Abstract
Activation of human interleukin 3 (IL-3) and granulocyte-macrophage colony-stimulating factor (GM-CSF) receptors, ectopically expressed in FDCP-mix multipotent cells, stimulates self-renewal or myeloid differentiation, respectively. These receptors are composed of unique α subunits that interact with common βc subunits. A chimeric receptor (hGM/βc), comprising the extracellular domain of the hGM-CSF receptor α subunit (hGM Rα) fused to the intracellular domain of hβc, was generated to determine whether hβc activation is alone sufficient to promote differentiation. hGM-CSF activation of hGM/βc, expressed in the presence and absence of the hβc subunit, promoted maintenance of primitive phenotype. This indicates that the cytosolic domain of the hGM Rα chain is required for differentiation mediated by activation of the hGM Rα, βc receptor complex. We have previously demonstrated that the α cytosolic domain confers signal specificity for IL-3 and GM-CSF receptors. Bioinformatic analysis of the IL-3 Rα and GM Rα subunits identified a tripeptide sequence, adjacent to the conserved proline-rich domain, which was potentially a key difference between them. Cross-exchange of the equivalent tripeptides between the α subunits altered receptor function compared to the wild-type receptors. Both the mutant and the corresponding wild-type receptors promoted survival and proliferation in the short-term but had distinct effects on developmental outcome. The mutated hGM Rα promoted long-term proliferation and maintenance of primitive cell morphology, whereas cytokine activation of the corresponding hIL-3 Rα mutant promoted myeloid differentiation. We have thus identified a region of the α cytosolic domain that is of critical importance for defining receptor specificity.
Introduction
Hematopoietic stem and progenitor cells can undergo self-renewal or commitment to lineage-specific differentiation. In the stem cell compartment, the basis for the decision to self-renew or differentiate is complex and still poorly understood. Recent data on the PAX-5 protein suggest that transcription factors can suppress certain developmental options.1 Several other transcription factors have been shown to be critical for hemopoietic development on the basis of the effects of their overexpression or deletion.2,3 These include PU.1, SCL, and GATA 1-3.4-8 Cytokines regulate the activity of transcription factors, via interaction with specific receptors, and can influence the developmental fate of hematopoietic stem and progenitor cells.2 For example, interleukin 3 (IL-3) and granulocyte-macrophage colony-stimulating factor (GM-CSF) bind to type I cytokine receptors composed of α and βc subunits. These receptors are expressed on a range of hematopoietic progenitor cells and their activation is associated with myeloid development.9,10 The GM-CSF receptor (GM-CSF R) can specifically instruct for myeloid differentiation.11 The IL-3 receptor (IL-3 R) is expressed earlier in myeloid cell development than the GM-CSF R, promotes expansion of CD34+ progenitor cell populations to a greater extent than GM-CSF, and has the ability to promote development of a greater range of lineages than GM-CSF.9,12 The α subunits bind IL-3 or GM-CSF with low affinity and interact with a common β subunit (βc) to form a functional high-affinity receptor complex. IL-3 and GM-CSF cannot bind to the βc subunit independently of the α subunits that are unique to each receptor. The βc subunit is believed to elicit many of the signaling events emanating from these receptors.13 14
Knock-in mutations of the GM-CSF R reveal a complex regulation of hematopoiesis in vivo by the α and βcsubunits.15-17 Furthermore, in vitro studies demonstrate that there are distinct differences in the signals emanating from the GM-CSF, IL-3, and IL-5 receptor complexes.18-20 We have previously demonstrated that the α subunits act in combination with the hβc to govern developmental fate.18Here, we further analyze the role of the βc subunit and the cytosolic domain of the human GM-CSF receptor α (hGM Rα) subunit in developmental regulation using the murine FDCP-mix cell line as a model system.
Ectopic expression of wild-type and mutant human IL-3 receptor (hIL-3 R) and granulocyte-macrophage receptor (GM R) in the murine hematopoietic cell line, FDCP-mix, facilitates their analysis in the context of a multipotent hematopoietic progenitor cell line and allows an unambiguous description of their biologic effects. IL-3 and GM-CSF are species specific; thus, hIL-3 and hGM-CSF selectively activate the transfected human IL-3 and GM-CSF receptors. Our previous data have shown that coexpression of hGM Rα and hβc in murine FDCP-mix cells promotes myeloid differentiation, whereas the hIL-3 Rα, hβc promotes maintenance of multipotential cells. Furthermore, transfection and activation of a hIL-3 Rα (extracellular domain), hGM Rα, (cytosolic domain) chimeric receptor demonstrated that signal specificity resided in the cytosolic domain.18 The features of the hIL-3 and hGM receptors that signal for self-renewal and differentiation are further characterized by analysis of the function of receptor mutants in FDCP-mix cells.
Materials and methods
The FDCP-mix cell line
The FDCP-mix (clone A4) cells were routinely cultured in Fischer medium supplemented with 5% (vol/vol) medium conditioned by the X63-Ag-653 cell line (used as a source of murine IL-3),2120% (vol/vol) horse serum. For granulocyte-macrophage differentiation (G/M Diff) cells were cultured in Iscove modified Dulbecco medium supplemented with fetal calf serum (20% vol/vol), recombinant murine (rm) GM-CSF (5 ng/mL), rmG-CSF (5 ng/mL), and rmIL-3 (0.1 ng/mL), as previously described.22 The effects of activation of hIL-3 R or hGM R were assessed by culturing the receptor transfects with hGM-CSF or hIL-3, respectively, as the single cytokine present. They were thus studied in the absence of murine cytokines. Murine cytokines were supplied by Calbiochem (Nottingham, United Kingdom). Recombinant hIL-3 and hGM-CSF were gifts from Sandoz Pharma (Basel, Switzerland) and Glaxo (Greenford, United Kingdom), respectively, as previously described.18
Transfection of FDCP-mix cells with hIL-3 and hGM-CSF receptor mutants
Retroviral transfections were performed using the pM5 vector containing the receptor subunit gene and an antibiotic-resistance gene as a selectable marker.18 Neomycin phosphotransferase (neo) and hygromycin phosphatases (hgr) were used to select for the mutated receptor α subunits and hβc, respectively. The chimeric receptor α subunit was produced by introducing a NheI restriction site (position 330 for the hGM Rα and 446 for the hβc) into the transmembrane domain by site-directed mutagenesis. The mutagenesis was carried out on dsDNA using the Chameleon ds mutagenesis kit (Stratagene, Amsterdam, The Netherlands) in accordance with the manufacturer's instructions. The mutant hGM Rα (hGM (M) Rα) and the corresponding hIL-3 (M) Rα were generated by site-directed mutagenesis using the Quickchange mutagenesis kit (Stratagene). All mutations and constructs were confirmed by DNA sequencing. The receptor constructs were transfected into the Phoenix fibroblast packaging cell line23 using calcium phosphate precipitation (Gibco, Paisley, United Kingdom). Retroviral transfection was performed by culturing FDCP-mix cells in viral supernatant for 48 hours. FDCP-mix cells were harvested, washed, and selected for antibiotic resistance by culturing in medium supplemented with 1 mg/mL G418 or 0.15 mg/mL hygromycin B (or both) as appropriate. Polyclonal cell populations were labeled for flow cytometric analysis using antibodies directed against the extracellular domain of the receptor subunits (see below). Cells were sorted on the basis of receptor subunit expression using the cell sorting facility of the FACS Vantage flow cytometer and 3 polyclonal populations obtained for analysis (Becton Dickinson, Oxford, United Kingdom). As evaluated by flow cytometry, the expression level of mutant receptors was found to be similar in the populations generated and equivalent to more than 10 000 receptors/cell.
Analysis of hGM-CSF R and hIL-3 R subunit expression
Ectopic receptor expression was confirmed by flow cytometry using a 2-step antibody labeling procedure as previously described.18
Measurement of proliferation
DNA synthesis was used as a measure of proliferation and was performed by determining incorporation of [3H]-thymidine as previously described.24 Briefly, cells were washed and incubated with cytokines (5 × 104 cells/sample) for 16 hours. Samples were then pulsed for 4 hours with 10 μCi/mL [3H]-thymidine (0.37 MBq), harvested using a Hewlett Packard Top Count cell harvester and the incorporated radioactivity measured by scintillation counting.
Morphologic analysis
Morphologic analysis of cells in liquid culture was performed as described.25 Slides were prepared using a Shandon cytospin centrifuge and stained with May-Grünwald-Giemsa stain. At least 100 cells were scored for each slide.
Analysis of differentiation markers
Cell surface expression of Sca-1, Gr-1, and Mac-1 (CD11b) differentiation markers were analyzed by flow cytometry as previously described.18
Results
Generation of hIL-3 R and hGM R mutants
To further characterize the role of the α and βc subunits in hGM-CSF and hIL-3 receptor-mediated signaling, we have constructed and evaluated the function of 2 types of mutant GM R and hIL-3 R. A schematic representation of the mutants tested is shown in Figures 1 and2. The chimeric hGM/βcreceptor is composed of the extracellular domain of the α subunit and a βc cytosolic domain (Figure 1). This was used to study the biologic response elicited by activation of hβc in the absence of the α cytosolic domain. The hIL-3 (M) Rα and hGM (M) Rα mutants were generated based on sequence comparison of the α cytosolic domains that indicated unique tripeptide sequences, PIG and KLN, in IL-3 Rα and GM Rα, respectively (shown as underlined sequences in Figure 2). These tripeptides were different between the receptors but conserved across species and therefore postulated to perhaps play a role in conferring receptor specificity. For the hIL-3 (M) Rα, the tripeptide PIG344–346 was replaced with the corresponding KLN365–367 tripeptide from the hGM Rα subunit using site-directed mutagenesis. Replacing the KLN sequence with PIG of hIL-3 Rα generated the corresponding mutant hGM Rα, designated hGM (M) Rα (Figure 2).
Sequence of transmembrane region of the hGM/βc chimera.
The transmembrane domain sequence of the chimeric hGM/βcreceptor mutant is shown with the adjacent amino acids. The sequences from wild-type hGM Rα and hβc subunits are shown in plain and bold text, respectively. The putative transmembrane region is underlined and is based on sequence data for the hGM Rα and h/βc.51 52
Sequence of transmembrane region of the hGM/βc chimera.
The transmembrane domain sequence of the chimeric hGM/βcreceptor mutant is shown with the adjacent amino acids. The sequences from wild-type hGM Rα and hβc subunits are shown in plain and bold text, respectively. The putative transmembrane region is underlined and is based on sequence data for the hGM Rα and h/βc.51 52
Sequence comparison of cytosolic domains of hGM Rα, IL –3 Rα, and IL-5 Rα.
Aligned cytosolic sequences of hGM Rα, IL-3 Rα, and the related IL-5 Rα are shown. Amino acids present in all 3 Rα subunits or that are subject to conservative change are shown in bold. Those conserved between species are boxed and those present in 2 α subunits are marked with an asterisk (*). The PIG and KLN sequences of hIL-3 Rα and hGM Rα are indicated by underlining.
Sequence comparison of cytosolic domains of hGM Rα, IL –3 Rα, and IL-5 Rα.
Aligned cytosolic sequences of hGM Rα, IL-3 Rα, and the related IL-5 Rα are shown. Amino acids present in all 3 Rα subunits or that are subject to conservative change are shown in bold. Those conserved between species are boxed and those present in 2 α subunits are marked with an asterisk (*). The PIG and KLN sequences of hIL-3 Rα and hGM Rα are indicated by underlining.
FDCP-mix cells are nonleukemic, karyotypically normal and their survival, proliferation, and development are subject to regulation by cytokines.26 27 The cell line is maintained in murine IL-3, in which cells maintain a primitive phenotype and multipotential differentiation capacity. Cells were cultured in murine IL-3 during transfection and antibiotic selection of cells to preserve the multipotential differentiation capacity. Cells were then analyzed for human receptor gene expression by flow cytometry, using antibodies directed against the extracellular domain of the hIL-3 and hGM R subunits. The cells maintained multipotency following transfection and selection (see below). Human cytokine receptor function was assessed in the panel of cell lines generated by determining the effect of culturing the cells with either hIL-3 or hGM-CSF as the only cytokine present.
The effects of chimeric hGM/βc
To perform a complete analysis of the potential of βc to promote myeloid differentiation, we determined the effects of expression of a chimeric receptor hGM/βcexpressed in the presence and absence of full-length hβcin FDCP-mix cells. The chimeric hGM/βc receptor represents a “minimal” form of the receptor with the ligand-binding domain of the α subunit and the cytosolic signaling domain of the hβc.
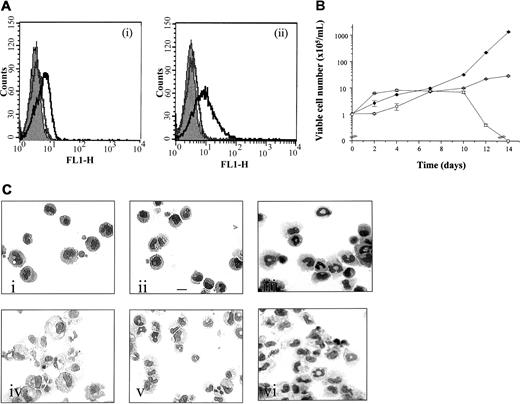
As previously reported, no specific fluorescence labeling of the parental FDCP-mix cells was detected with anti–hIL-3 Rα, anti-hGM Rα, or anti-hβc antibodies.18 Cells positive for hβc and hGM/βc coexpression were identified and sorted by flow cytometry using anti-hGM Rα and anti-hβc antibodies, respectively, following antibiotic selection (Figure 3A). The expression profiles of hGM/βc and hβc in a representative populations are shown in Figure 3, panels Ai and Aii, respectively, where a log increase in fluorescence was observed relative to the control. The level of receptor subunit expression was similar to that previously obtained when the hβc was coexpressed with the hGM Rα or hIL-3 Rα.18
Effect of expression of hGMβcchimera with or without hβc in FDCP-mix cells.
(A) Analysis of receptor subunit expression. Parental and hGM/βc transfected cells were analyzed for expression of the extracellular domain of the (i) hGM Rα and (ii) hβcsubunit using a 2-step labeling procedures and flow cytometry. The solid gray histogram represents the cell autofluorescence obtained in the absence of antibody staining. Representative histograms are shown for labeling obtained in the presence of secondary antibody only (gray) and in the presence of both primary and secondary antibody (black). (B) Effects on cellular proliferation. Cells expressing hGM/βc (○) and hGM/βc, hβc(●) were cultured in hGM-CSF (1 ng/mL) for 14 days. Cellular viability was assessed at intervals and the results are expressed as viable cell number (× 105/mL) and are mean ± SEM from 3 experiments. The results obtained with the wild-type hGM Rα,βc are shown for comparison (■). (C) Effect of activation of hGM/βc expressed alone or in combination with the hβc subunit on cell morphology. (i) hGM/βc cells, (ii) hGM/βc,hβc, and (iii) wild-type hGM Rα, hβc were cultured in hGM-CSF (1 ng/mL) for 7 days and photomicrographs were prepared following May-Grünwald-Giemsa staining of cytospin preparations. The morphology of cells cultured in the presence of murine cytokines (G/M Diff conditions), which promote myeloid development, are shown for comparison for (iv) hGM/βccells and (v) hGM/βc,βc and (vi) wild-type hGM Rα, hβc. Results are shown from an experiment representative of 3. Bar is 10 μm.
Effect of expression of hGMβcchimera with or without hβc in FDCP-mix cells.
(A) Analysis of receptor subunit expression. Parental and hGM/βc transfected cells were analyzed for expression of the extracellular domain of the (i) hGM Rα and (ii) hβcsubunit using a 2-step labeling procedures and flow cytometry. The solid gray histogram represents the cell autofluorescence obtained in the absence of antibody staining. Representative histograms are shown for labeling obtained in the presence of secondary antibody only (gray) and in the presence of both primary and secondary antibody (black). (B) Effects on cellular proliferation. Cells expressing hGM/βc (○) and hGM/βc, hβc(●) were cultured in hGM-CSF (1 ng/mL) for 14 days. Cellular viability was assessed at intervals and the results are expressed as viable cell number (× 105/mL) and are mean ± SEM from 3 experiments. The results obtained with the wild-type hGM Rα,βc are shown for comparison (■). (C) Effect of activation of hGM/βc expressed alone or in combination with the hβc subunit on cell morphology. (i) hGM/βc cells, (ii) hGM/βc,hβc, and (iii) wild-type hGM Rα, hβc were cultured in hGM-CSF (1 ng/mL) for 7 days and photomicrographs were prepared following May-Grünwald-Giemsa staining of cytospin preparations. The morphology of cells cultured in the presence of murine cytokines (G/M Diff conditions), which promote myeloid development, are shown for comparison for (iv) hGM/βccells and (v) hGM/βc,βc and (vi) wild-type hGM Rα, hβc. Results are shown from an experiment representative of 3. Bar is 10 μm.
The functional capability of βc to promote self-renewal/differentiation in the absence of the α subunit cytosolic domain was assessed and the results are shown in Figure 3, panels B and C. Cells expressing hGM/βc alone or in combination with hβc survived and proliferated in response to hGM-CSF (at ≥ 0.01 ng/mL). There was significantly greater proliferation of cells coexpressing the hGM/βcplus hβc compared to hGM/βc alone in response to hGM-CSF (Figure 3B) and both could promote proliferation for more than 10 days, unlike the wild-type hGM Rα, hβc. These data suggest that the formation of higher order βc complexes stimulates proliferation more effectively. Interestingly, chimeric receptors, composed of the extracellular and transmembrane domains of the hGM Rα (or IL-5 Rα) and the intracellular domain of the hβc, have been shown to form functional oligomeric receptor complexes in combination with hβc.28-30
We compared the morphology of hGM-CSF–treated cells expressing hGM/βc with or without hβc to cells expressing the wild-type hGM Rα, hβc. The results are shown in Figure 3, panel Ci-iii, and Table1. Cells were cultured at a dose of 1 ng/mL since there is clear receptor activation at this dose and it avoids activation of endogenous mGM-CSF R subunit, which would occur at higher doses (100 ng/mL). The wild-type hGM Rα, hβcpromotes formation of granulocytes and macrophages in response to hGM-CSF (Figure 3Ciii and Table 118). In contrast, the hGM/βc with or without hβc cells cultured in hGM-CSF for 7 days exhibited primitive cell morphology, composed mainly of blast cells and early granulocytes. The cells, however, retained their ability to undergo differentiation into granulocytes and macrophages in response to murine cytokines (Figure 3Civ-vi). This infers that the hβc alone does not elicit a differentiation stimulus equivalent to that of wild-type hGM Rα, hβc and further supports our hypothesis that it is the cytosolic domain of the GM-CSF Rα that acts, in combination with the βc, to promote differentiation.
Effect of activation of hGM/βc chimera with or without hβc on cell morphology
| Receptor transfect . | Blasts . | Early granulocytes . | Late granulocytes . | Macrophages . |
|---|---|---|---|---|
| Wild-type hGM Rα, hβc | 7.5 ± 0.7 | 13 ± 2.8 | 38 ± 4.2 | 41.5 ± 7.8 |
| Chimeric hGM/βc | 67.5 ± 10.7 | 30 ± 12.3 | 1.5 ± 2.1 | 1 ± 1.4 |
| hGM/βc, hβc | 80.5 ± 1 | 18.5 ± 1 | 0 ± 0 | 0 ± 0 |
| Receptor transfect . | Blasts . | Early granulocytes . | Late granulocytes . | Macrophages . |
|---|---|---|---|---|
| Wild-type hGM Rα, hβc | 7.5 ± 0.7 | 13 ± 2.8 | 38 ± 4.2 | 41.5 ± 7.8 |
| Chimeric hGM/βc | 67.5 ± 10.7 | 30 ± 12.3 | 1.5 ± 2.1 | 1 ± 1.4 |
| hGM/βc, hβc | 80.5 ± 1 | 18.5 ± 1 | 0 ± 0 | 0 ± 0 |
Cells were cultured in hGM-CSF (1 ng/mL) for 7 days. Morphology was then assessed using May-Grünwald-Giemsa–stained cytospin preparations. Cell morphology was then scored and the results are presented as percentage of total cells. The results are pooled data from 3 experiments ± SEM. Prior to culture in hGM-CSF cell morphology was ≥ 94% blast.
Bioinformatic analysis of the primary sequence of the IL-3 Rα and GM-CSF Rα subunits
The differences between the IL-3 Rα– and GM Rα–mediated maintenance of pluripotentiality and development signals lie in the cytosolic domain sequence18 and are potentially revealing about the molecular mechanisms of cytokine receptor activation. Sequence comparisons were made between the cytosolic domains of the α subunits of the GM-CSF, IL-3, and IL-5 receptor subfamily (Figure 2). Areas of sequence that are divergent between receptor cytosolic domains but conserved between species were deemed to be important in the functional differences between the receptors. A further consideration was the potential existence of functional domains. There is no evidence of tyrosine or serine/threonine phosphorylation of the α cytosolic domains and there are no apparent WW, SH3, or SH2 domains present. However, previous work on the α subunit has shown that the first 29 cytosolic amino acids (from the membrane region) are sufficient for cell signaling, proliferation, and differentiation.31,32 This encompasses the proline-rich juxtamembrane region and adjacent amino acids. Figure 2 shows that there is cross-species and cross-receptor conservation of this region (QRLFP**P). However, there is a divergent region close to the proline-rich domain, which may be important in that a proline residue in the IL-3 Rα is replaced by lysine in the hGM Rα. Predictions of hydrophobicity and secondary structure revealed that the proline residue was likely to be exposed to the cytosol, making it a candidate for mediating differential signaling. Furthermore, proline residues have the potential to affect secondary structure by introducing kinks in the helices.33
We therefore exchanged the tripeptide KLN365-367 in GM Rα for PIG from the IL-3Rα sequence to generate a mutant hGM Rα subunit (hGM (M) Rα). The corresponding hIL-3 Rα mutant was also generated for comparison (Figure 2). The aim was to define a regionof the receptor that may regulate GM Rα–mediated differentiation.
Substitution of the PIG region of IL-3 Rα with the KLN sequence from GM Rα generates a receptor that promotes myeloid differentiation
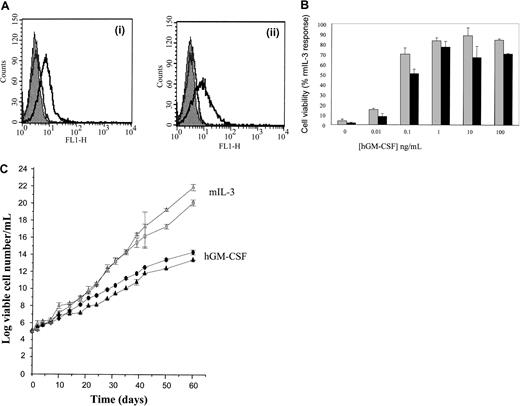
FDCP-mix cells were cotransfected with the hIL-3 (M) Rα and hβc subunits and compared to the wild type hIL-3 Rα, hβc cells. The expression of the hIL-3 (M) Rα and hβc subunits was analyzed by flow cytometry (Figure4Ai,ii). These expression levels were similar to those obtained for the wild-type hIL-3 R.18 We have previously determined that culture of wild-type receptor transfects with hIL-3 promoted maintenance of blast cell phenotype and proliferative potential.18 Addition of hIL-3 promoted survival and proliferation of both the wild-type and mutant hIL-3 R (Figure 4Bi,ii). However, whereas activation of the wild-type hIL-3 receptors promotes self-renewal (Figure 4Ci), addition of hIL-3 to hIL-3 (M) Rα, hβc cells generated cells with a much more differentiated phenotype (Table2). The hIL-3 (M) Rα, hβc cells therefore more closely resembled the response of wild-type hGM R-expressing cells (Figure 4Cii, Table118). The dose of hIL-3 used (0.1-100 ng/mL) did not influence the outcome observed when mutated IL-3 Rα, hβc was activated. In all cases mature myeloid cells were formed in a 7-day period. The hIL-3 (M) Rα, hβc cells show an increased expression of Gr-1 and Mac-1 compared to the wild-type cells in response to hIL-3 (Table 3). These data show that PIG→KLN substitution thus transforms a signal for proliferation and self-renewal into a differentiation signal.
Effects of hIL-3 (M) Rα activation.
(A) Cell surface expression. hIL-3 (M) Rα, hβc cell transfects were analyzed for expression of the extracellular domains of the (i) hIL-3 Rα and (ii) hβc by flow cytometry. Cells were sequentially incubated with anti–hIL-3 Rα antibody and fluorescein isothiocyanate (FITC)–conjugated antimouse secondary antibody. The solid gray histogram represents the cell autofluorescence obtained in the absence of antibody staining. Representative histograms are shown for labeling obtained in the presence of secondary antibody only (gray) and in the presence of both primary and secondary antibody (black). (B) Cell survival proliferation. Cells expressing wild-type hIL-3 Rα, hβc (░) or hIL-3 (M) Rα, hβc (▪) were cultured in hIL-3 (0-100 ng/mL). (i) Cell viability was assessed at 48 hours using trypan blue. The results are expressed as cell viability (percent rmIL-3 response) and are mean values ± SEM from at least 3 experiments. (ii) [3H]-thymidine incorporation was assessed after 16 hours in culture. The results are expressed as percentage of the response obtained with rmIL-3 (10 ng/mL). (C) Cell morphology. Cells expressing (i) wild-type hIL-3 Rα, hβc or (ii) hIL-3 (M) Rα, hβc were cultured in hIL-3 (1 ng/mL) for 7 days. Photomicrographs were prepared from May-Grünwald-Giemsa–stained cytospin preparations. Results are representative of 3 experiments. Bar is 10 μm.
Effects of hIL-3 (M) Rα activation.
(A) Cell surface expression. hIL-3 (M) Rα, hβc cell transfects were analyzed for expression of the extracellular domains of the (i) hIL-3 Rα and (ii) hβc by flow cytometry. Cells were sequentially incubated with anti–hIL-3 Rα antibody and fluorescein isothiocyanate (FITC)–conjugated antimouse secondary antibody. The solid gray histogram represents the cell autofluorescence obtained in the absence of antibody staining. Representative histograms are shown for labeling obtained in the presence of secondary antibody only (gray) and in the presence of both primary and secondary antibody (black). (B) Cell survival proliferation. Cells expressing wild-type hIL-3 Rα, hβc (░) or hIL-3 (M) Rα, hβc (▪) were cultured in hIL-3 (0-100 ng/mL). (i) Cell viability was assessed at 48 hours using trypan blue. The results are expressed as cell viability (percent rmIL-3 response) and are mean values ± SEM from at least 3 experiments. (ii) [3H]-thymidine incorporation was assessed after 16 hours in culture. The results are expressed as percentage of the response obtained with rmIL-3 (10 ng/mL). (C) Cell morphology. Cells expressing (i) wild-type hIL-3 Rα, hβc or (ii) hIL-3 (M) Rα, hβc were cultured in hIL-3 (1 ng/mL) for 7 days. Photomicrographs were prepared from May-Grünwald-Giemsa–stained cytospin preparations. Results are representative of 3 experiments. Bar is 10 μm.
Effect of activation of hIL-3 (M) Rα, hβc compared to wild-type hIL-3 Rα, hβcon cell morphology
| Receptor transfect . | Blasts . | Early granulocytes . | Late granulocytes . | Macrophages . |
|---|---|---|---|---|
| Wild-type hIL-3 Rα, hβc | 95.3 ± 0.9 | 4.3 ± 0.7 | 0 ± 0 | 0.3 ± 0.3 |
| hIL-3 (M) Rα, hβc | 25.5 ± 4.5 | 39 ± 0.9 | 35.5 ± 3.5 | 0 ± 0 |
| Receptor transfect . | Blasts . | Early granulocytes . | Late granulocytes . | Macrophages . |
|---|---|---|---|---|
| Wild-type hIL-3 Rα, hβc | 95.3 ± 0.9 | 4.3 ± 0.7 | 0 ± 0 | 0.3 ± 0.3 |
| hIL-3 (M) Rα, hβc | 25.5 ± 4.5 | 39 ± 0.9 | 35.5 ± 3.5 | 0 ± 0 |
Cells were cultured in hIL-3 (1 ng/mL) for 7 days. Morphology was then scored using May-Grünwald-Giemsa–stained cytospin preparations. The results are presented as percentage of total cells and are pooled data from 3 experiments ± SEM. Prior to culture in hIL-3, cell morphology was ≥ 94% blasts.
Effect of activation of hIL-3 (M) Rα, hβc compared to wild-type hIL-3 Rα, hβcon expression of differentiation markers
| Receptor transfect . | Sca-1 . | Gr-1 . | Mac-1 . |
|---|---|---|---|
| Wild-type hIL-3 Rα, hβc | 1.04 ± 0.08 | 1.12 ± 0.1 | 1.19 ± 0.11 |
| hIL-3 (M) Rα, hβc | 1.28 ± 0.19 | 2.75 ± 1.2 | 3.24 ± 1.54 |
| Receptor transfect . | Sca-1 . | Gr-1 . | Mac-1 . |
|---|---|---|---|
| Wild-type hIL-3 Rα, hβc | 1.04 ± 0.08 | 1.12 ± 0.1 | 1.19 ± 0.11 |
| hIL-3 (M) Rα, hβc | 1.28 ± 0.19 | 2.75 ± 1.2 | 3.24 ± 1.54 |
Changes in differentiation marker expression are shown as a fold increase in fluorescence intensity compared to levels expressed on control cells cultured in rmIL-3 (10 ng/mL), which are assigned a value of 1. Data are the mean of at least 3 experiments ± SEM.
Effect of reciprocal KLN→PIG substitution on hGM R function
A key question arising from these results obtained is the nature of the effect of the reciprocal substitution of the KLN tripeptide in the hGM Rα cytosolic domain with the PIG tripeptide region from the hIL-3 Rα. If this region is of critical importance, it would be predicted that such a mutant hGM receptor would exhibit a perturbation of the hGM-CSF–mediated differentiation response. Retroviral vectors containing the mutant hGM (M) Rα with KLN365-367 replaced by PIG and the hβc were used to generate cotransfected populations of FDCP-mix cells expressing both the GM (M) Rα and hβc subunits. A profile of receptor subunit expression as determined by flow cytometry is shown in Figure5A. The expression levels detected by anti-hGM Rα and hβc antibodies were similar to those previously obtained for the hGM/βc, hβctransfects (Figure 3A) and also the wild-type receptor.18
Coexpression of hGM (M) Rα and hβc in FDCP-mix cells.
(A) Cell surface expression. Cells transfected with hGM (M) Rα in combination with hβc cells were analyzed for expression of the extracellular domains of the hGM Rα and hβc by flow cytometry. Cells were sequentially incubated with (i) anti-hGM Rα antibody or (ii) anti-hβc antibody and FITC-conjugated antimouse secondary antibody. The solid gray histogram represents the cell autofluorescence obtained in the absence of antibody staining. Representative histograms are shown for labeling obtained in the presence of secondary antibody only (gray) and in the presence of both primary and secondary antibody (black). (B) Effects on cell survival. Cells coexpressing hGM (M) Rα, hβc (░) were cultured in the presence of hGM-CSF (0-100 ng/mL) for 48 hours prior to assessment of cell viability based on trypan blue exclusion. The data obtained for the chimeric hGM/βc, hβc (▪) are shown for comparison. Results are expressed as a percentage of the rmIL-3 (10 ng/mL) response and are mean values ± SEM of 3 experiments. (C) Long-term proliferation. Cells coexpressing either hGM/βc, hβc (●) or hGM (M) Rα, hβc (▴) were cultured for 60 days in the presence of hGM-CSF (1 ng/mL). The growth rate was determined from the initial and subsequent cultures of cells seeded at 1 × 105/mL and resuspended in fresh media when the cell number was more than 5 × 105/mL. The results are expressed as log viable cell number/mL. The growth rates of hGM/βc, hβc( ) and hGM (M) Rα, hβc(
) and hGM (M) Rα, hβc( ) cells in response to rmIL-3 (10 ng/mL) are also shown. Results are mean ± SEM of 3 experiments.
) cells in response to rmIL-3 (10 ng/mL) are also shown. Results are mean ± SEM of 3 experiments.
Coexpression of hGM (M) Rα and hβc in FDCP-mix cells.
(A) Cell surface expression. Cells transfected with hGM (M) Rα in combination with hβc cells were analyzed for expression of the extracellular domains of the hGM Rα and hβc by flow cytometry. Cells were sequentially incubated with (i) anti-hGM Rα antibody or (ii) anti-hβc antibody and FITC-conjugated antimouse secondary antibody. The solid gray histogram represents the cell autofluorescence obtained in the absence of antibody staining. Representative histograms are shown for labeling obtained in the presence of secondary antibody only (gray) and in the presence of both primary and secondary antibody (black). (B) Effects on cell survival. Cells coexpressing hGM (M) Rα, hβc (░) were cultured in the presence of hGM-CSF (0-100 ng/mL) for 48 hours prior to assessment of cell viability based on trypan blue exclusion. The data obtained for the chimeric hGM/βc, hβc (▪) are shown for comparison. Results are expressed as a percentage of the rmIL-3 (10 ng/mL) response and are mean values ± SEM of 3 experiments. (C) Long-term proliferation. Cells coexpressing either hGM/βc, hβc (●) or hGM (M) Rα, hβc (▴) were cultured for 60 days in the presence of hGM-CSF (1 ng/mL). The growth rate was determined from the initial and subsequent cultures of cells seeded at 1 × 105/mL and resuspended in fresh media when the cell number was more than 5 × 105/mL. The results are expressed as log viable cell number/mL. The growth rates of hGM/βc, hβc( ) and hGM (M) Rα, hβc(
) and hGM (M) Rα, hβc( ) cells in response to rmIL-3 (10 ng/mL) are also shown. Results are mean ± SEM of 3 experiments.
) cells in response to rmIL-3 (10 ng/mL) are also shown. Results are mean ± SEM of 3 experiments.
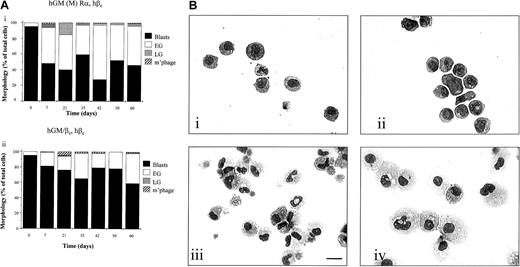
Effects of coexpression of hGM (M) Rα and hβc were determined by culturing cells in the presence of hGM-CSF. Cell viability, proliferation, and differentiation were assessed and compared to those obtained by activation of the hGM/βc, hβc receptor. Both types of mutant receptor promoted cell survival and proliferation (Figure 5B,C). In contrast to the wild-type hGM Rα,βc receptor, however, the hGM (M) Rα, hβc and hGM/βc, hβc promoted the long-term proliferation of FDCP-mix cells, which were continuously cultured in hGM-CSF for a period of 60 days (Figure 5C).
The morphology of the cells was assessed at intervals during this time period and the cells maintained a primitive morphology, with the cultures being composed of mainly blasts and early granulocytes (Figure6Ai,ii and Bi,ii). The dose of hGM-CSF (0.1-100 ng/mL) did not influence the outcome observed when mutated GM R was activated. The cells remained with a blast cell or early granulocytic morphology over the time course of the experiment. They did not acquire factor independence during this time as they remained responsive to mIL-3 and hGM-CSF, undergoing apoptosis following cytokine removal (data not shown). The differentiation potential of the cells was maintained because cells washed free of hGM-CSF after 60 days differentiated into granulocytes and macrophages after 7 days of culture with murine cytokines, which promote G/M Diff (Figure 6Biii,iv).
Effects of activation of hGM (M) Rα, hβcand hGM/βc, hβc on cell development.
(A) Cell morphology. (i) hGM (M) Rα, hβc and (ii) hGM/βc, hβc cells were cultured in hGM-CSF (1 ng/mL) for 60 days and cytospin samples prepared at intervals during this time. Morphology was assessed following May-Grünwald-Giemsa staining. The morphology results are expressed as percentage of the total cells scored and pooled data from 3 experiments. The SEMs were less than 10%. (B) Ability to differentiate following long-term culture in hGM-CSF. Photomicrographs are shown of (i) hGM (M) Rα, hβc cells and (ii) hGM/βc, hβc cells cultured in hGM-CSF (1 ng/mL) for 60 days. After this time, (iii) hGM (M) Rα, hβc cells and (iv) hGM/βc, hβc cells were harvested, washed, and cultured in murine cytokines that promote myeloid development for a further 7 days. Cytospin preparations were May-Grünwald-Giemsa stained and are representative of 3 separate experiments. Bar is 10 μm.
Effects of activation of hGM (M) Rα, hβcand hGM/βc, hβc on cell development.
(A) Cell morphology. (i) hGM (M) Rα, hβc and (ii) hGM/βc, hβc cells were cultured in hGM-CSF (1 ng/mL) for 60 days and cytospin samples prepared at intervals during this time. Morphology was assessed following May-Grünwald-Giemsa staining. The morphology results are expressed as percentage of the total cells scored and pooled data from 3 experiments. The SEMs were less than 10%. (B) Ability to differentiate following long-term culture in hGM-CSF. Photomicrographs are shown of (i) hGM (M) Rα, hβc cells and (ii) hGM/βc, hβc cells cultured in hGM-CSF (1 ng/mL) for 60 days. After this time, (iii) hGM (M) Rα, hβc cells and (iv) hGM/βc, hβc cells were harvested, washed, and cultured in murine cytokines that promote myeloid development for a further 7 days. Cytospin preparations were May-Grünwald-Giemsa stained and are representative of 3 separate experiments. Bar is 10 μm.
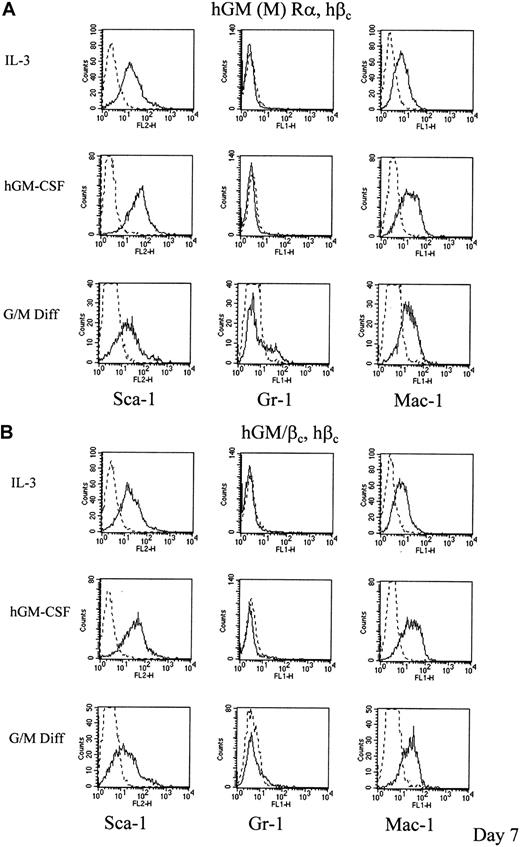
These data demonstrate that the PIG→KLN mutation fundamentally alters the response to hGM-CSF compared to wild-type hGM Rα, βc. Differentiation status was also assessed by the expression of cell surface maturation markers. Expression levels of stem cell antigen-1 (Sca-1, Ly-6) and mature myeloid marker antigens recognized by Gr-1 (Ly6/G) and Mac-1 were measured by flow cytometry. At day 7, control cultures of the hGM R mutants in murine cytokines, which promote G/M Diff, expressed increased levels of Gr-1 and Mac-1 with the level of Sca-1 being decreased (hGM/βc, hβc) or similar (hGM (M) Rα, hβc) to that obtained for cells cultured in murine IL-3 (Figure7A,B). Wild-type hGM Rα, βc cells increase expression of the differentiation markers Gr-1 and Mac-1 but not Sca-118 (Table4). This is consistent with myeloid differentiation. For the hGM/βc, hβc and hGM (M) α, hβc cells cultured with hGM-CSF the situation with respect to expression of biochemical markers of differentiation was more complex. While these cells were primitive in terms of morphology and proliferative potential, there was increased expression of not only a primitive marker, Sca-1, but also the differentiation marker, Mac-1, at day 7. Interestingly, cells expressing hIL-3 Rα, hβc do not show changes in differentiation marker expression and undergo self-renewal (Tables 3and 4). The results obtained for the GM R mutants indicate that there is uncoupling of the regulation of differentiation marker expression from clonal suppression that is regulated by the wild-type hGM R.
hGM-CSF mediated changes in expression of differentiation markers of cells expressing mutant hGM-CSF receptors.
Cells expressing (A) hGM (M) Rα, hβc and (B) hGM/βc, hβc cells were cultured in 1 ng/mL hGM-CSF for 7 days. The levels of expression of the differentiation markers are shown as representative histograms from at least 3 experiments (____). Nonspecific labeling was determined using the corresponding isotype control (- - -). The results obtained for cells cultured in murine cytokines that promote G/M Diff for 7 days are also shown for comparison.
hGM-CSF mediated changes in expression of differentiation markers of cells expressing mutant hGM-CSF receptors.
Cells expressing (A) hGM (M) Rα, hβc and (B) hGM/βc, hβc cells were cultured in 1 ng/mL hGM-CSF for 7 days. The levels of expression of the differentiation markers are shown as representative histograms from at least 3 experiments (____). Nonspecific labeling was determined using the corresponding isotype control (- - -). The results obtained for cells cultured in murine cytokines that promote G/M Diff for 7 days are also shown for comparison.
Effects of activation of wild-type and mutant hIL-3 and hGM-CSF receptors in FDCP-mix cells
| Receptor . | Morphology in rmIL-3 . | Morphology in human cytokine . | Change in Sca-1 expression . | Change in Gr-1 expression . | Change in Mac-1 expression . | Proliferate in culture > 10 d? . |
|---|---|---|---|---|---|---|
| Wild-type hGM Rα, hβc | Blasts | Granulocytes and macrophages | = | ⇑ | ⇑ | No |
| Chimeric hGM/βc, hβc | Blasts | Blasts, EG | ⇑ | = | ⇑ | Yes |
| Wild-type hIL-3 Rα, hβc | Blasts | Blasts | = | = | = | Yes |
| Mutant HGM (M) Rα, hβc | Blasts | Blasts, EG | ⇑ | = | ⇑ | Yes |
| Mutant hIL-3 (M) Rα, hβc | Blasts | Blasts, granulocytes, and macrophages | = | ⇑ | ⇑ | No |
| Receptor . | Morphology in rmIL-3 . | Morphology in human cytokine . | Change in Sca-1 expression . | Change in Gr-1 expression . | Change in Mac-1 expression . | Proliferate in culture > 10 d? . |
|---|---|---|---|---|---|---|
| Wild-type hGM Rα, hβc | Blasts | Granulocytes and macrophages | = | ⇑ | ⇑ | No |
| Chimeric hGM/βc, hβc | Blasts | Blasts, EG | ⇑ | = | ⇑ | Yes |
| Wild-type hIL-3 Rα, hβc | Blasts | Blasts | = | = | = | Yes |
| Mutant HGM (M) Rα, hβc | Blasts | Blasts, EG | ⇑ | = | ⇑ | Yes |
| Mutant hIL-3 (M) Rα, hβc | Blasts | Blasts, granulocytes, and macrophages | = | ⇑ | ⇑ | No |
Data were obtained for the FDCP-mix cell cytokine receptor transfects cultured with either hIL-3 or hGM-CSF for 7 days. The effects on proliferation, morphology, and differentiation markers Sca-1, Gr-1, and Mac-1 are summarized. Differentiation marker expression remained at similar levels (=) increased (⇑) or decreased (⇓).
EG indicates early granulocytes.
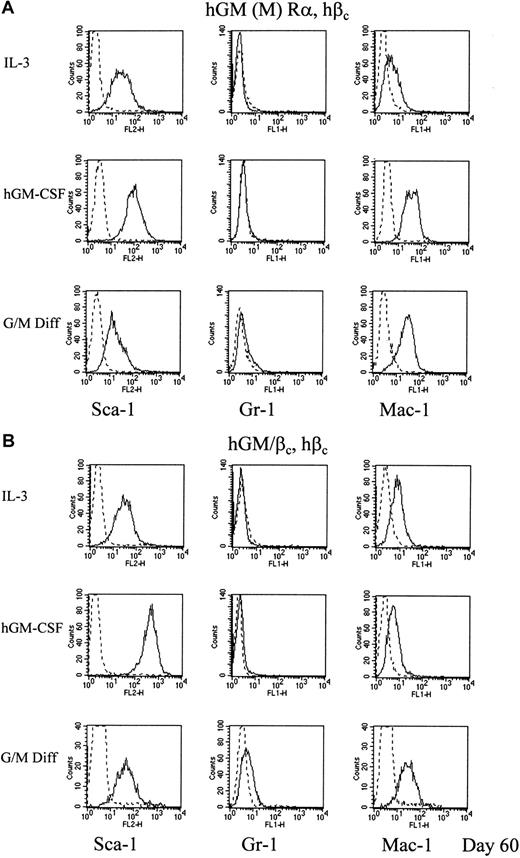
Cells washed to remove the cytokine after 60 days, and subsequently cultured with murine cytokines that promote G/M Diff, differentiated morphologically into granulocytes and macrophages (Figure 6Biii,iv) and showed decreased levels of Sca-1 and increased levels of Gr-1 and Mac-1 (Figure 8A,B). This confirms the maintenance of differentiation potential when cultured in hGM-CSF, even after a prolonged period. Culture of hGM (M) Rα, hβccells and hGM/βc, hβc cells with hGM-CSF resulted in continued perturbation of differentiation marker expression; in particular, there was increased Sca-1 expression relative to time-matched controls of cells cultured in murine IL-3, the cytokine in which cells are maintained routinely.
hGM-CSF mediated changes in expression of differentiation markers of cells expressing mutant hGM-CSF receptors.
Cells expressing (A) hGM (M) Rα, hβc and (B) hGM/βc, hβc cells were cultured in 1 ng/mL hGM-CSF for 60 days. The levels of expression of the differentiation markers are shown as representative histograms from at least 3 experiments (____). Nonspecific labeling was determined using the corresponding isotype control (- - -). The results obtained for cells cultured in murine cytokines that promote G/M Diff for 7 days after 60 days culture in hGM-CSF are also shown.
hGM-CSF mediated changes in expression of differentiation markers of cells expressing mutant hGM-CSF receptors.
Cells expressing (A) hGM (M) Rα, hβc and (B) hGM/βc, hβc cells were cultured in 1 ng/mL hGM-CSF for 60 days. The levels of expression of the differentiation markers are shown as representative histograms from at least 3 experiments (____). Nonspecific labeling was determined using the corresponding isotype control (- - -). The results obtained for cells cultured in murine cytokines that promote G/M Diff for 7 days after 60 days culture in hGM-CSF are also shown.
The data obtained for the wild-type and mutant hIL-3 and hGM-CSF receptors after 7 days in culture are summarized in Table 4.
Discussion
Analysis of the role of the cytosolic domain of the hGM Rα and the functional capability of the hβc subunit
The GM-CSF receptor consists of a ligand-specific α subunit and the βc subunit that is shared in common with the IL-3 and IL-5 receptors. The βc subunit becomes tyrosine phosphorylated on receptor activation, and βchas been proposed to be the primary signal transduction protein in the receptor α, βc subunit complex.34,35 However, our data, and those of others, shows the α subunit cytosolic domains are essential for receptor function and stimulate different signaling pathways.18,19,28,31,32,36,37 More important, perhaps, is the fact that these α subunits can influence differentiation.18 However, the significance of βc cannot be understated. Constitutively activated βc can promote myeloproliferative disorders when expressed in marrow reconstitution systems or transgenic mice.38 39 It is not clear, however, if activation of βc alone is sufficient to promote myeloid cell development.
Experiments with a chimeric hGM/βc receptor transgenic mouse have addressed this question.17 This receptor was composed of the extracellular and transmembrane domain of the α subunit and the cytoplasmic domain of hβc. Administration of hGM-CSF to these mice results in a phenotype equivalent to that of transgenic mice expressing wild-type hGM-CSF receptor in which white blood cell counts are elevated and splenomegaly is observed. The cytoplasmic region of the α subunit would, thus, appear to be functionally redundant and it was proposed that the box 1 region of the cytosolic domain of βc functionally compensates for the lack of the α cytosolic domain. However, analysis of the signals emanating from the wild-type and chimeric receptors revealed that they promote different patterns of JAK kinase phosphorylation.17 This indicates that the α cytosolic domain is key in regulating signal transduction pathways activated by the receptor complex. It also suggests that the subtleties of this signaling system are unapproachable by the use of whole animal experiments at present and the careful use of multipotent cell lines can better facilitate this type of analysis and identify the key events which promote the commitment to differentiation.
The use of the FDCP-mix cell line as a model for the functional consequences of hGM R complex formation overcomes many difficulties in this respect. We have analyzed the potential of βc to promote differentiation by generation of a chimeric hGM/βc receptor composed of the extracellular ligand-binding domain of the hGM Rα subunit and the intracellular domain of the hβc. Addition of hGM-CSF will promote formation of the hGM/βc complexes that will signal via the cytoplasmic domain of βc because it has been demonstrated that GM-CSF can promote homodimerization of hGM Rα subunits, without a requirement for the presence of a βc(hβc or endogenous murine βcsubunits).40 There has been some controversy over the ability of chimeric hGM/βc constructs to signal. The construct of others17,25 is unable to function unless βc is coexpressed. The construct of Eder et al, however, functions in the absence of hβc due to the insertion of an additional glutamate residue adjacent to the fusion point.29 41 The fusion point of our hGM/βcconstruct was in the mid transmembrane domain to avoid interference with membrane proximal sequences of the cytoplasmic domain of hβc, those of other groups is located in the juxta(cytoplasmic) membrane region of βc , which probably accounts for its activity in FDCP-mix cells.
This hGM/βc construct was expressed in the presence and absence of full-length hβc in FDCP-mix. The activation of the βc by homodimerization (promoted by agonist-stimulated activation of chimeric GM/βc) was not equivalent to activation of the wild-type hGM R for either population. Coexpression of hβc with GM/βc chimera enhanced the hGM-CSF–mediated proliferative response of the cells compared to GM/βc alone. The morphology of the cells is primitive compared to that obtained for the wild-type hGM Rα, hβc that undergo myeloid differentiation in response to hGM-CSF18 (Table 1 and Figure 3C). Our data show that “pure” hβc signaling in this multipotent cell background is insufficient to promote terminal myeloid differentiation. Addition of hGM-CSF to FDCP-mix cell hGM/βc, hβc transfects increases expression of the primitive cell marker, Sca-1, relative to levels expressed when cultured in murine IL-3, an effect that is particularly noticeable after prolonged culture in hGM-CSF (Figures 7B and 8B). This perhaps reflects the myeloproliferative disorders observed in mice expressing constitutively activated mutant forms of hβc mutant.38 39
A model for hGM R complex formation has been put forward based on results obtained with several different classes of activating hβc subunit mutations.42 It is proposed that hβc complexes can exist in at least 2 active states, each of which can promote survival, proliferation, and some differentiation signals. In the resting, that is, unstimulated state, hβccan exist in a pre-formed complex with either hGM Rα or an additional βc. Binding of hGM-CSF alters the conformation to generate functional receptor complexes. Activation of the JAK2 kinase, which is constitutively associated with the box 1 region of βc, results in tyrosine phosphorylation. This kinase may not, however, be solely responsible for hβcphosphorylation43 but is critical for hGM-CSF receptor function.44 The hGM R is thought to function as an α2βc2 complex14,40 with GM-CSF binding inducing oligomerization of hβc.45It is possible, however, that a higher order complex can be generated to promote an amplified/modified signal, perhaps by recruitment of additional signaling molecules.42 Within the appropriate cellular context, therefore, the α subunit can influence developmental fate. Interestingly, a splice variant of hβc with a truncation in the intracellular domain has recently been shown to inhibit survival and mitogenic signaling by the hGM R, leading to the proposal that recruitment of alternatively spliced receptor subunits may regulate the receptor function.46 The hGM R may also contain additional distinct components.47,48 and a GM Rα associating protein (GRAP) has been identified by yeast 2 hybrid technology.48
The identification of a key region of hGM Rα, which regulates differentiation
The cytosolic domain of the hGM Rα subunit is important for receptor function.19,31,32,36 37 The α subunit cytosolic domains of hGM R and hIL-3 R stimulate different developmental responses. We aimed to identify structural features of the α cytosolic domain that promote differentiation and maintenance of primitive phenotype, respectively. Analysis of the amino acid sequence of the IL-3, GM-CSF, and related IL-5 receptor cytosolic domains identified a candidate tripeptide region.
The data obtained using mutant and wild-type hIL-3 and hGM-CSF receptors, in which there is a cross-exchange of a specific tripeptide region, indicate that all can promote short-term proliferation, as assessed by thymidine incorporation (Figures 4B and 5B). There are specific differences in terms of cell fate (Figures 3C and4C). The effects of hIL-3 or hGM-CSF on the morphology of the wild-type and mutant hIL-3 R and hGM R cells, respectively, were similar at all concentrations of human cytokine tested (0.1-100 ng/mL). This indicated that the differential effects of the hIL-3 and hGM-CSF receptor activation were not simply due to the dose of the cytokine used. The demonstration that the IL-3 Rα (PIG→KLN) signals for development of FDCP-mix (Figure 4) suggests that PIG tripeptide is critical for self-renewal signaling from IL-3. Replacing KLN in GM-CSF Rα with PIG, the equivalent tripeptide from IL-3Rα, we generated a receptor that signals predominantly for maintenance of the primitive cell phenotype (Figure 6), which resembles the wild-type IL-3 Rα more than hGM Rα subunit.18
Activation of the wild-type hIL-3 Rα, hβc promotes self-renewal and allows the long-term maintenance of FDCP-mix cells. The mutated GM-CSF R resembled the wild-type hIL-3 R in promoting long-term survival and proliferation. However, cultures of hGM (M) Rα, hβc and hGM/βc, hβc contain cells of early granulocyte morphology in addition to blast cells in response to hGM-CSF. There is also a significant up-regulation in the cell surface expression of Mac-1, a biochemical marker of differentiation (Figure 7). Thus there is some degree of granulocytic differentiation in response to activation by hGM-CSF with an apparent uncoupling of the differentiation pathway from clonal suppression compared to the wild-type hGM R. These data are reminiscent of results obtained using βc truncation mutants to study hGM-CSF receptor function. This work demonstrated that different aspects of the differentiated phenotype could be dissociated, leading to the model that there are distinct regions of the receptor βc subunit that act cooperatively, and perhaps with the α subunit, to produce full differentiation.49 This is also similar to the situation with the G-CSF R where several domains contribute to the developmental response following receptor activation.50
Here we show critical differences in IL-3 R and GM-CSF R in the governing cell fate. This is further evidence that the α cytosolic domains can be ascribed specific function, not simply an ability to activate βc. We demonstrate for the first time that such “instructions” are implicitly associated with receptor structure. Interestingly, it has recently been shown that hGM R activation can promote cell-fate conversion of murine lymphoid-committed progenitors: transfection of this cell population with the hGM R resulted in G/M Diff in the presence of hGM-CSF.11 Our next goal is to identify the differential signaling events associated with cytokine-mediated differentiation or maintenance of primitive cell phenotype using our model system.
Prepublished online as Blood First Edition Paper, July 12, 2002; DOI 10.1182/blood-2001-12-0235.
Supported by Leukaemia Research Fund (United Kingdom). S.A. is supported by a government grant from Universiti Kebangsaan, Malaysia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anthony D. Whetton, Leukaemia Research Fund Cellular Development Unit, Department of Biomolecular Sciences, UMIST, Sackville St, Manchester, United Kingdom M60 1QD; e-mail:tony.whetton@umist.ac.uk.

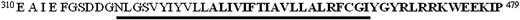

![Fig. 4. Effects of hIL-3 (M) Rα activation. / (A) Cell surface expression. hIL-3 (M) Rα, hβc cell transfects were analyzed for expression of the extracellular domains of the (i) hIL-3 Rα and (ii) hβc by flow cytometry. Cells were sequentially incubated with anti–hIL-3 Rα antibody and fluorescein isothiocyanate (FITC)–conjugated antimouse secondary antibody. The solid gray histogram represents the cell autofluorescence obtained in the absence of antibody staining. Representative histograms are shown for labeling obtained in the presence of secondary antibody only (gray) and in the presence of both primary and secondary antibody (black). (B) Cell survival proliferation. Cells expressing wild-type hIL-3 Rα, hβc (░) or hIL-3 (M) Rα, hβc (▪) were cultured in hIL-3 (0-100 ng/mL). (i) Cell viability was assessed at 48 hours using trypan blue. The results are expressed as cell viability (percent rmIL-3 response) and are mean values ± SEM from at least 3 experiments. (ii) [3H]-thymidine incorporation was assessed after 16 hours in culture. The results are expressed as percentage of the response obtained with rmIL-3 (10 ng/mL). (C) Cell morphology. Cells expressing (i) wild-type hIL-3 Rα, hβc or (ii) hIL-3 (M) Rα, hβc were cultured in hIL-3 (1 ng/mL) for 7 days. Photomicrographs were prepared from May-Grünwald-Giemsa–stained cytospin preparations. Results are representative of 3 experiments. Bar is 10 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/9/10.1182_blood-2001-12-0235/4/m_h82123321004.jpeg?Expires=1767750163&Signature=GRD28fNqzUznyjeB-EzYLK7neP-f1JrW0urmoUa8CFuW9NMVS-pxJ4B2Sg6u-WRfxLGrMMeyK2is~Mq05gWkDCCac299VgkETjTdHaW2uroOr3gZsgU7sA~aCgUvnsL~Cuzvo~JE~vFqS9ktYT4hFhsPsQJWWJ5d7N13m2lF5q4b6SKfWaJ19D1NxaJ86StdYuKjObfrEdNnwRjTFM5heoifMxFt5Y~dbE2HtyIwfmRe3zX4OJj-ZoAUoqJTyT-biD-oKBkGIzJNiVYGdnKAs8patTtRjkFgEPJs4fpRJv1~DKtscoqY8DkXH~B8xwJ6qjkddRFe2pAyigDegQm9KQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal