Abstract
Severe congenital neutropenia (SCN) is a syndrome characterized by an isolated block in granulocytic differentiation and an increased risk of developing acute myeloid leukemia (AML). Recent studies have demonstrated that the majority of patients with SCN and cyclic neutropenia, a related disorder characterized by periodic oscillations in the number of circulating neutrophils, have heterozygous germline mutations in the ELA2 gene encoding neutrophil elastase (NE). To test the hypothesis that these mutations are causative for SCN, we generated transgenic mice carrying a targeted mutation of theirEla2 gene (“V72M”) reproducing a mutation found in 2 unrelated patients with SCN, one of whom developed AML. Expression of mutant NE mRNA and enzymatically active protein was confirmed. Mice heterozygous and homozygous for the V72M allele have normal numbers of circulating neutrophils, and no accumulation of myeloid precursors in the bone marrow was observed. Serial blood analysis found no evidence of cycling in any of the major hematopoietic lineages. Rates of apoptosis following cytokine deprivation were similar in wild-type and mutant neutrophils, as were the frequency and cytokine responsiveness of myeloid progenitors. The stress granulopoiesis response, as measured by neutrophil recovery after cyclophosphamide-induced myelosuppression, was normal. To define the leukemogenic potential of V72M NE, a tumor watch was established. To date, no cases of leukemia have been detected. Collectively, these data suggest that expression of V72M NE is not sufficient to induce an SCN phenotype or leukemia in mice.
Introduction
Severe congenital neutropenia (SCN) is a disorder characterized by severe neutropenia present from birth. Absolute neutrophil counts (ANCs) are usually less than 200 cells/mm3, with the remainder of the blood counts relatively normal. The bone marrow invariably shows an arrest in myeloid maturation with an accumulation of promyelocytes or myelocytes. Kostmann first reported the disease in 1956 as a familial dysgranulopoietic disorder that appeared to be inherited in an autosomal recessive fashion.1 Since then, sporadic, autosomal dominant, and X-linked forms of the disease have also been described.2,3 Cyclic neutropenia (CN) is an autosomal dominant disorder characterized by periodic oscillations in the numbers of circulating neutrophils and other peripheral blood cells.4 Although SCN and CN have different presentations, recent studies have found evidence of cycling in patients classified as having SCN, suggesting that these 2 diseases are related and may fall along a continuum of hematopoietic disorders.5 Patients with SCN and CN suffer from recurrent opportunistic infections. Furthermore, approximately 9% of patients with SCN develop myelodysplasia or acute myeloid leukemia (AML). A recent report from the Severe Congenital Neutropenia International Registry6found that 35 of 388 patients enrolled in the registry as of December 31, 1999, had developed AML or myelodysplastic syndrome (MDS). Currently, the only cure for SCN is bone marrow transplantation,7 although most patients achieve a significant increase in the numbers of circulating neutrophils in response to high doses of granulocyte colony-stimulating factor (G-CSF).8
In an effort to characterize the block in granulocytic differentiation, several groups have studied the growth and differentiation of hematopoietic progenitors isolated from patients with SCN. These studies showed that the number and cytokine responsiveness of myeloid progenitors from patients with SCN are reduced.9-12 In particular, G-CSF–induced proliferation and granulocytic differentiation is markedly impaired. Collectively, these studies suggest a cell intrinsic defect in granulocytic differentiation of myeloid progenitors in patients with SCN. Consistent with this observation, a recent preliminary report suggested that CD34+, CD33+/CD34−, and CD15+/CD33− cells from patients with SCN display an increased susceptibility to apoptosis.13Likewise, increased rates of apoptosis have been observed in myeloid cells from patients with CN14 and Shwachman-Diamond syndrome.15 16
Acquired mutations of the G-CSF receptor (G-CSFR) are present in a subset of patients with SCN.17-20 In a recent series, G-CSFR mutations were detected in 34 of 97 patients with SCN.21 Intriguingly, these mutations are nearly always nonsense C→T transitions, which introduce a premature stop codon resulting in the truncation of the distal cytoplasmic portion of the G-CSFR. G-CSFR mutations are associated with the development of AML/MDS in patients with SCN. A recent study found that 13 of 34 patients with G-CSFR mutations developed MDS or AML, whereas only 2 of 63 patients without G-CSFR mutations developed leukemia.21A role for G-CSFR mutations in the pathogenesis of SCN has been postulated. However, recent data have shown that G-CSFR mutations are acquired and have no apparent effect on the severity of neutropenia or response to G-CSF.22 Transgenic mice expressing a G-CSFR with a mutation derived from several patients with SCN have been generated.23-25 These mice, although modestly neutropenic, do not have an accumulation of myeloid precursors in their bone marrow. Indeed, hematopoietic progenitors from these mice display a hyperproliferative response to G-CSF. Thus, G-CSFR mutations are not responsible for the block in myeloid maturation in SCN, although their contribution to leukemia in patients with SCN is still unclear.
Genome-wide genetic linkage analysis and positional cloning determined that CN is a genetically homogeneous disease associated with theELA2 gene on chromosome 19p13.3.26 TheELA2 gene encodes neutrophil elastase (NE), a serine protease found in the primary granules of neutrophils. Further studies have found that a subset of patients with SCN also have mutations to the ELA2 gene.27,28 A recent review identified mutations in ELA2 in 43 of 46 patients with SCN,29 and a second report found mutations in 9 of 18 patients.27 To date, a total of 27 mutations causing 25 distinct alterations in the NE protein have been reported.26-28,30 None of the DNA sequence changes have been identified in large control populations, confirming that these mutations are not polymorphisms. The recent description of a case of paternal mosaicism in a family with SCN provides further evidence implicating ELA2 gene mutations in the pathogenesis of SCN. The father of a patient with SCN and an ELA2 mutation was found to have the same mutation but was not neutropenic. Although approximately 50% of his T lymphocytes are heterozygous for theELA2 mutation, it is nearly absent in his peripheral blood neutrophils. These results suggest that this individual is a mosaic who acquired a mutation in one of his ELA2 alleles at the 2-cell stage of development.30 Because NE expression is normally restricted to myeloid cells,31 and because no toxic paracrine effects on wild-type neutrophils were seen, these observations suggest that expression of mutant NE inhibits granulopoiesis in a cell intrinsic manner. Collectively, these data support the hypothesis that mutations to the ELA2 gene are causative for some cases of SCN.
To test this hypothesis, we generated mice carrying a targeted (knock-in) mutation of their NE gene, reproducing a mutation found in patients with SCN. Specifically, a 298G>A (G→A substitution at nucleotide 298) of the murine cDNA was introduced into exon 3 of the murine Ela2 gene by homologous recombination, resulting in the substitution of methionine for valine at amino acid 72 of the active protease. This amino acid is conserved in human and murine NE. The rationale for choosing this mutation (called V72M NE) is 2-fold. First, the Val72Met mutation has been identified in 2 unrelated families with SCN but not in any healthy controls; therefore, it is unlikely to represent a polymorphism. Second, one of the patients with the V72M NE mutation developed AML, suggesting that the V72M NE mutation may be leukemogenic. In this study, we show that expression of V72M NE is not sufficient to cause an SCN phenotype or leukemia in mice.
Materials and methods
Generation of transgenic mice
The d715 G-CSFR mutant mice were generated in our laboratory as described previously.25 These mice have been backcrossed for 10 generations onto a C57BL/6 background. NE−/−/CG−/− mice lacking NE and cathepsin G32 (CG) were generated from NE−/−mice33 and CG−/− mice34 and maintained on a 129/SvJ background (a gift of Timothy Ley, Washington University, St Louis, MO). NE−/−/CG−/− mice have normal granulopoiesis.35
The Val72Met mutation was introduced into the murineEla2 gene by homologous recombination in embryonic stem (ES) cells. A 298G>A of the murine NE cDNA (corresponding to G43362A, GenBank accession no. AC087114) was introduced by site directed mutagenesis into exon 3 of the Ela2 gene using the following primers: forward 5′-CCTTCTCTATGCAGCGGATCTTCGAG-3′, reverse 5′-CCGCTGCATAGAGAAGGTCTGTCGAG-3′. The underlined nucleotides represent the G-to-A substitution, which replaces the valine at amino acid 72 (numbered from the first amino acid of the active protease) with methionine. The replacement-type targeting vector contains 4 kb of 5′ targeting sequence, a neomycin phosphotransferase gene driven by the phosphoglycerate kinase I promoter flanked byloxP sites (PGK-neo), and 3 kb of 3′ targeting sequence (Figure 1). RW4 ES cells were transfected with the NotI linearized V72M targeting vector, and G418-resistant clones were isolated as described.36 Two independent clones that had undergone homologous recombination were identified by Southern blot analysis. The loxP-flanked PGK-neo gene was subsequently removed bycre-recombinase-mediated excision. C57BL/6 blastocysts were microinjected with ES cells from each of these clones and implanted into pseudopregnant Swiss Webster foster females, as described previously.36 Chimeric mice derived from both ES clones were capable of germline transmission of the V72M allele. The wild-type and heterozygous V72M mice studied are C57BL/6 × 129/SvJ F1 hybrids; homozygous V72M mice generated by F1 intercrosses are outbred on a C57BL/6 × 129/SvJ background. All mice were housed in a specific-pathogen–free environment and examined twice weekly by veterinary staff for signs of illness. All studies were approved by the Animal Studies Committee at Washington University.
Targeting strategy and Southern blot analysis.
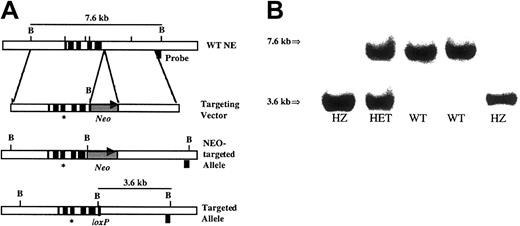
(A) Targeting strategy. The genomic organization of the murineEla2 gene is shown in the upper panel. Exons are indicated by solid boxes. The targeting vector is shown in the second panel. The asterisk represents the G→A mutation at nucleotide 298 of the cDNA. The neomycin phosphotransferase gene driven by the phosphoglycerate kinase I (PGK) promoter and flanked by loxP sites is denoted as Neo. Cre-recombinase–mediated excision of theNeo resistance cassette generated the final recombined allele, shown in the bottom panel. The location of the external probe used in the Southern blot analysis and the size of the expectedBamHI fragments are shown. (B) Southern blot analysis ofBamHI-digested genomic tail DNA isolated from the progeny of a heterozygous intercross. The wild-type allele (7.6 kb) and recombined allele (3.6 kb) are shown.
Targeting strategy and Southern blot analysis.
(A) Targeting strategy. The genomic organization of the murineEla2 gene is shown in the upper panel. Exons are indicated by solid boxes. The targeting vector is shown in the second panel. The asterisk represents the G→A mutation at nucleotide 298 of the cDNA. The neomycin phosphotransferase gene driven by the phosphoglycerate kinase I (PGK) promoter and flanked by loxP sites is denoted as Neo. Cre-recombinase–mediated excision of theNeo resistance cassette generated the final recombined allele, shown in the bottom panel. The location of the external probe used in the Southern blot analysis and the size of the expectedBamHI fragments are shown. (B) Southern blot analysis ofBamHI-digested genomic tail DNA isolated from the progeny of a heterozygous intercross. The wild-type allele (7.6 kb) and recombined allele (3.6 kb) are shown.
Real-time quantitative RT-PCR
Total cellular RNA was isolated from 107 bone marrow cells using the High Pure RNA Isolation Kit (Roche Diagnostics, Indianapolis, IN) and eluted in 50 μL buffer. Real-time quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) was performed using the TaqMan One-step RT-PCR Master Mix Reagents Kit (Applied Biosystems, Foster City, CA) on a GeneAmp 5700 Sequence Detection System (Applied Biosystems). The reaction mix consisted of 1 μL RNA, 12.5 μL RT-PCR reaction mix, 10 μM forward primer (5′-GTAGTGCTGGGAGCCCATGAC-3′), 10 μM reverse primer (5′-ACATGGAGTTCTGTCACCCA-3′), 10 μM internal probe (5′-CCAACGTGCAGGTGGCCCAG-3′), and 5 μL Multiscribe reverse transcriptase and RNase inhibitor in a total reaction volume of 25 μL. Reactions were repeated in the absence of reverse transcriptase to confirm that DNA contamination was not present. RNA content was normalized to murine glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the TaqMan Rodent GAPDH Control Reagent Kit (Applied Biosystems). PCR conditions were 48°C for 30 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
NE activity assay
Approximately 107 bone marrow cells were pelleted and resuspended in 200 μL HEBS buffer (20 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 137 mM NaCl, 5 mM KCl, 0.7 mM Na2PO4, 6 mM glucose). An equal volume of lysis buffer (2 M NaCl,50 mM Tris [tris(hydroxymethyl)aminomethane], pH 7.5,1 mM EDTA [ethylenediaminetetraacetic acid], 2% Triton X-100) was added, and total protein was harvested following sonication and centrifugation. Elastase activity was assayed using the chromogenic, NE-specific substrate N-methoxysuccinyl Ala-Ala-Pro-ValP-nitroanilide (Sigma, St Louis, MO)37reconstituted to a concentration of 1 mM in 0.5M HEPES buffer (0.5 M HEPES, 2.5 M NaCl, 0.5% Brij 35, pH 7). Then, 40 μL cleared cell lysate was added to wells containing 40 μL Hanks buffered salt solution (Sigma) and 20 μL 1 mM substrate. Optical density measurements at 405 nm were taken at 0, 5, 10, 15, 30, and 60 minutes using a Molecular Devices Emax Precision Microplate Reader (Molecular Devices, Sunnyvale, CA).
Peripheral blood and bone marrow analysis
Blood was obtained by retro-orbital venous plexus sampling in polypropylene tubes containing EDTA. Complete blood counts were determined using a Hemavet 850FS automated cell counter (CDC Technologies, Oxford, CT). Bone marrow was harvested by flushing both femurs with phosphate-buffered saline with 0.1% bovine serum albumin (BSA). Manual leukocyte differentials were performed on Wright-stained blood smears (minimum 200 cells) or cytospin preparations of bone marrow cells (minimum 500 cells).
Apoptosis assay
Neutrophils were purified from the bone marrow of mice using a discontinuous Percoll gradient exactly as described.38Neutrophil purity (as assessed by leukocyte differentials) was at least 70% for all cell preparations. Cells were suspended in Opti-MEM media (Gibco, Grand Island, NY) with 10 μg/mL ciprofloxacin and cultured in a humidified chamber with 6% CO2 at 37°C for the indicated period of time in the presence or absence of 100 ng/mL human G-CSF. Absolute cell numbers were determined using a hemacytometer. To assay for apoptosis, cells were washed once in binding buffer (20 mM HEPES, pH 7.4, 132 mM NaCl, 6 mM KCl, 2.5 mM CaCl2, 1 mM MgCl2, 1.2 mM KH2PO4, 5.5 mM glucose, and 0.5% BSA), incubated with fluorescein isothiocyanate (FITC)–conjugated annexin V (Nexins Research, Roermond, The Netherlands) for 30 minutes at 4°C, washed twice in binding buffer, and stained with 7-aminoactinomycin D (7-AAD; Calbiochem, La Jolla, CA). Analyses were performed on a FACScan flow cytometer (Becton Dickinson Immunocytometry Systems, Mansfield, MA). Baseline percentages of apoptotic cells were determined after 1 hour of culture to reduce nonspecific binding of annexin V.
Hematopoietic progenitor cell assays
A total of 2.5 to 5 × 104 bone marrow mononuclear cells were plated in 3.0 mL methylcellulose media (MethoCult 3231; Stem Cell Technologies, Vancouver, BC, Canada) supplemented with human G-CSF (Amgen, Thousand Oaks, CA) or murine granulocyte-macrophage colony-stimulating factor (GM-CSF; R & D Systems, Minneapolis, MN) at the indicated concentrations. Cultures were plated in duplicate and placed in a humidified chamber with 6% CO2 at 37°C. Colonies containing at least 50 cells were scored on day 7 of culture.
Stress granulopoiesis assay
Cyclophosphamide (Sigma) was reconstituted in sterile water and given as a single intraperitoneal injection at a dose of 200 mg/kg. Peripheral blood analysis was performed prior to injection and on days 3, 6, 8, and 10 (cohort 1) or on days 4, 6, and 8 (cohort 2) after cyclophosphamide treatment.
Tumor watch
No published data on the rate of development of AML in C57BL/6 × 129/SvJ F1 hybrids are available. A search of the Jackson Labs Mouse Tumor Biology Database (http://tumor.informatics.jax.org) found no reports of spontaneous myeloid leukemia in 129/SvJ inbred mice and one published report involving C57BL/6 inbred mice in which no mice developed leukemia. Assuming that hybrids will not be at increased risk for the development of AML relative to either parental strain, we conservatively estimated the background rate of AML to be less than 0.1%. Based on this assumption, we calculated that a minimum of 27 wild-type and heterozygous V72M NE mice would be needed to detect a 9% difference in rates of leukemia with probabilities of type I and type II errors of 0.05 and 0.10, respectively. Accordingly, a tumor watch with 37 heterozygous V72M NE and 27 wild-type strain-matched mice was established.
Statistical analysis
Data are presented as the mean ± SEM. Statistical significance was assessed by 2-tailed Student ttest.
Results
Generation of V72M NE mice
To generate the Val72Met mutation in mice, 298G>A of the murine NE cDNA was introduced into the murine Ela2 gene by homologous recombination in ES cells. Two correctly targeted ES clones of a total of 144 clones were identified. Because retention of selectable markers can have unpredictable neighborhood effects on nearby genes, the loxP-flanked PGK-Neo cassette was removed by cre-recombinase–mediated excision.39Transgenic mouse lines carrying the V72M NE allele were derived from both ES clones. Because the phenotype of these mice was similar, the data have been combined throughout this paper. The V72M NE allele is inherited in a mendelian fashion. Animals heterozygous and homozygous for the V72M NE allele display normal growth, development, and fertility, and they are grossly indistinguishable from wild-type littermates.
Wild-type and V72M NE alleles are expressed in a similar manner
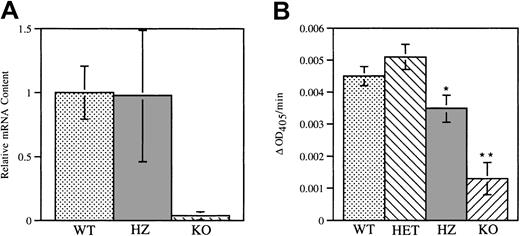
Expression of V72M NE mRNA in bone marrow cells was assessed using a real-time quantitative RT-PCR assay. RNA isolated from mice carrying targeted disruptions of their NE and CG genes, which do not express NE, served as a negative control. Similar levels of NE mRNA transcripts were detected in wild-type and homozygous V72M bone marrow cells (Figure 2A), indicating that mRNA expression from the V72M NE allele is comparable to that of the wild-type NE allele. The fidelity of the V72M NE allele was confirmed by direct sequencing of the RT-PCR product from homozygous mice (data not shown).
V72M NE expression.
(A) NE mRNA was measured in wild-type and homozygous V72M NE bone marrow cells using a real-time quantitative RT-PCR assay. The amount of NE mRNA transcripts from wild-type (WT) and homozygous (HZ) mice (n = 5, each) were quantified relative to the amount of GAPDH mRNA. NE/CG-deficient mice (KO, n = 2) were used as a negative control. Data are normalized relative to the amount of NE mRNA transcripts in wild-type mice. (B) Functional NE protein in bone marrow cells from wild-type (n = 7), heterozygous (HET, n = 5), homozygous (n = 5), and NE/CG-deficient mice (n = 2) was quantified using a chromogenic, NE-sensitive substrate. The change in optical density at 405 nm (ΔOD405) per minute is shown. Data represent the mean ± SEM. *P < .05 compared with heterozygous V72M NE mice. **P < .05 compared to wild-type, heterozygous, and homozygous mice.
V72M NE expression.
(A) NE mRNA was measured in wild-type and homozygous V72M NE bone marrow cells using a real-time quantitative RT-PCR assay. The amount of NE mRNA transcripts from wild-type (WT) and homozygous (HZ) mice (n = 5, each) were quantified relative to the amount of GAPDH mRNA. NE/CG-deficient mice (KO, n = 2) were used as a negative control. Data are normalized relative to the amount of NE mRNA transcripts in wild-type mice. (B) Functional NE protein in bone marrow cells from wild-type (n = 7), heterozygous (HET, n = 5), homozygous (n = 5), and NE/CG-deficient mice (n = 2) was quantified using a chromogenic, NE-sensitive substrate. The change in optical density at 405 nm (ΔOD405) per minute is shown. Data represent the mean ± SEM. *P < .05 compared with heterozygous V72M NE mice. **P < .05 compared to wild-type, heterozygous, and homozygous mice.
Expression of functional NE protein in the bone marrow cells of wild-type and heterozygous and homozygous V72M NE mice was assessed using an NE-specific assay (Figure 2B). Compared with NE/CG-deficient cells, significant elastolytic activity was detected in bone marrow cells isolated from all 3 genotypes. However, elastolytic activity in homozygous V72M NE cells was modestly reduced to approximately 70% of that observed in wild-type cells. Of note, a previous report showed that the elastolytic activity of protein extracts from RBL-1 and 32D cells expressing human V72M NE is approximately 40% that of extracts from cells expressing wild-type human NE.40 Although the lack of an antibody specific for murine NE precludes precise measurements of V72M NE-specific activity, these data suggest that the Val72Met mutation may have a similar effect on the elastolytic activity of human and murine NE.
Mice expressing V72M NE have normal basal granulopoiesis
The hallmarks of SCN are neutropenia and an accumulation of granulocytic precursors in the bone marrow. Analysis of peripheral blood from mice 4 to 6 weeks of age revealed no significant differences in red blood cell (RBC) counts, white blood cell (WBC) counts, platelets, or ANCs between wild-type mice and mice heterozygous or homozygous for the V72M allele (Table 1). Peripheral blood also was analyzed in 1-week-old mice. At 1 week of age, ANCs (× 106/mL blood) were 0.84 ± 0.10 for wild-type mice (n = 7), 1.03 ± 0.11 for heterozygous mice (n = 5), and 1.07 ± 0.09 for homozygous mice (n = 14), suggesting that heterozygous and homozygous mice have normal neutrophil counts from birth. Examination of bone marrow revealed normal cellularity and found that the morphology and frequency of granulocytic precursors in the bone marrow is similar in mice of all 3 genotypes. Importantly, no accumulation of granulocytic precursors at the promyelocyte stage indicative of a block in myeloid maturation was observed.
Peripheral blood and bone marrow analysis
| . | WT . | HET . | HZ . |
|---|---|---|---|
| Peripheral blood | |||
| No. of mice | 14 | 19 | 11 |
| WBC (× 106/mL) | 8.59 ± 0.93 | 8.35 ± 0.43 | 7.27 ± 0.66 |
| RBC (× 109/mL) | 9.33 ± 0.67 | 9.05 ± 0.31 | 9.96 ± 0.66 |
| Platelets (× 106/mL) | 566 ± 32 | 610 ± 28 | 561 ± 34 |
| Neutrophils (%) | 7.8 ± 1.0 | 8.1 ± 0.9 | 8.6 ± 1.0 |
| Neutrophils (× 106/mL) | 0.63 ± 0.10 | 0.67 ± 0.07 | 0.58 ± 0.06 |
| Bone marrow | |||
| No. of mice | 5 | 5 | 5 |
| Blasts and promyelocytes (%) | 1.5 ± 0.2 | 1.6 ± 0.3 | 2.4 ± 0.2 |
| Myelocytes (%) | 9.3 ± 1.0 | 8.4 ± 0.4 | 7.8 ± 0.9 |
| Metamyelocytes (%) | 10.2 ± 0.3 | 10.1 ± 0.6 | 9.2 ± 0.9 |
| Bands and neutrophils (%) | 24.0 ± 2.9 | 27.0 ± 4.0 | 22.7 ± 1.0 |
| Eosinophils (%) | 3.5 ± 0.6 | 3.5 ± 0.4 | 3.3 ± 0.5 |
| Erythroid lineage (%) | 36.8 ± 2.4 | 39.0 ± 3.7 | 39.1 ± 1.4 |
| Lymphoid lineage (%) | 14.6 ± 1.5 | 10.6 ± 1.2 | 15.5 ± 1.1 |
| Myeloid-erythroid ratio | 1.2 ± 0.1 | 1.2 ± 0.3 | 1.1 ± 0.1 |
| . | WT . | HET . | HZ . |
|---|---|---|---|
| Peripheral blood | |||
| No. of mice | 14 | 19 | 11 |
| WBC (× 106/mL) | 8.59 ± 0.93 | 8.35 ± 0.43 | 7.27 ± 0.66 |
| RBC (× 109/mL) | 9.33 ± 0.67 | 9.05 ± 0.31 | 9.96 ± 0.66 |
| Platelets (× 106/mL) | 566 ± 32 | 610 ± 28 | 561 ± 34 |
| Neutrophils (%) | 7.8 ± 1.0 | 8.1 ± 0.9 | 8.6 ± 1.0 |
| Neutrophils (× 106/mL) | 0.63 ± 0.10 | 0.67 ± 0.07 | 0.58 ± 0.06 |
| Bone marrow | |||
| No. of mice | 5 | 5 | 5 |
| Blasts and promyelocytes (%) | 1.5 ± 0.2 | 1.6 ± 0.3 | 2.4 ± 0.2 |
| Myelocytes (%) | 9.3 ± 1.0 | 8.4 ± 0.4 | 7.8 ± 0.9 |
| Metamyelocytes (%) | 10.2 ± 0.3 | 10.1 ± 0.6 | 9.2 ± 0.9 |
| Bands and neutrophils (%) | 24.0 ± 2.9 | 27.0 ± 4.0 | 22.7 ± 1.0 |
| Eosinophils (%) | 3.5 ± 0.6 | 3.5 ± 0.4 | 3.3 ± 0.5 |
| Erythroid lineage (%) | 36.8 ± 2.4 | 39.0 ± 3.7 | 39.1 ± 1.4 |
| Lymphoid lineage (%) | 14.6 ± 1.5 | 10.6 ± 1.2 | 15.5 ± 1.1 |
| Myeloid-erythroid ratio | 1.2 ± 0.1 | 1.2 ± 0.3 | 1.1 ± 0.1 |
Mice were analyzed at 4 to 6 weeks of age. Peripheral blood differentials are based on 200 cell counts.
Bone marrow differentials are based on 500 cell counts. Data represent the mean ± SEM.
HET indicates heterozygous; HZ, homozygous; WT, wild-type.
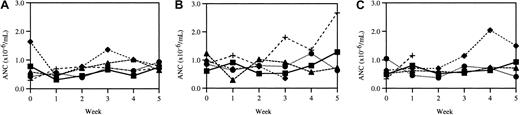
Because mutations of the NE gene have been associated with CN as well as SCN, 3 cohorts of age-matched mice were bled weekly for 6 weeks to determine if there was obvious cycling in any of the hematopoietic lineages. Whereas counts in patients with CN have been observed to cycle with an average periodicity of 21 days, no such cycling of the ANCs (Figure 3), WBC counts, RBC counts, or platelet counts (data not shown) was seen. To measure the degree of cycling of peripheral blood neutrophils more quantitatively, we plotted the difference between the maximum and minimum neutrophil count for each mouse obtained during the 5-week observation period. For wild-type mice, the average maximum difference in ANCs (× 106/mL blood) was 0.65 ± 0.14 (n = 5); for heterozygous mice, the average was 0.96 ± 0.24 (n = 5); and for homozygous mice, the average was 0.68 ± 0.26 (n = 4). Collectively, these data suggest that basal granulopoiesis is normal in mice expressing V72M NE.
Weekly neutrophil counts.
Three cohorts of age-matched wild-type mice (A) and mice heterozygous (B) and homozygous (C) for the V72M NE allele were bled weekly for 6 weeks and the number of neutrophils quantified. Each line represents a single animal. One heterozygous mouse died after week 4 (♦); one homozygous mouse developed an eye infection after week 2 (+).
Weekly neutrophil counts.
Three cohorts of age-matched wild-type mice (A) and mice heterozygous (B) and homozygous (C) for the V72M NE allele were bled weekly for 6 weeks and the number of neutrophils quantified. Each line represents a single animal. One heterozygous mouse died after week 4 (♦); one homozygous mouse developed an eye infection after week 2 (+).
Neutrophils expressing V72M NE do not have higher rates of apoptosis in response to serum deprivation than wild-type neutrophils
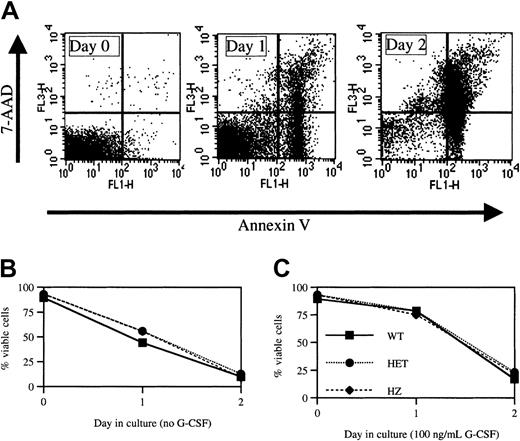
A previous study showed an increased rate of apoptosis in myeloid progenitors and granulocytic precursors isolated from patients with CN.14 In addition, preliminary data suggest an increased susceptibility to apoptosis is also present in myeloid cells obtained from patients with SCN.13 To test whether the expression of V72M NE results in increased rates of apoptosis, neutrophils were isolated from the bone marrow of wild-type and heterozygous and homozygous V72M NE mice (n = 3, each) and cultured in serum-free media. Cells were then analyzed for viability at 24 and 48 hours using a flow cytometry–based assay utilizing annexin-V and 7-AAD staining (Figure 4A). At the time of harvest, more than 90% of neutrophils isolated from mice of each genotype were viable (Figure 4B). The percentage of viable cells decreased to 50% by 24 hours and to 10% by 48 hours in cultures of wild-type neutrophils. Similar rates of apoptosis were observed in neutrophils isolated from heterozygous or homozygous V72M NE mice.
Apoptosis assay.
(A) Histogram plots from a representative experiment in which bone marrow neutrophils isolated from a heterozygous V72M NE mouse were cultured in the absence of G-CSF and harvested at the indicated times. Cells were stained for Gr-1, annexin V, and 7-AAD. Shown is annexin V and 7-AAD staining after gating for Gr-1+ (granulocytic) cells. The percentages of viable (annexin V−, 7-AAD−) cells within the Gr-1+ population were 91.8%, 64.5%, and 9.8% on days 0, 1, and 2, respectively. (B,C) Neutrophils isolated from the bone marrow of mice of each genotype (n = 3, each) were cultured in the absence (B) or presence (C) of 100 ng/mL G-CSF. The percent of viable cells is shown. Data represent mean ± SEM.
Apoptosis assay.
(A) Histogram plots from a representative experiment in which bone marrow neutrophils isolated from a heterozygous V72M NE mouse were cultured in the absence of G-CSF and harvested at the indicated times. Cells were stained for Gr-1, annexin V, and 7-AAD. Shown is annexin V and 7-AAD staining after gating for Gr-1+ (granulocytic) cells. The percentages of viable (annexin V−, 7-AAD−) cells within the Gr-1+ population were 91.8%, 64.5%, and 9.8% on days 0, 1, and 2, respectively. (B,C) Neutrophils isolated from the bone marrow of mice of each genotype (n = 3, each) were cultured in the absence (B) or presence (C) of 100 ng/mL G-CSF. The percent of viable cells is shown. Data represent mean ± SEM.
It is known that G-CSF suppresses apoptosis in serum-deprived neutrophils.41 We therefore assessed the ability of G-CSF to suppress apoptosis in neutrophils expressing V72M NE (Figure 4C). As expected, G-CSF suppressed apoptosis in cultures of wild-type cells such that more than 75% of neutrophils were viable at 24 hours. Importantly, a similar antiapoptotic effect of G-CSF was observed in cultures of heterozygous and homozygous V72M NE neutrophils. Collectively, these data suggest that neutrophils expressing V72M NE do not have higher rates of apoptosis in response to serum deprivation nor is G-CSF–induced suppression of apoptosis blunted in these mice.
G-CSF responsiveness of hematopoietic progenitors expressing V72M NE is normal
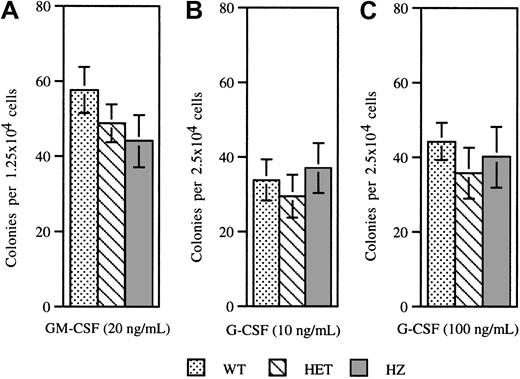
Previous studies have shown that the frequency and cytokine responsiveness of myeloid progenitors from patients with SCN are decreased. In particular, these cells display an impaired responsiveness to G-CSF. To determine whether expression of V72M NE has similar effects on hematopoietic progenitors, colony-forming cell assays were performed on bone marrow cells isolated from mice of each genotype (Figure 5). No significant difference was observed in the numbers of colonies formed in response to GM-CSF (Figure 5A). Moreover, the numbers of colonies formed in response to G-CSF at 10 ng/mL or 100 ng/mL were also similar (Figure5B,C). These data show that both the frequency and G-CSF responsiveness of myeloid progenitors are unaffected by expression of V72M NE.
Colony-forming assays.
Bone marrow cells isolated from wild-type, heterozygous, and homozygous V72M NE mice were plated in methylcellulose in the presence of 20 ng/mL GM-CSF (A), 10 ng/mL G-CSF (B), or 100 ng/mL G-CSF (C). A minimum of 4 mice were used for each genotype in each condition. Data represent the mean ± SEM.
Colony-forming assays.
Bone marrow cells isolated from wild-type, heterozygous, and homozygous V72M NE mice were plated in methylcellulose in the presence of 20 ng/mL GM-CSF (A), 10 ng/mL G-CSF (B), or 100 ng/mL G-CSF (C). A minimum of 4 mice were used for each genotype in each condition. Data represent the mean ± SEM.
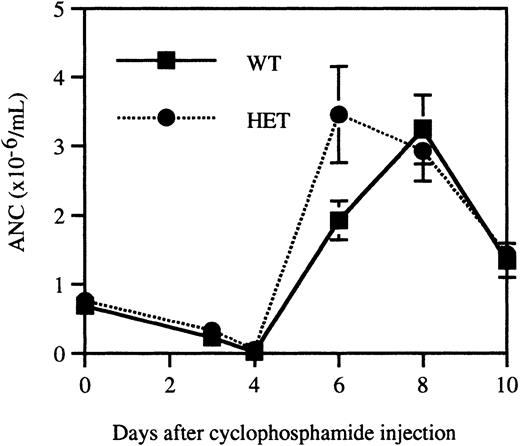
Neutrophil recovery following cyclophosphamide treatment is normal in mice expressing V72M NE
It is possible that granulopoiesis in heterozygous V72M NE mice is sustained through compensatory mechanisms. To test this possibility, a stress granulopoiesis assay was performed in which age, sex, and strain-matched wild-type and heterozygous mice (n = 10, each) were injected with the myeloablative agent cyclophosphamide. Because significant differences in cyclophosphamide responsiveness may exist between mouse strains, outbred homozygous V72M NE mice were not studied in this assay. Consistent with previous reports,42cyclophosphamide-treated mice displayed a transient neutropenia with a nadir on day 4, followed by a sharp increase in neutrophil counts, peaking at day 8 and falling off slightly by day 10 (Figure6). The response of wild-type and heterozygous mice was essentially indistinguishable, suggesting that granulopoiesis in heterozygous V72M NE mice is not being sustained through compensatory mechanisms.
Recovery from cyclophosphamide-induced myelosuppression.
Age, sex, and strain-matched wild-type and heterozygous mice (n = 10, each) were injected with 200 mg/kg cyclophosphamide intraperitoneally. Blood counts were obtained immediately prior to injection and on days 3, 4, 6, 8, and 10. Data represent the mean ± SEM.
Recovery from cyclophosphamide-induced myelosuppression.
Age, sex, and strain-matched wild-type and heterozygous mice (n = 10, each) were injected with 200 mg/kg cyclophosphamide intraperitoneally. Blood counts were obtained immediately prior to injection and on days 3, 4, 6, 8, and 10. Data represent the mean ± SEM.
Mice heterozygous for the d715 GCSFR and V72M NE have normal neutrophil counts
We previously described the generation of transgenic mice carrying a targeted mutation of their G-CSFR reproducing a mutation found in a patient with SCN. This mutation (termed d715 G-CSFR) results in the truncation of the carboxy-terminal 96 amino acids of the G-CSFR. Although granulocytic differentiation is normal in these mice, a modest reduction in the level of circulating neutrophils is observed.23 25 To determine whether coexpression of the d715 G-CSFR and V72M NE results in impaired granulopoiesis, d715 G-CSFR mice inbred on a C57BL/6 background were intercrossed with V72M NE chimeric mice. Mice doubly heterozygous for the d715 G-CSFR and V72M NE (on a C57BL/6 × 129/SvJ F1 background) were produced with the expected frequency. Importantly, the ANC in these mice (0.61 ± 0.07 × 106/ mL, n = 6) was similar to that observed in strain-matched wild-type or heterozygous V72M NE mice (Table 1), illustrating that these mutations do not cooperate to impair granulopoiesis in mice.
No cases of leukemia have been detected in V72M NE mice
Patients with SCN are at increased risk for the development of AML, with a crude malignant rate of transformation of 9%.6 Of note, AML has developed in a patient with SCN and the V72M NE mutation.28 To test whether mice expressing the V72M allele are at an increased risk for the development of leukemia, a tumor watch containing 37 heterozygous and 27 strain-matched wild-type mice was established to detect a 9% difference in the rates of leukemia (see “Materials and methods”). To date, none of the mice have developed leukemia (mean observation period of 9.3 months). Peripheral blood counts were performed on cohorts (n = 6, each) of wild-type (mean age, 8.5 months) and heterozygous V72M NE mice (mean age, 10.1 months). ANCs (× 106/mL) for wild-type and heterozygous mice were 1.06 ± 0.18 and 1.08 ± 0.20, respectively. Furthermore, no abnormalities in white cell morphology were observed. These results indicate that expression of V72M NE is not sufficient to induce leukemia in mice.
Discussion
Recent studies provide strong genetic evidence that mutations in the ELA2 gene encoding NE play a causative role in the pathogenesis of CN and SCN. Published reports show that mutations of NE are present in 69 of 69 patients from 13 families with CN26 and in 51 of 65 patients with SCN.27 28In contrast, only 2 coding sequence changes have been observed in 165 healthy individuals and are presumed to represent polymorphisms. A remarkable feature of these data is the diversity of mutations. To date, 27 distinct mutations of the ELA2 gene have been reported and are predicted to result in single amino acid substitutions, in-frame deletions or insertions, or premature stop codons. Although the mechanisms by which mutant NE proteins contribute to the pathogenesis of SCN and CN are currently unknown, 2 observations provide significant clues. First, the ELA2 mutations in patients with SCN and CN are heterozygous, suggesting a dominant, gain-of-function mechanism. Second, the case report of paternal mosaicism for an ELA2 mutation provides evidence that expression of mutant NE inhibits granulopoiesis in a cell-intrinsic fashion because no toxic paracrine effects of mutant NE protein on wild-type granulocytic cells in this mosaic individual were observed.
In vitro characterization of the biochemical properties of a large number of NE mutations found in patients with SCN identified no consistent effect of these mutations on NE proteolytic activity, substrate specificity, or serpin inhibition.40Surprisingly, a modest dominant-negative effect of mutant NE was observed when cell lines were contransfected with wild-type and mutant NE cDNAs, suggesting that a loss of NE function may contribute to the pathogenesis of SCN. However, it is unlikely that a simple loss of NE activity is responsible for the block in granulocytic differentiation. Mice lacking NE have normal granulopoiesis, whereas mice deficient in dipeptidyl peptidase-1 (DPP1 or cathepsin C), a cysteine protease required for the activation of NE,43 also have normal granulopoiesis.44 Furthermore, humans patients lacking cathepsin C (Papillon-Lefebvre syndrome) are not neutropenic.45
To test the hypothesis that mutations to NE cause SCN, we generated mice carrying a targeted mutation in the murine Ela2 gene that recapitulates a mutation found in 2 unrelated patients with SCN. The targeted transgenic, or knock-in, approach is currently the best way to model the effects of human gene mutations in mice because it maintains normal tissue- and development-specific expression of the gene under study. Indeed, we show, using a quantitative RT-PCR assay, that the numbers of NE transcripts in bone marrow cells from wild-type and homozygous V72M NE mice are similar, indicating that the transcriptional regulation of the V72M NE allele is normal. Mice heterozygous or homozygous for the V72M allele do not display the hallmarks of SCN, namely, neutropenia and an accumulation of promyelocytes or myelocytes (or both) in the bone marrow, nor is there evidence of abnormal cycling of the number of neutrophils in the blood. Expression of V72M NE did not result in impaired G-CSF responsiveness of myeloid progenitor cells nor did it result in an increased susceptibility of granulocytic cells to apoptosis, both features of SCN. Neutrophil recovery following cyclophosphamide-induced myelosuppression was normal, indicating that stress granulopoiesis in heterozygous V72M NE mice is normal. Finally, coexpression of the d715 G-CSFR with V72M NE did not result in neutropenia. Collectively, these data provide compelling evidence that expression of the V72M NE is not sufficient to induce an SCN phenotype in mice.
Patients with SCN have a markedly increased risk for the development of AML. It is possible that the mechanisms by which mutant NE impairs granulopoiesis and contributes to leukemogenesis are distinct. Accordingly, we established a tumor watch to ascertain whether the V72M NE mutation, which is associated with AML, can induce leukemia in the absence of overt neutropenia. With a mean observation period of 9.3 months, no cases of leukemia have been observed, indicating that V72M NE is not sufficient to induce leukemia in mice. However, because other murine models of leukemia have long latencies, observation of the V72M NE mice for 18 months will be required to definitively assess the leukemogenic potential of V72M NE. The presence of G-CSFR mutations in patients with SCN significantly increases their risk of developing AML. Studies are in progress to determine whether the d715 G-CSFR can cooperate with V72M NE to induce leukemia in mice.
There are several potential explanations for our observation that expression of V72M NE is not sufficient to induce an SCN phenotype in mice. The Val72Met mutation may not have the same effect on murine NE structure or function as it does on human NE. The human and murine NE genes share a similar genomic organization and exonic structure,46 and human and murine NE are 76% identical at the amino acid level. Importantly, 25 of the 26 amino acids mutated in human NE in patients with SCN or CN, including Val72, are conserved in murine NE, suggesting that these amino acids are important for normal NE structure and function. The only known effect of the Val72Met mutation in human NE is reduced proteolytic activity. Interestingly, we show that elastolytic activity (as measured by cleavage of the methoxysuccinyl ala-ala-pro-val pNa substrate) in homozygous V72M NE bone marrow cells is reduced compared with wild-type cells, suggesting that the Val72Met mutation may have a similar effect on murine NE proteolytic activity. Nonetheless, significant differences in the effects of the Val72Met mutation on human versus murine NE activity may exist. Studies are under way to characterize the effects of other NE mutants on granulopoiesis.
The murine hematopoietic cell environment may not present the necessary NE target protein(s) for the development of an SCN phenotype. It is possible that significant differences in the expression or sequence of proteins that interact with NE may exist between humans and mice. It follows that putative NE target proteins required for the development of an SCN phenotype may not be present or accessible in murine hematopoietic cells. Considerable evidence suggests that numerous, and potentially important, differences exist between human and murine hematopoiesis. For example, murine neutrophils are generally hypogranular in comparison with human neutrophils, and human neutrophil primary granules contain defensins and azurocidin, whereas murine primary granules lack both.47 Furthermore, attempts to model other human bone marrow failure syndromes in mice, including Fanconi anemia, have not been entirely successful in reproducing the hematopoietic phenotype seen in patients.48-50
Finally, though the genetic evidence implicating NE mutations in the pathogenesis of SCN is compelling, expression of mutant NE may not be sufficient to cause SCN. There are at least 2 mechanisms by which the genetic changes observed in SCN may contribute to the pathogenesis of the disease independent of their effects on NE. One possibility is that the genetic changes observed in SCN are affecting another gene (or genes) in the locus. As an example of this phenomenon, thep16INK4a and p19ARFgenes share genomic sequence such that a single mutation can affect the function of both genes.51 However, the observation that all of the mutations described are predicted to alter the amino acid sequence of NE argues against this possibility. Moreover, an examination of the 50-kb region in which ELA2is located finds no expressed sequence tags or hypothetical genes on either the sense or complementary strands. A second possibility is that mutations in the ELA2 gene may disrupt regulatory elements required for the expression of neighboring genes. In this regard it is interesting to note that other genes coordinately regulated with NE, namely, azurocidin and proteinase 3, are contained within this locus.52 Moreover, a number of myeloid-specific DNase I hypersensitivity sites have been observed in this region.53 Thus, it is possible that the mutations are disrupting a locus control region (LCR), which regulates a number of genes expressed during myeloid development.
In summary, we generated transgenic mice carrying a targeted mutation of their Ela2 gene reproducing the V72M NE mutation found in SCN. Despite evidence of regulated expression of V72M NE, no effects on basal or stress granulopoiesis and no cases of leukemia were observed. Collectively, these data show that expression of V72M NE is not sufficient to induce an SCN phenotype or leukemia in mice.
Note added in proof. A competitive repopulation assay was performed with strain-matched wild-type and heterozygous V72M NE mice. We found that wild-type and V72M NE cells contributed equally to B lymphocytic and granulocytic lineages as determined by peripheral blood analysis at 12 weeks after transplantation. These observations show that the repopulating potential is not affected by the expression of V72M NE and further support the conclusion that the Val72Met mutation is not sufficient to impair granulopoiesis in vivo.
Prepublished online as Blood First Edition Paper, July 12, 2002; DOI 10.1182/blood-2002-05-1372.
D.C.L. was supported by American Cancer Society grant RSG LIB-104122. D.S.G. is a Howard Hughes Medical Institute Medical Student Research Training Fellow.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Daniel C. Link, Division of Oncology, Department of Medicine, 660 S Euclid Ave, Campus Box 8007, Saint Louis, MO 63110; e-mail: dlink@im.wustl.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal