Hematopoietic bone marrow stem cells generate differentiated blood cells and, when transplanted, may contribute to other organs, such as the brain, heart, and liver. An understanding of in vivo clonal behavior of stem cells will have important implications for cellular and gene therapy. For the first time, we have directly demonstrated the derivation of circulating peripheral blood cells from individual stem cell clones. We analyzed the clonal composition of retrovirus-marked peripheral blood leukocyte populations in 2 different primate models by a novel direct genomic sequencing technique allowing the identification of vector insertion sites. More than 80 contributing long-term hematopoietic clones were identified in individual rhesus macaque peripheral blood transplant recipients and more than 25 different clones in a baboon marrow transplant recipient. Up to 5 insertion sequences from each animal were used to trace the long-term contribution of stem cell clones in these primate models. Continuous and mostly pluripotent contributions of peripheral blood leukocytes from each of the traced clones could be detected for the entire follow-up period of 23 to 33 months. Our study provides direct molecular evidence for a polyclonal, multilineage, and sustained contribution of individual stem cells to primate hematopoiesis.
Introduction
Insights into the number and the contribution of individual pluripotent hematopoietic stem cells (HSCs) to the formation of blood lineages would have great significance for the understanding of normal and abnormal hematopoiesis and the design of therapeutic interventions such as gene therapy and transplantation.1It was previously demonstrated that murine repopulating HSCs have long-term pluripotent activity that may fluctuate when cells are transplanted at limiting dilution.2-6 Their frequency was estimated at 1 to 8 per 105 nucleated marrow cells.7,8 However, a comparative computer simulation analysis between murine and feline autologous transplantation data indicates that the in vivo behavior of mouse stem cells cannot be extrapolated to large animals or humans.8 To estimate clonal stem cell contributions to human hematopoiesis in vivo, allelic expression of X-linked genetic polymorphisms was analyzed in allogeneic transplant recipients.9 10 These studies suggested that hematopoietic stem cell activity after transplantation in humans could be either oligoclonal or polyclonal. However, their indirect results allowed neither enumeration of individual HSC clones nor determination of their lifespan or multipotentiality.
Because of their random insertion into the genome, retrovirus and lentivirus vectors can be used as molecular markers to trace a transduced cell's progeny at the clonal level.5,11,12Previous analyses of the clonal activity of primitive human cells have employed xenotransplantation of human cord blood cells into immunodeficient BNX or NOD/SCID mice. Using Southern blot or inverse polymerase chain reaction (PCR) to distinguish retroviral integration sites, these studies have established that fewer than 10 human hematopoietic cell clones engrafted in these animals.13,14Since only a small fraction of human marrow or cord blood was transferred in these experiments, the number of clones active in humans could be substantially higher. It is difficult to trace pluripotent stem cell activity in such xenotransplantation models because seeding inefficiency leads to oligoclonal engraftment, and the lifespan of mice that underwent transplantation is short.14-16 Furthermore, the techniques hitherto used for insertion site analysis did not permit an understanding of the contributions of individual clones in highly complex polyclonal circulating cell populations.17
It is widely known that the genetic and biologic similarity between higher primates and humans makes nonhuman primates the best available model in which to study in vivo hematopoiesis. Recent progress in achieving high gene marking efficiency of repopulating cells for the first time permits the study of normal hematopoiesis using insertion site analysis in peripheral blood leukocytes.18 19 In addition, the extended lifespan of these animals (up to 30 years) allows long-term observation and assessment of the impact of cytokine treatment, chemotherapy, or other clinically relevant interventions.
In a previous study, we tried to define whether multiple repopulating cells were present in gene-marked primates. We found several different integration sites in marrow-derived colony-forming unit cell (CFU-C) colonies and purified lymphocytes within the first 9 months after autologous transplantation. However, technical limitations of the inverse PCR method and reliance on an analysis of marrow-derived colonies20 did not allow an accurate assessment of the number of clones contributing to hematopoiesis over time or a rigorous analysis of their lineage contributions. In contrast to marrow-derived colonies, peripheral blood granulocytes, as short-lived circulating descendants of more primitive cells, would be particularly useful as sentinels for stem cell activity and would provide an excellent measure of ongoing contributions to hematopoiesis over time.1 21
In the present study, we succeeded in directly analyzing the clonal contributions of retrovirus-marked primitive cells to peripheral blood granulocytes and other blood lineages of rhesus macaques and baboons. With a newly developed, highly sensitive and robust assay, more than 80 different clones could be detected in the leukocyte populations of primates. The identification of specific clones in purified granulocytes and lymphocytes by their unique molecular markers allowed the assessment of how individual clones contribute to specific lineages, indicating that they have continuously contributed to peripheral blood lineages for more than 2 years to date.
Methods
Rhesus macaque transplantation model
All animal experiments were performed after approval of the institutional animal care and use committee in complete compliance with all relevant laws and institutional guidelines. Stem-cell factor (SCF)/granulocyte colony-stimulating factor (G-CSF)–mobilized and CD34-enriched peripheral blood cells from each animal were exposed to retrovirus vectors G1NA or LNL6 for 4 days. Cells were transduced in the presence of interleukin-3 (IL-3), IL-6, SCF, Flt-3 ligand (Flt-3L) on fibronectin CH-296 fragment or autologous stroma (animals RC501 and 95E003) or in the presence of Flt-3L, SCF, megakaryocyte growth and development factor (MGDF) on retronectin (RC706 and 96E019). Reinfusion after 1000 cGy of total body irradiation, density gradient purification, antibody staining, fluorescence-activated cell sorting, and DNA extraction were performed as previously described.20 22
Baboon transplantation model and integration site analysis
A juvenile baboon (T94433) received a transplant of G-CSF/SCF–primed CD34+ marrow cells after transduction on CH-296, using a gibbon ape leukemia virus (GALV) pseudotype retroviral vector. Density gradient purification, antibody staining, fluorescence-activated cell sorting, and DNA extraction were performed as previously described.18 19
Transgene detection by semiquantitative PCR and Southern blot analysis
We amplified a 589-bp fragment of the 5′ vector sequence in each primate sample of DNA (10 ng) in comparison with a limiting dilution (10 ng, 1 ng, 100 pg) of DNA from single-copy LN-transduced HeLa cells. Taq polymerase (2.5 U; Qiagen, Valencia, CA) was used in 35 PCR cycles (denaturation at 95°C for 60 seconds, annealing at 60°C for 45 seconds, extension at 72°C for 60 seconds) after initial denaturation for 5 minutes and before final extension for 10 minutes with primers LTR6 (5′-GTGGTCTCGCTGTTCCTT-3′; Roth, Karlsruhe, Germany) and MISC (5′-CGCTCGACATCTTTCCAGT-3′; Roth). Of each PCR product, 20% were separated on a 2% agarose gel, transferred to a nylon membrane (Hybond-N; Amersham, Piscataway, NJ) by pressure blot (Stratagene, La Jolla, CA), probed with a digoxigenin-labeled long terminal repeat (LTR)/extended packaging signal probe, and documented by chemiluminescence exposure to x-ray film (Roche Diagnostics, Mannheim, Germany).
5′-LTR integration site analysis using linear amplification-mediated (LAM) PCR
The genomic-proviral junction sequence was preamplified by repeated primer extension using 0.25 pmol of vector-specific, 5′-biotinylated primer LTR1 (5′-AGCTGTTCCATCTGTTCTTGGCCCT-3′; Roth) with Taq polymerase (2.5 U; Qiagen) from 100 ng of each sample DNA. As described above, 100 cycles of amplification were performed with addition of fresh Taq polymerase (2.5 U) after 50 cycles. Selection of biotinylated extension products was performed with 200 μg of magnetic beads according to the manufacturer's instructions (Dynal, Oslo, Norway). The samples were incubated with Klenow polymerase (2 U; Roche), dNTPs (300 μM; Pharmacia, Uppsala, Sweden), and random hexanucleotide mixture (Roche) in a volume of 20 μL for 1 hour at 37°C. Samples were washed on the magnetic particle concentrator (Dynal) and incubated with Sse9I endonuclease (4 U in 20 μL; Hybaid-AGS, Middlesex, United Kingdom) for 1 hour at 55°C. After an additional wash step, 100 pmol of a double-stranded asymmetric linker cassette and T4 DNA Ligase (6 U; New England Biolabs, Beverly, MA) were incubated with the beads in a volume of 10 μL at 16°C overnight. Denaturing was performed with 5 μL of 0.1N NaOH for 10 minutes at room temperature. Each ligation product was amplified with Taq polymerase (5 U; Qiagen), 25 pmol of vector-specific primer LTR2 (5′-GACCTTGATCTGAACTTCTC-3′; Roth), and linker cassette primer LC1 (GACCCGGGAGATCTGAATTC), using the amplification conditions described above. Of each PCR product, 0.2% served as a template for a second, nested PCR with internal primers LTR3 (5′-TCCATGCCTTGCAAAATGGC-3′; Roth) and LC2 (5′-GATCTGAATTCAGTGGCACAG-3′; Roth) at identical conditions. Of this final product, 80% were separated on a Spreadex high-resolution gel (Elchrom Scientific, Cham, Switzerland). Specific DNA bands were excised and reamplified with primers LC3 (5′-AGTGGCACAGCAGTTAGG-3′; Roth) and LTR4 (5′-CCTTGCAAAATGGCGTTACT-3′; Roth) for the total of 45 cycles. PCR products were cycle-sequenced directly or after cloning into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA).
3′Integration flank direct primer walking analysis
For each of 3 randomly selected 5′integration site sequences, a second linear amplification-mediated (LAM) PCR was performed as described above on a 20-ng DNA template of nontransduced primate DNA. Primer extension oligonucleotides were used in forward (5′ to 3′) orientation homologous to the respective 5′LTR flanking DNA sequence obtained in the initial LAM-PCR reaction. The following primers were used for the linear (primer /1) and the 2 exponential nested amplifications (primers /2 and /3): 2082/1 (5′-TGTGTCATCCATGTTTGTGT-3′; Roth), 2082/2 (5′-GAAGGGGCATGGGCAAGTGA-3′; Roth), and 2082/3 (5′-GCTGTGCAGGAGTGAAACC-3′; Roth) for clone 2082, 2861/1 (5′-CATGTCTGGCGTGGAGTAAG-3′; Roth), 2861/2 (5′-ATTCAATACATGTGGCTACTA-3′; Roth), and 2861/3 (5′-ATGTGGCTACTATGACTCTC-3′; Roth) for clone 2861 (Figure A, middle panel). To verify that the following new sequence should be the 3′ genomic flank of each retroviral integration locus, a pair of 3′flanking genomic DNA-nested reverse primers in reverse (3′ to 5′) orientation (Figure 3, right panel, 3′ Fl) was designed for each clone: 2082/1rev (5′-CACTGTCTCTATGGCTGTTCC-3′; Roth) and 2082/2rev (5′-TCACCGTGACAGCTCTGGT-3′; Roth), 2861/1rev (5′-TAGCGTATTGACACGTGGAG-3′; Roth) and 2861/2rev (5′-AGGCAGCCAGGTTTGACTTC -3′; Roth).
PCR tracing of individual clones
Of 9 primers designed for the sensitive and specific seminested detection of individual hematopoietic cell clones, 4 (rhesus) and 5 (baboon) were sufficiently sensitive to trace the integration site in the complex DNA background of the day 339 sample. Tracing was focused on 3 clones per animal because the amount of DNA available from different time points and from purified hematopoietic lineages was limited. PCR reactions used genomic DNA (100 ng) isolated from different cell lineages and time points after transplantation. A 35-cycle PCR reaction with an annealing temperature of 56°C was performed as described above. Of this product, 0.02% was used as a template for the second, seminested PCR. Except for a 60°C annealing temperature, PCR conditions were identical to the first PCR. The primers used were 2082/3 (see above), 3392/1 (5′-TGAAGTCAAAGAGGGAAGTC-3′; Roth), 3397/1 (5′-CGCGCAGTGGAGTTAT-3′; Roth) with LTR4 (see above) for the first PCR and LTR5 (5′-CAAACCTACAGGTGGGGTCT-3′; Roth) for the second, seminested PCR. The flanking primers used in the baboon model were 11/1 (5′-ATGTAGCCATGATTTGCACC-3′), 14/1 (5′-CTGAACTTGAAGATGGGTCT-3′), 29/1 (5′-TAGGAGAAAACGCATGTGGA-3′).
Results and discussion
Gene-marked pluripotent long-term hematopoiesis in primate models
To analyze the clonal contributions of primitive HSCs and progenitors to peripheral blood lineages in the rhesus macaque model, we initially analyzed an animal in which we had achieved stable high level retroviral marking in all lineages.22 After full myeloablation, rhesus macaque RC501 received a transplant of 7 million/kg autologous peripheral blood CD34+ cells, transduced with retrovirus marking vectors that had previously been mobilized by SCF and G-CSF. Blood was collected at 16 different time points after transplantation (between 1 and 23 months), and marked progeny cells were analyzed by limiting-dilution semiquantitative PCR and Southern blot. Marked cells were found to represent between 5% and 20% of peripheral blood leukocytes at all time points sampled after leukocyte regeneration as previously described (also see below).22
To verify our approach and conclusions in an independent primate model, we analyzed the clonal composition of transduced peripheral blood cells after transplantation of gene-marked SCF- and G-CSF–prestimulated marrow cells in a baboon model. We previously showed efficient gene transfer into baboon marrow repopulating cells,18 19 using a GALV pseudotype vector and recombinant human fibronectin fragment CH-296 for the ex vivo transduction of SCF- and G-CSF–prestimulated bone marrow CD34+ cells.
In vivo clonality analysis of peripheral blood hematopoiesis by LAM-PCR
To detect multiple different retrovirus integration sites within populations of marked peripheral blood cells with sufficient sensitivity and specificity, we devised a novel LAM-PCR. For this purpose, we combined repeated linear primer extension, primer tag selection, and asymmetric oligonucleotide cassette ligation23 24 with nested exponential PCR amplification (Figure 1).
Outline of linear amplification-mediated (LAM)–PCR.
A new combination of linear amplification of target DNA with solid-phase second-strand synthesis, followed by ligation of an oligonucleotide cassette and then nested exponential PCR, was devised for the detection and direct genomic sequencing of unknown retroviral vector integration sites. (A) Linear PCR with a long terminal repeat (LTR)–specific biotinylated primer was performed by repeated primer extension. Subsequently, the amplified fragments of target DNA were enriched by magnetic tag selection of extension primers. (B) A second DNA strand of each enriched target sequence was synthesized by random hexanucleotide priming. (C) Resulting double-stranded DNA was specifically digested with the restriction enzyme Sse9I, which cuts within genomic DNA approximately every 256 bp. The length of each fragment is thus dependent on the distance of the vector insertion site from the next Sse9I recognition sequence. (D) An asymmetric oligonucleotide ligation cassette (LC) was ligated to the end of the Sse9I-digested fragments. (E) Nested exponential PCR amplifications were then performed with LC-specific forward primers (LC 1 followed by LC 2) and LTR-specific reverse primers (LTR II followed by LTR III).
Outline of linear amplification-mediated (LAM)–PCR.
A new combination of linear amplification of target DNA with solid-phase second-strand synthesis, followed by ligation of an oligonucleotide cassette and then nested exponential PCR, was devised for the detection and direct genomic sequencing of unknown retroviral vector integration sites. (A) Linear PCR with a long terminal repeat (LTR)–specific biotinylated primer was performed by repeated primer extension. Subsequently, the amplified fragments of target DNA were enriched by magnetic tag selection of extension primers. (B) A second DNA strand of each enriched target sequence was synthesized by random hexanucleotide priming. (C) Resulting double-stranded DNA was specifically digested with the restriction enzyme Sse9I, which cuts within genomic DNA approximately every 256 bp. The length of each fragment is thus dependent on the distance of the vector insertion site from the next Sse9I recognition sequence. (D) An asymmetric oligonucleotide ligation cassette (LC) was ligated to the end of the Sse9I-digested fragments. (E) Nested exponential PCR amplifications were then performed with LC-specific forward primers (LC 1 followed by LC 2) and LTR-specific reverse primers (LTR II followed by LTR III).
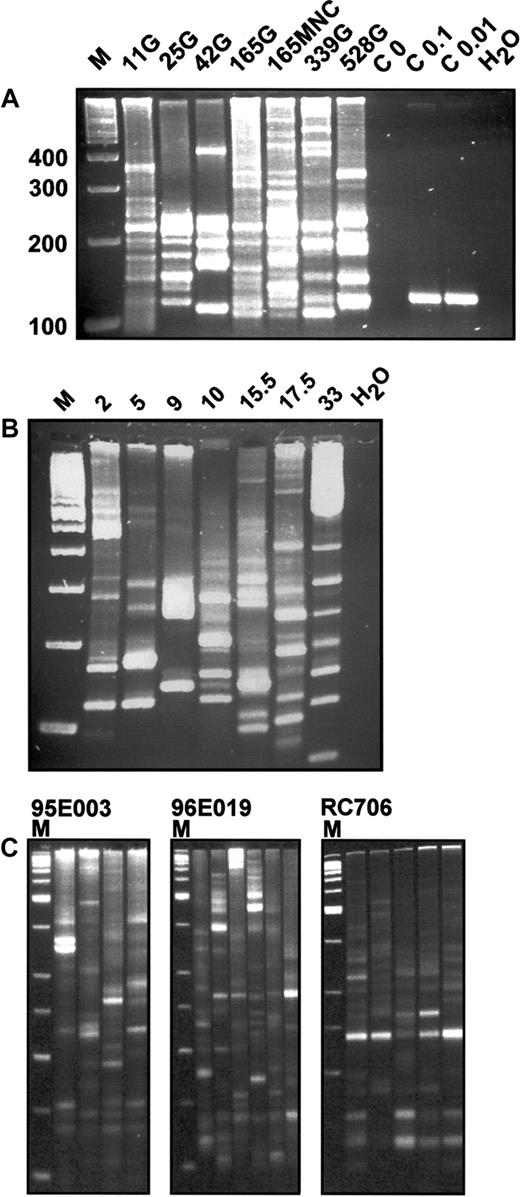
With this strategy, we found unprecedented sensitivity that allowed the identification and direct genomic sequencing of proviral-genomic fusion sequences. Analysis of rhesus granulocyte DNA from 6 time points after transplantation demonstrated a highly polyclonal composition of vector-containing cells (Figure2A).
In vivo integration site analysis of primate leukocytes.
(A) LAM-PCR analysis in a rhesus macaque. DNA samples of 100 ng from FACS-sorted granulocytes (G) or mononuclear cells (MNCs) of rhesus macaque RC501 were analyzed at different time points after transplantation. Numbers indicate days after transplantation. M indicates a 100-bp DNA ladder; 100, 200, 300, 400, size of molecular weight marker in base pairs; C 0, negative control analysis on 1.4 μg nontransduced simian DNA; C 0.1, C 0.01, positive control analysis on 0.1 and 0.01 ng of DNA from a HeLa cell clone with a known single retrovirus insertion site corresponding to 15 and 1.5 genomic DNA equivalents, respectively; and H2O, water control. (B) LAM-PCR analysis in a baboon. DNA samples (100 ng) from FACS-sorted mononuclear cells (MNCs) of baboon T94433 were analyzed at different time points after transplantation. Note that the integration site signal is polyclonal throughout the observation period. Numbers indicate months after transplantation. M indicates a 100-bp DNA ladder; H2O, water control. (C) Reproducibility of LAM-PCR analysis in highly polyclonal rhesus macaque samples. From 4 to 6 repetitions of LAM-PCR analysis on individual 200-ng aliquots of DNA from purified granulocytes (G) obtained from rhesus macaques 95E003, 96E019, and RC706 were analyzed. Animal 95E003 was 40 months after transplantation, animal RC706 was 21 months after transplantation, and animal 96E019 was 27 months after transplantation at the time of sampling. For each animal, the separate amplifications are shown in adjacent lanes. M indicates a 50-bp DNA ladder.
In vivo integration site analysis of primate leukocytes.
(A) LAM-PCR analysis in a rhesus macaque. DNA samples of 100 ng from FACS-sorted granulocytes (G) or mononuclear cells (MNCs) of rhesus macaque RC501 were analyzed at different time points after transplantation. Numbers indicate days after transplantation. M indicates a 100-bp DNA ladder; 100, 200, 300, 400, size of molecular weight marker in base pairs; C 0, negative control analysis on 1.4 μg nontransduced simian DNA; C 0.1, C 0.01, positive control analysis on 0.1 and 0.01 ng of DNA from a HeLa cell clone with a known single retrovirus insertion site corresponding to 15 and 1.5 genomic DNA equivalents, respectively; and H2O, water control. (B) LAM-PCR analysis in a baboon. DNA samples (100 ng) from FACS-sorted mononuclear cells (MNCs) of baboon T94433 were analyzed at different time points after transplantation. Note that the integration site signal is polyclonal throughout the observation period. Numbers indicate months after transplantation. M indicates a 100-bp DNA ladder; H2O, water control. (C) Reproducibility of LAM-PCR analysis in highly polyclonal rhesus macaque samples. From 4 to 6 repetitions of LAM-PCR analysis on individual 200-ng aliquots of DNA from purified granulocytes (G) obtained from rhesus macaques 95E003, 96E019, and RC706 were analyzed. Animal 95E003 was 40 months after transplantation, animal RC706 was 21 months after transplantation, and animal 96E019 was 27 months after transplantation at the time of sampling. For each animal, the separate amplifications are shown in adjacent lanes. M indicates a 50-bp DNA ladder.
Granulocyte, lymphocyte, and mononuclear cell fractions at each time point showed at least 20 and up to more than 50 different retrovirus integration site amplification products by LAM-PCR. The band frequency was confirmed by use of fluorescent primers and separation of the products via an automated sequencer capillary system (ABI 9600) and GeneScan software (data not shown). A similar polyclonality of hematopoiesis was confirmed in 3 additional rhesus macaques and the baboon (Figure 2B,C).
Repeated analyses on DNA aliquots from the same blood samples showed the majority of bands present in all samples, but also additional bands present in each sample, reflecting, we believe, the presence of some clonal contributions at close to the threshold for detection in a 100-ng test sample (Figure 2C).
Because the reproducibility of the single amplification steps and defined control mixtures was excellent,24 25this observation suggests that the small samples analyzed, containing between 103 and 104 nucleated blood cells each, are randomly selected, partial representations of the total population of clones present in each animal. Thus, the number of detectable bands at an individual time point is likely an underestimate of the total number of contributing clones.
Verification of integration sites by direct sequencing
To verify that genomic retrovirus integration sites were detected, we sequenced 83 randomly selected LAM-PCR amplification products from samples of rhesus macaque RC501 at different time points from various lineages. In the baboon T94433, 29 unique integration sites were identified and sequenced by LAM-PCR. Each contained the expected order of oligonucleotide cassette, restriction enzyme recognition sequence, genomic sequence, and retrovirus 5′LTR (data submitted but not shown). We further verified genomic integration for 3 randomly selected 5′ integration site sequences by sequencing each corresponding 3′ genomic flank. In all 3, the sequence of the 5′ integration flank was contiguous with the corresponding 3′ flank sequence in nontransduced cells. The 4-bp genomic DNA direct repeat generated by the murine leukemia retrovirus integrase adjacent to each proviral LTR26 could be correctly predicted from this sequence, confirming the origin of the amplification products from retrovirus insertion sites (Figure 3).
Sequencing of complete integration loci.
To verify the genomic origin of the integration sequences obtained by LAM-PCR, a direct genomic primer walking from 5′ to 3′ was performed by repeated application of the LAM-PCR direct genomic sequencing protocol in different directions. 5′ Fusion: the 5′ genomic DNA/5′ LTR-cDNA fusion sequences were obtained by initial LAM-PCR reactions. Integration Site: an amplification product spanning the complete 5′ and 3′ integration locus was produced by the second LAM-PCR reaction for each of the analyzed clones. All bands obtained were isolated, purified, and sequenced. In each of the 3 clones analyzed (shown: 2 clones), this second LAM-PCR reaction obtained a sequence that was identical to the sequence obtained in the first LAM-PCR reaction from the 5′ end of the amplification product to the 5′ genomic proviral fusion sequence (arrows), then identical to the 3′ integration flank to the 3′ end of the amplification product. 3′ Fusion: the 3′ LTR-cDNA/3′ genomic DNA fusion sequences were obtained by conventional PCR sequencing. Together with a 3′ LTR U5 forward primer (LTR), the 3′ flanking genomic primer pairs (3′ Fl) produced a clone-specific amplification product by nested amplification on 1 μg of transduced genomic DNA from day 339 peripheral blood granulocytes. Sequencing confirmed that the amplification products consisted of genomic 3′ flanking sequence, the 4-bp repeat identical to the 4 bp flanking the 5′ LTR U3 of the respective clone and the expected 3′ LTR U5 vector sequence. The 4-bp genomic sequence direct repeat reduplicated by the retrovirus integrase is enlarged. The first 4 proviral DNA nucleotides are shown in underlined italics. C indicates unrelated DNA controls; LAM1, 5′ LTR extension primer of the initial LAM-PCR; LAM2, 5′ integration site–specific genomic DNA extension primers for the second LAM-PCR based on the 5′ genomic DNA sequences identified in the initial LAM-PCRs.
Sequencing of complete integration loci.
To verify the genomic origin of the integration sequences obtained by LAM-PCR, a direct genomic primer walking from 5′ to 3′ was performed by repeated application of the LAM-PCR direct genomic sequencing protocol in different directions. 5′ Fusion: the 5′ genomic DNA/5′ LTR-cDNA fusion sequences were obtained by initial LAM-PCR reactions. Integration Site: an amplification product spanning the complete 5′ and 3′ integration locus was produced by the second LAM-PCR reaction for each of the analyzed clones. All bands obtained were isolated, purified, and sequenced. In each of the 3 clones analyzed (shown: 2 clones), this second LAM-PCR reaction obtained a sequence that was identical to the sequence obtained in the first LAM-PCR reaction from the 5′ end of the amplification product to the 5′ genomic proviral fusion sequence (arrows), then identical to the 3′ integration flank to the 3′ end of the amplification product. 3′ Fusion: the 3′ LTR-cDNA/3′ genomic DNA fusion sequences were obtained by conventional PCR sequencing. Together with a 3′ LTR U5 forward primer (LTR), the 3′ flanking genomic primer pairs (3′ Fl) produced a clone-specific amplification product by nested amplification on 1 μg of transduced genomic DNA from day 339 peripheral blood granulocytes. Sequencing confirmed that the amplification products consisted of genomic 3′ flanking sequence, the 4-bp repeat identical to the 4 bp flanking the 5′ LTR U3 of the respective clone and the expected 3′ LTR U5 vector sequence. The 4-bp genomic sequence direct repeat reduplicated by the retrovirus integrase is enlarged. The first 4 proviral DNA nucleotides are shown in underlined italics. C indicates unrelated DNA controls; LAM1, 5′ LTR extension primer of the initial LAM-PCR; LAM2, 5′ integration site–specific genomic DNA extension primers for the second LAM-PCR based on the 5′ genomic DNA sequences identified in the initial LAM-PCRs.
Long-term activity of individual stem cell clones
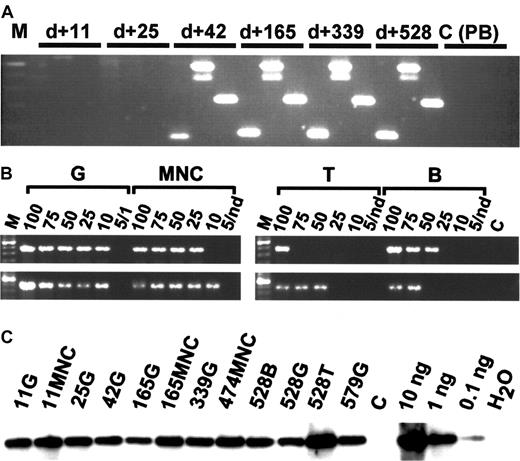
Because granulocytes have a half-life of only 2 to 5 days and derive from stem cells and primitive progenitors in close succession, the presence of a clonal marker in purified granulocytes indicates the activity of a repopulating stem cell clone. To trace the presence of clonal progeny from specific stem cells in different peripheral blood lineages over time, we designed specific PCR primer sets to specifically amplify different randomly selected integration site sequences by conventional PCR. In the rhesus macaque, the integration site sequences of the 4 selected clones had initially been identified in the granulocyte populations at 6 months after transplantation. None of the clones examined (clones 2082, 3393, 3397) were detectable until 6 to 8 weeks after transplantation, suggesting that a different population of repopulating cells is responsible for hematopoiesis immediately following transplantation (Figures 4,5).
In vivo clone tracing by conventional PCR and limiting-dilution analysis.
(A) Conventional PCR. DNA isolated from FACS-purified granulocytes 11, 25, 42, 165, 339, and 528 days after transplantation of rhesus macaque 501 shows evidence for the presence of clones 3392, 2082, and 3397 (from left to right in each sample). The entire analysis from 16 time points is summarized in Figure 5A. M indicates molecular marker (100-bp ladder); C (PB), unrelated transduced peripheral blood control. (B) Limiting-dilution analysis. Note the presence of approximately equal copy numbers of clone 3392 in different lineages on days 240 (top lane) and 528 (bottom lane). Numbers indicate nanograms of DNA submitted to PCR analysis. Nd indicates not done; C, unrelated template control. (C) Total vector DNA. Per sample analyzed in Figures 2 and 3 (A, B), 10 ng DNA was analyzed for the presence of vector cDNA by semiquantitative PCR. Numbers indicate days after transplantation. G indicates DNA of purified peripheral blood granulocytes; MNC, purified peripheral blood mononuclear cells; T, T-cell DNA; B, B-cell DNA; DNA, transduced MNC DNA control; and H2O, no template control.
In vivo clone tracing by conventional PCR and limiting-dilution analysis.
(A) Conventional PCR. DNA isolated from FACS-purified granulocytes 11, 25, 42, 165, 339, and 528 days after transplantation of rhesus macaque 501 shows evidence for the presence of clones 3392, 2082, and 3397 (from left to right in each sample). The entire analysis from 16 time points is summarized in Figure 5A. M indicates molecular marker (100-bp ladder); C (PB), unrelated transduced peripheral blood control. (B) Limiting-dilution analysis. Note the presence of approximately equal copy numbers of clone 3392 in different lineages on days 240 (top lane) and 528 (bottom lane). Numbers indicate nanograms of DNA submitted to PCR analysis. Nd indicates not done; C, unrelated template control. (C) Total vector DNA. Per sample analyzed in Figures 2 and 3 (A, B), 10 ng DNA was analyzed for the presence of vector cDNA by semiquantitative PCR. Numbers indicate days after transplantation. G indicates DNA of purified peripheral blood granulocytes; MNC, purified peripheral blood mononuclear cells; T, T-cell DNA; B, B-cell DNA; DNA, transduced MNC DNA control; and H2O, no template control.
Detection and tracing over time of different retrovirus-marked hematopoietic clones in a rhesus macaque model and a baboon model.
Part A shows a rhesus macaque model and part B shows a baboon model. Days indicates days after transplantation; BM, bone marrow; G, peripheral blood granulocytes; MNC, peripheral blood mononuclear cells; T, peripheral blood T cells; B, peripheral blood B cells; dark gray shading, positive time points; light gray shading, negative time points; and *, verified in 1 μg DNA.
Detection and tracing over time of different retrovirus-marked hematopoietic clones in a rhesus macaque model and a baboon model.
Part A shows a rhesus macaque model and part B shows a baboon model. Days indicates days after transplantation; BM, bone marrow; G, peripheral blood granulocytes; MNC, peripheral blood mononuclear cells; T, peripheral blood T cells; B, peripheral blood B cells; dark gray shading, positive time points; light gray shading, negative time points; and *, verified in 1 μg DNA.
However, from 6 to 8 weeks on, all 3 traced rhesus clones continuously contributed to granulocyte and other peripheral cell populations for the entire 23 months analyzed (Figure 5A). Clones 2082 and 3393 could be identified in both sorted T and B lymphoid and myeloid cells, and therefore represent true multipotent transduced clones with long-term repopulating stem cell activity. In contrast, clone 3397 was detected only in granulocytes and unsorted mononuclear cells, including lymphocytes and monocytes. This suggests the presence of a long-term repopulating clone with primarily or only myeloid potential. The fourth clone (clone 2861) could be detected in another granulocyte fraction at 6 months after initial identification. In the baboon, peripheral blood mononuclear cells and granulocytes (11 time points) were isolated up to 33 months after transplantation. Two (clones 11 and 14) of 3 randomly selected retroviral integration site clones were detectable in both mononuclear cells (MNCs) and granulocytes (Figure 5B) at all time points analyzed between 2 and 33 months after transplantation. Two additional clones (clones 10 and 15) could repeatedly be detected by LAM-PCR in granulocyte fractions at 2 or more time points after transplantation.
The continuous presence of individual integration sites in myeloid cells of high purity is the first demonstration of sustained long-term hematopoiesis originating from individual stem cell clones, which we observed in 2 completely independent primate models. A detailed analysis of early posttransplantation hematopoiesis in the rhesus model further indicates that these clones did not significantly contribute to hematopoiesis in the first 6 to 8 weeks after transplantation. This confirms the hypothesis derived from human xenograft models that repopulation early after engraftment is phenotypically different from stable long-term repopulation.14 27-29
Pluripotency of stem cell clones with long-term activity
To investigate the lineage contributions of these clones, we performed PCR analysis, using the same tracing by clone-specific PCR primers, on granulocytes, T cells, and B cells highly (> 99%) purified by fluorescence-activated cell sorter (FACS). In both models, 2 of these 3 clonal integration sites that were analyzed in detail were detectable in both myeloid and lymphoid cells simultaneously. This points to a presence of repopulating stem cells or their progeny with lymphomyeloid pluripotency. In rhesus macaque RC501, limiting-dilution PCR demonstrated that highly purified myeloid and lymphoid populations contained similar copy numbers of individual integration sites over time (Figure 3B), suggesting that the activity of the originally marked stem cells remains pluripotent. The limiting-dilution analysis also ruled out contamination of granulocytes into purified T or B cells or vice versa, proving that the signals indicate true multilineage clonal contributions. The signal intensity of an individual clone in the different populations was similar, despite the less than 1% to 2% of cellular contamination possible during sorting or granulocyte purification. It is interesting to note that the signal is stronger in the myeloid population, which further excludes a retrovirus-marking artifact by transduced long-lived lymphocytes.
The quantitative contribution of traced individual stem cell clones
A brief calculation of sampling statistics indicates that all clones that could be traced at multiple time points must have contributed significant cell numbers to in vivo hematopoiesis. The amounts of DNA (100 ng) that were analyzed from each sample in our study represent the leukocytes (15 000) from about 3 μL of peripheral blood. The limit of detection of each PCR assay was established at approximately 4 to 10 copies of the integration site sequences per microliter of peripheral blood (data not shown). Considering that leukocytes circulated in approximately 1 L of body volume in these animals, each of the clones has consistently contributed at least 4 to 10×106 myeloid cells to the peripheral circulation, that is, approximately 0.7% to 2% of retrovirus-marked repopulating cells.
Clonal composition of in vivo hematopoiesis
Sequencing of less than a quarter of randomly selected and clearly visible insertion site amplification products from granulocytes resulted in the identification of 52 (rhesus model) and 16 (baboon model) integration sites. The number of visible vector insertion site amplification products in lymphocyte fractions was similar. Since retroviral insertion in these models is confined to one copy per cell on average,20 we conclude that marked myeloid/lymphoid hematopoiesis was repopulated by the progeny of at least 50 to 100 initially transduced progenitors per animal. Assuming that the proportion of marked leukocytes represents approximately 10% of the total competitively repopulated hematopoiesis in both models, our study suggests that up to 10-fold more unmarked long-term stem cell clones may have simultaneously contributed to in vivo circulating hematopoietic lineages in these primate models. Animal RC501 originally received a transplant of 40 million CD34+ cells, representing approximately 1% of the mobilized blood MNC population, and therefore we would estimate the frequency of long-term repopulating stem cells in rhesus mobilized peripheral blood to be about 3 or 4 per 10 million MNCs, in agreement with estimates obtained using a completely different approach in the cat, a large animal of similar size and, presumably, hematopoietic demand.30
Our study not only provides the first molecular evidence for long-term activity and pluripotency in adult primate hematopoietic stem cells but also has several implications for cell and gene therapy. First, the stem cell population responsible for the long-term production of marked blood was highly polyclonal. In contrast to earlier observations of oligoclonal fluctuation in small animal models,2 in the large animal models we studied, the contribution of the pluripotent long-term repopulating clones traced was remarkably stable. This is encouraging in terms of predicting a stable phenotype following successful genetic modification of human HSCs. Second, the lack of a significant contribution of these clones to hematopoiesis observed early after engraftment indicated that, as in mice,14,28,31,32 stem cells with long-term activity differ from transiently engrafted short-term repopulating cells. This finding complements the discovery of human short-term repopulating cells in xenotransplants of human bone marrow.27,29 Third, the stable long-term activity and pluripotency of individual clones also prove that ex vivo–induced cell cycling required for retrovirus integration33 does not, by itself, eliminate stem cell function. Our study demonstrates that stem cells with integrated virus vectors can be enumerated and traced in vivo, enabling better control of the long-term success of genetic modifications. The sustained contribution of these ex vivo–manipulated stem cells strongly suggests that retrovirus and lentivirus vectors are suitable tools for stem cell gene transfer aimed at the long-term correction of human genetic diseases.
We wish to thank Silke Klingenberg, Stephanie Sellers, Maasaki Takatoku, Julie Morris, and Connie Peterson for their technical assistance. We are grateful to Hyeoung Joon Kim for his contribution and helpful discussion. We also wish to thank Mike Gough and the staff of the University of Washington Regional Primate Research Center for assistance with the baboons, and Robert Donahue and the staff at the NHLBI 5 Research Court primate facility for assistance with rhesus macaques. We gratefully acknowledge the helpful discussion and support of Dr C. Peters and Dr Roland Mertelsmann.
Prepublished online as Blood First Edition Paper, June 21, 2002; DOI 10.1182/blood-2002-02-0407.
Supported by grants Ka 976/4-1 and SFB 364/7 awarded by the Deutsche Forschungsgemeinschaft; grant 01KV9527/7 awarded by the German Minister for Education and Research, Bundesministerium für Bildung und Forschung; and National Institutes of Health grants HL 54881, HL 53750, DK 47754, DK 56465, and RR 00166.
P.Z., G.H., and S.H. contributed equally to this article.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Cynthia E. Dunbar, Hematology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bldg 10/7C103, 9000 Rockville Pike, Bethesda, MD 20892; e-mail:dunbarc@nhlbi.nih.gov.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal