69 | 66.39 | 1.88 | 1.84 | 1.68 | 1.48 | 1.05 | 0.61 | 0.20 |
| D16S409 | 62 310 | 53.69 | 67.33 | 3.08* | 3.02* | 2.77 | 2.45 | 1.78 | 1.06 | 0.36 |
| D16S419 | 63 668 | 56.83 | 78.35 | -infini | 1.32 | 1.75 | 1.71 | 1.31 | 0.77 | 0.25 |
| D16S3039 | 65 124 | 56.83 | 92.29 | -infini | 1.32 | 1.73 | 1.68 | 1.26 | 0.72 | 0.23 |
| D16S3096 | 91 611 | 69.66 | 120.05 | -infini | −1.39 | −0.17 | 0.20 | 0.34 | 0.24 | 0.08 |
Values for 2-point LOD score (MLINK) for linkage between familial multiple coagulation factor deficiency (FMFD) and selected chromosome 16 markers based on the 2 families, obtained at different recombination fractions. Male and female map distances are given in centimorgans (McM and FcM, respectively). Comp refers to a composite location in megabases according to the LDB database.
Indicates LOD scores greater than 3.0, providing an odds ratio of linkage versus free recombination of greater than 1000:1.
Haplotype analysis provided additional evidence against the potential candidate gene NAD(P)H:menadione oxidoreductase 1. NMOR 1 is located between markers D16S514 and D16S515 and microsatellite analysis revealed heterozygosity for these markers in the severely affected individual II:8 (data not shown). Therefore, we were able to exclude mutations in potentialcis-regulatory elements in the surrounding regions of NMOR1.
The presence of an identical haplotype for the interval between D16S261 and D16S419 in the parents of family B (Figure 2B) suggests that these chromosomal segments are identical by descent (IBD) and that the parents are likewise related. An association by chance of the alleles of 26 markers spanning a genetic distance of at least 3 cM is extremely unlikely.34
Discussion
We have assigned a second gene for hereditary combined multiple coagulation factor deficiency (FMFD) to chromosome 16p12-q21 after exclusion of several other potential candidate genes. Homozygosity mapping revealed the pericentromeric region of chromosome 16 as the most likely location of this hereditary bleeding disorder. Missense mutations in another gene involved in γ-carboxylation, namelyγ-glutamyl carboxylase, have already been described.11,12 Cain et al20 suggested that a warfarin-sensitive member of the glutathione-S-transferase (GST) superfamily of proteins is a component of the multienzyme complex VKOR. We have scanned several available gene maps, based on the current working draft (July 2001) of the human genome. Among the approximately 300 annotated genes in the candidate region no novel GST family member has been predicted so far, but presumably there are a number of gaps in the current working draft due to large and complex duplications of genomic material on 16p.35 It is conceivable that mutations in the gene for this unknown GST enzyme would also lead to symptoms comparable to multiple congenital coagulation factor deficiency. It remains to be established whether the FMFD phenotype in the families we studied is based on gene mutations in a still unknown GST or another currently unknown gene product which is involved in vitamin K recycling.
A phenotype related to human coagulation factor deficiency has been described in rats and mice as resistance to the anticoagulant drug warfarin. The genes responsible for warfarin resistance have been mapped in mice (war) to chromosome 7 at a position of 62.5 cM from pter (mouse genome informatics) and in rats (Rw) to chromosome 1q35-42. Chromosome 7 of mouse and chromosome 1 of rat share extensive areas of synteny (Mouse to Rat Homology Data; see “”). Fine mapping studies for Rw performed by Kohn and Pelz36 revealed several homologous regions on human chromosomes 10q, 12q, and 16p11-13. Similarly, the Human to Mouse Homology Data (see “”) predict homology of mouse chromosome 7 to human chromosomes 10, 11, 15, 16, and 19. The corresponding LOD scores for all these chromosomes (except for 16) in family A are clearly below −2 (data not shown), which excludes linkage of FMFD to these segments of the human genome. Several anchor loci flanking the locus for warfarin resistance, both in mouse and rat (ie,interleukin 4 receptor, alpha (IL4R), andsialophorin [SPN]) are located on the short arm of human chromosome 16. Colocalization of FMFD in humans and warfarin resistance in mouse and rat on homologous chromosomal segments implies that both phenotypes may be allelic. The fact that a warfarin-sensitive GST is part of the VKOR complex provides further evidence for this notion.20 Alterations in the warfarin binding site of the GST could lead to a phenotype of warfarin resistance, while alterations in another part of the enzyme (influencing the associated catalytic epoxide reductase component) would result in the FMFD phenotype with bleeding tendency due to a lack of recycled vitamin K1H2. Assuming a single gene for both phenotypes, mouse-human homology maps would restrict the candidate gene region to the short arm of chromosome 16. Alternatively, there may be 2 genes leading to FMFD and warfarin resistance, which are located on chromosome 16p12-q21.
Since warfarin is a widely used oral anticoagulant in humans and rodents, the isolation of the FMFD gene would be an important step toward elucidation of the vitamin K cycle and associated abnormalities, including defects of the multienzyme complex vitamin K 2,3-epoxide reductase.
We thank A. Reis for the whole-genome scan data and A. Baumer for her invaluable help with preparing the manuscript.
electronic database information
Online Mendelian Inheritance in Man (OMIM),http://www.ncbi.nlm.nih.gov./Omim (for FMFD [MIM #277 450])
Human to Mouse Homology Data, http://www.ncbi.nlm.nih.gov/Omim/Homology
Mouse to Rat Homology Data, http://gapp.gen.gu.se/index.html
Mouse Genome Informatics, http://www.informatics.jax.org/
Order of markers (Genethon),http://www.genlink.wustl.edu/genethon_frame/chr16/
Order of markers (Marshfield),http://research.marshfieldclinic.org/genetics/
Order of markers (LDB), http://cedar.genetics.soton.ac.uk/pub/chrom16
Prepublished online as Blood First Edition Paper, July 5, 2002; DOI 10.1182/blood-2002-03-0698.
Supported by grants of the DFG (OI 100/3-1), Baxter Germany, the Stiftung Hämotherapie-Forschung, and the Gesellschaft für Thrombose und Hämostaseforschung. The Gene Mapping Centre was supported by a grant-in aid from the German Genome Project.
A.F., S.R., and W.W. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Johannes Oldenburg, Institute of Transfusion Medicine and Immune Haematology of the DRK Blood Donor Service, Sandhofstr 1, 60526 Frankfurt, Germany; e-mail:joldenburg@bsdhessen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal