We hypothesized that the CXC chemokine receptor-4 (CXCR4)–stromal-derived factor-1 (SDF-1) axis may be involved in metastasis of CXCR4+ tumor cells into the bone marrow and lymph nodes, which secrete the α-chemokine SDF-1. To explore this hypothesis, we phenotyped by fluorescence-activated cell sorter analysis various human tumor cell lines for expression of CXCR4 and found that it was highly expressed on several rhabdomyosarcoma (RMS) cell lines. We also observed that cell lines derived from alveolar RMS, which is characterized by recurrent PAX3- andPAX7-FKHR gene fusions and is associated with a poor prognosis, expressed higher levels of CXCR4 than lines derived from embryonal RMS. Furthermore, transfer of a PAX3-FKHRgene into embryonal RMS cell activates CXCR4 expression. Because alveolar RMS frequently metastasizes to the bone marrow and lymph nodes, it seems that the CXCR4–SDF-1 axis could play an important role in this process. These findings prompted us to determine whether SDF-1 regulates the metastatic behavior of RMS cells. Accordingly, we found that, although SDF-1 did not affect proliferation or survival of these cell lines, it induced in several of them (1) phosphorylation of mitogen-activated protein kinase p42/44; (2) locomotion; (3) directional chemotaxis across membranes covered by laminin, fibronectin, or Matrigel; (4) adhesion to laminin, fibronectin, and endothelial cells; and (5) increased MMP-2 and diminished tissue inhibitors of metalloproteinases secretion. The small-molecule CXCR4-specific inhibitor, T140, effectively blocked the in vitro responses of RMS cells to SDF-1. On the basis of these observations we suggest that the CXCR4–SDF-1 axis may play an important role in tumor spread and metastasis of RMS cells to bone marrow and that molecular strategies aimed at inhibiting this axis could thus prove to be useful therapeutic measures.
Introduction
The α-chemokine stromal-derived factor-1 (SDF-1) secreted by bone marrow stromal cells chemoattracts normal CD34+ CXC chemokine receptor-4 (CXCR4)–positive early hematopoietic cells,1-3 and the CXCR4–SDF-1 axis is believed to play an important role in the homing of stem/progenitor cells into the hematopoietic niches of the bone marrow. This premise is in accord with the report that SDF-1 activates VLA-4 and LFA-1 integrins on the surface of CD34+ cells, allowing these cells to adhere to the bone marrow endothelium.4,5 Such adhesion is the first step in the migration of CD34+ cells across the endothelial barrier in the process of homing.6-9 We have reported that SDF-1 has no direct effect on the proliferation or survival of lymphohematopoietic cells but influences the interaction of these cells with the hematopoietic microenvironment by regulating their migration and adhesion as well as the secretion of vascular endothelial growth factor (VEGF)–1 and matrix metalloproteinases (MMPs).6-9 This evidence further supports the hypothesis of an important role for the CXCR4–SDF-1 axis in the trafficking of early hematopoietic cells.
Tumor cells expressing functional CXCR4 could use similar mechanisms for “homing” to the bone marrow to those used by CXCR4+hematopoietic cells. Involvement of the CXCR4–SDF-1 axis in the metastasis of breast cancer to the SDF-1–rich environment of the bone marrow and lymph nodes has been reported.10 To assess whether the CXCR4–SDF-1 axis plays a role in the metastasis of other tumor cells into bone marrow and lymph nodes, we phenotyped a number of human tumor cell lines for expression of CXCR4, using immunostaining and fluorescence-activated cell sorter (FACS) analysis. We found that it was not detectable in most cell lines derived from common human solid tumors (melanoma, breast and lung cancer), but was highly expressed on the surface of rhabdomyosarcoma (RMS) cells.
There are 2 major histologic subtypes of RMS, alveolar (ARMS) and embryonal (ERMS). Clinical evidence indicates that ARMS is more aggressive and associated with a significantly worse outcome than ERMS.11-20 Genetic characterization of RMS has identified markers that show excellent correlation with histologic subtype. Specifically, ARMS is characterized by the t(2;13)(q35;q14) translocation in 70% of cases14 or the variant t(1;13)(p36;q14) translocation in a smaller percentage of cases.17 These translocations disrupt the PAX3 or PAX7 genes on chromosome 2 or 1, respectively, and the FKHR gene on chromosome 13, and generate PAX3-FKHR or PAX7-FKHR fusion genes. These fusion genes encode fusion proteins, PAX3-FKHR and PAX7-FKHR, which function as novel transcription factors. The PAX3-FKHR and PAX7-FKHR fusion proteins demonstrate enhanced transcriptional activity compared with wild-type PAX3 and PAX7 and are postulated to play an important role both in survival and cell-cycle dysregulation of ARMS cells.
It is well known that RMS cells, particularly ARMS, may infiltrate the marrow and, because they can resemble hematologic blasts, may sometimes be misdiagnosed as acute leukemia cells.11-13 The “contamination” of bone marrow by these cells may compromise its use for autologous transplantation.21 In this study our hypothesis was that the metastasis of RMS cells into bone marrow is mediated by the CXCR4–SDF-1 axis.
We focused on the biologic responses of CXCR4+ ARMS and ERMS cell lines to stimulation by exogenous SDF-1, such as phosphorylation of signaling proteins, proliferation, survival, adhesion, expression of MMPs and their tissue inhibitors (TIMPs), chemotaxis, and chemoinvasion. Our findings implicate the CXCR4–SDF-1 axis in the metastatic behavior of RMS cells.
Materials and methods
Cell lines
We used 5 human breast cancer cell lines: HTB-26, T47D, BT-549, BT20, and MDA-175 (gift from Dr C. Clevenger, University of Pennsylvania, Philadelphia); 8 human melanoma cell lines: WM-39, WM-164, WM-1205, WM-451, WM-115, WM-278, HTB-140, and SBCL2 (gift from Dr M. Herlyn, Wistar Institute, Philadelphia, PA); 5 human lung cancer cell lines: HTB-177, HTB-183, CRL-2066, CRL-2062, and A549; and 2 human sarcoma cell lines: A673 (Ewing sarcoma) and A204 (undifferentiated sarcoma), all purchased from ATCC (Manassas, VA) .
We also investigated 7 RMS cell lines, including 5 ARMS lines (RH5, RH28, RH30, RHRKMP-4, and CW9019), and 2 ERMS lines (RD and SMS-CTR). RMS cells used for experiments were cultured in RPMI-1640 medium (Sigma, St Louis, MO), supplemented with 100 IU/mL penicillin, 10 μg/mL streptomycin, and 50 μg/mL neomycin (Gibco, Grand Island, NY) in the presence of 10% heat-inactivated fetal calf serum (Gibco). The ERMS cell line, RD, transfected with the PAX3-FKHR gene, was cultured in the presence of the selective agent geneticin (G-418) as described.14,16 19 The cells were plated at an initial cell density of 2.5 × 104 cells per Corning flask (Costar-Corning, Cambridge, MA). In cell cultures the medium was changed every 48 hours, and cells were cultured in a humidified atmosphere with 5% CO2 at 37°C.
FACS analysis
The expression of CXCR4 on various tumor cell lines was evaluated by fluorescence activated cell sorting (FACS) analysis as previously described.22 The CXCR4 antigen was detected with phycoerythrin (PE)–anti-CXCR4 monoclonal antibody (MoAb; R&D Systems, Minneapolis, MN), clone no. 12.G5. Briefly, the cells were stained in phosphate-buffered saline (PBS; Ca- and Mg-free) supplemented with 5% bovine calf serum (BCS; Hyclone, Logan, UT). After the final wash, cells were fixed in 1% paraformaldehyde prior to FACS analysis, performed using the FACscan (Becton Dickinson, San Jose, CA).
Evaluation of adhesion molecules
The expression of adhesion molecules on RMS cells was evaluated by FACS. Cells were stained with specific anti–PECAM-1, ICAM-1, VCAM-1, E-selectin, VLA-5, and VLA-4 antibodies detected with PE-conjugated secondary PE-goat antimouse MoAbs as described previously.23 The following antibodies were used for this study: 4G6 (immunoglobulin G2b [IgG2b], mouse antihuman PECAM-1) generously provided by Dr S. Albelda24; R6.5 (BIRR-1), a murine IgG2a MoAb directed against extracellular domain 2 of the ICAM-1 molecule, from Boehringer Ingelheim Pharmaceuticals (Ridgefield, CT25); 4B9, an IgG1 MoAb directed against human VCAM-1, from Dr R. Lobb, Biogen, Cambridge, MA26; and ES2 (IgG1-k mouse antihuman–E-selectin MoAb), provided by Dr R. McEver, University of Oklahoma.27 Antibodies against α6β1 integrin were purchased from Pharmingen (San Diego, CA).
Phosphorylation of intracellular pathway proteins
Western blots were done on extracts prepared from RMS cell lines (1 × 107 cells) that were kept in RPMI medium containing low levels of bovine serum albumin (BSA; 0.5%) to render the cells quiescent. The cells were divided and stimulated with optimal doses of SDF-1α or SDF-1β (500 ng/mL) for 1 minute to 2 hours at 37°C and then lysed (for 10 minutes) on ice in M-Per lysing buffer (Pierce, Rockford, IL) containing protease and phosphatase inhibitors (Sigma). Subsequently, the extracted proteins were separated on either a 12% or 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel, and the fractionated proteins were transferred to a nitrocellulose membrane (Schleicher & Schuell, Keene, NH) as previously described.23 Phosphorylation of the intracellular kinases, 44/42 mitogen-activated protein kinase (MAPK) (Thr 202/Tyr 204) and AKT, and STAT-1, -3, -5, and -6 proteins was detected by using commercial mouse phosphospecific MoAb (p44/42) or rabbit phosphospecific polyclonal antibodies for each of the remainder (all from New England Biolabs, Beverly, MA) with horseradish peroxidase (HRP)–conjugated goat antimouse IgG or goat antirabbit IgG as a secondary antibody (Santa Cruz Biotech, Santa Cruz, CA) as described.7 23 Equal loading in the lanes was evaluated by stripping the blots and reprobing with appropriate MoAbs: p42/44 anti-MAPK antibody clone no. 9102, anti-AKT antibody clone no. 9272, anti–STAT-3 no. 9132 (New England Biolabs), anti–STAT-1 no. sc-464 and anti–STAT-6 no. sc-1689 (Santa Cruz Biotech), and anti–STAT-5 no. 89 (Transduction Laboratories, Lexington, KY). The membranes were developed with an enhanced chemiluminescence (ECL) reagent (Amersham Life Sciences, Little Chalfont, United Kingdom), dried, and subsequently exposed to film (HyperFilm; Amersham Life Sciences).
Detection of SDF-1 by Western blot analysis
SDF-1 protein was detected by using specific rabbit polyclonal antibody (R&D Systems) that was detected with HRP-conjugated goat antimouse IgG or goat antirabbit IgG as a secondary antibody (Santa Cruz Biotech) as previously described.23
Isolation of mRNA and RT-PCR
The RMS cells were lysed in 200 μL RNAzol (Biotecx Labs, Houston, TX) plus 22 μL chloroform as described.28Briefly, mRNA (0.5 μg) was reverse-transcribed with 500 U Moloney murine leukemia virus reverse transcriptase and 50 pmol ODN primer complementary to the 3′ end of the reported chemokine sequences of SDF-1/CXCL12 (5′-CAC ATG TTG AAC CTC TTG TTT AAA AGC-3′). The resulting cDNA fragments were amplified by using 5 U Thermus aquaticuspolymerase and primers specific for the 5′ end of SDF-1/CXCL12 (5′-AAC GCC AAG GTC GTG GTC GTG CTG-3′). β-Actin mRNAs were amplified simultaneously by using specific primers as reported previously.28 29 The predicted size of the reverse transcriptase–polymerase chain reaction (RT-PCR) product for SDF-1/CXCL12 is 281 bp. Amplified products (10 μL) were electrophoresed on a 2% agarose gel and transferred to a nylon filter. The specificity of the amplified products was further confirmed by Southern blotting (data not shown).
RNase protection assay
For analysis of CXCR4 mRNA levels, total mRNA was isolated from cells as described previously.30 An aliquot of total cellular mRNA (10 μg) was hybridized with both [32P]UTP-labeled CXCR4 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) control antisense probes and digested with ribonucleases (RPAII Kit; Ambion Incorporated, Austin, TX). Following denaturing polyacrylamide gel electrophoresis, protected bands were quantified by phosphorimaging (Molecular Dynamics). After normalizing for the uridine content of each riboprobe, the ratio of CXCR4 mRNA to GAPDH mRNA was calculated for each sample as described.30
Cell proliferation and apoptosis
Cells were plated in culture flasks at an initial density of 104 cells/cm2 in the presence or absence of SDF-1β (300 ng/mL). The cell number was calculated at 12, 24, 36, 48, 60, and 72 hours after culture initiation. In some experiments the cultures were prolonged up to 7 days. At the indicated time points cells were harvested from the culture flasks by trypsinization, and the number of cells was determined with the use of a Bürker hemocytometer (Buffalo, NY). Apoptosis was evaluated by the Annexin-V binding assay and intracellular staining for activated caspase-3, as described.28
Calcium flux studies
Briefly, cells were incubated for 30 minutes at 30°C with 1 to 2 μM Fura-2/am (Molecular Probes, Eugene, OR). After incubation, the cells were washed once, resuspended in loading buffer without fetal bovine serum (FBS), stimulated with SDF-1β (500 ng/mL), and analyzed within 1 hour as described.28
Fluorescent staining of the actin cytoskeleton
For the visualization of the actin, cytoskeleton cells were cultured for 12 hours on glass coverslips in RPMI-1640 medium supplemented with 10% FBS in the presence (300 ng/mL SDF-1β) or absence of SDF-1β. Subsequently, the cells were fixed in 3.7% paraformaldehyde/Ca- and Mg-free PBS for 15 minutes, permeabilized by 0.1% Triton X-100 in PBS for 1 minute at RT, and stained with TRITC-phalloidin at a concentration of 500 ng/mL for 1 hour. The stained cells were examined by using confocal laser scanning microscopy. All measurements were performed with BioRad MRC 1024 (Hemel Hempstead, United Kingdom) based on an inverted Nikon Diaphot 300 microscope (Nikon, Canagawa, Japan). The confocal system was equipped with a 60 × PlanApo 1.4 NA oil immersion objective lens, an air-cooled 25 mW output Argon-Krypton laser, 3 fluorescence detection channels, and a 3-color transmitted nonconfocal light detector. Laser light (568 nm) attenuated to 1% of the maximum intensity was introduced into the sample. TRITC fluorescence was collected by using 568 nm excitation. Fluorescence light passed through a 488/568/645 dichromic filter (T1 BioRad filter block), a 560 dichromic long pass filter (T2A BioRad filter block), and a 580LP filter (Croma, VT) and was detected by a photomultiplier (Biorad PMT2). For each image 3 scans in slow acquisition mode or photon-counting mode were collected with the use of Kalman filtering.
Time-lapse monitoring of the locomotion of individual cells
The images of human RMS cells migrating on plastic at 37°C were evaluated with an inverted Hund Wetzlar microscope by using phase-contrast optics (Wetzlar, Germany). Analysis of cell migration was begun 2 hours after seeding, when the cells were already dispersed over the plastic. The locomotion images were recorded with a Hitachi CCD camera (sensitivity, 10−3 1 ×; Tokyo, Japan) as described.31-34 CW9019, RH30, SMS-CTR, or A204 cells were plated to Corning flasks at a density of 2 to 2.5 × 104 cells/cm2 and were mock-treated or prestimulated by SDF-1β (300 ng/mL) 30 minutes before recording. The cell trajectories were constructed from 40 subsequent cell centroid positions recorded for 200 minutes at 5-minute time intervals. The cell trajectories were presented in circular diagrams, and the length of the cell tracks was calculated in addition to the final displacement. Cell tracks (100) were recorded under the conditions described earlier for each cell line.
The parameters of cell movement were calculated for each cell by using procedures written in the Mathematica language as described.31-34 The following parameters were determined: (1) the total length of the cell trajectory (in micrometers); the trajectory was a sequence of n straight-line segments, each corresponding to cell centroid translocation within one time interval between 2 successive images; (2) the total length of cell displacement from the starting point to the final cell position (in micrometers); (3) the ratio of cell displacement length to cell trajectory length, here called the coefficient of movement efficiency (CME); (4) the average speed of cell movement defined as total length of cell trajectory/time of recording; and (5) the average velocity of cell displacement calculated from the final cell displacement in a given time.31-34
Transmembrane chemotaxis
Cells were seeded into 6-well plates in RPMI medium containing 10% FBS. After adhesion to the dish bottom, the medium was changed and cells were made quiescent for 48 hours with 0.5% BSA in RPMI. The directional movement of cells toward the gradient of SDF concentration across an 8-μm pore polycarbonate membrane was evaluated. The membrane was covered with 50 μL fibronectin (50 μg/mL) and laminin (20 μg/mL) for 2 hours at 37°C and overnight at +4°C. The solution was discarded before assay. Cells were detached with 0.5 mM EDTA (ethylenediaminetetraacetic acid), washed in RPMI medium, resuspended in RPMI medium with 0.5% BSA, and seeded into the upper chamber of a Transwell insert (Costar Transwell; Costar-Corning) at a density of 105 in 200 μL. The lower chamber was filled with SDF at 300 ng/mL concentration, and the 0.5% BSA RPMI medium was used as a negative control. To determine whether the migration was stimulated by the gradient of the chemoattractant, in some samples SDF was also added into the upper chamber to equalize the difference in concentration between chambers. After 48 hours, the insert was removed from the transwell, cells remaining in the upper chamber were scraped off with cotton wool, and the cells that had transmigrated were counted either on the lower side of the membrane or on the bottom of the transwell.
Adhesion of RMS cells to fibronectin and laminin
Cells were made quiescent for 48 hours with 0.5% BSA in RPMI before incubation with SDF-1β for 24 hours (300 ng/mL). The cells were metabolically labeled overnight with 35S-methionine; detached with 0.5 mM EDTA; washed twice and suspended in 50 mM Tris (tris(hydroxymethyl)aminomethane)–Cl buffer, pH 7.4, containing 150 mM NaCl, 0.5 mM CaCl2, 0.1% glucose, and 1% BSA; and added directly onto the protein-coated wells (5 × 104/well) for 30 minutes. The wells were coated with BSA (4 μg/mL), fibronectin (10 μg/mL), and laminin (20 μg/mL) for 2 hours at 37°C and overnight at +4°C and blocked with BSA for 2 hours before the experiment. Following incubation at 37°C, the plates were vigorously washed 3 times, and adherent cells were dissolved by using 2% SDS. The SDS solutions were then counted for 35S in a liquid scintillation counter. The results were normalized against 10% input for each cell line.
Adhesion to HUVECs
RMS cells were labeled before assay with the fluorescent dye calcein-am and were subsequently added (for 5 minutes) to the 96-well plates covered by human umbilical vein endothelial cells (HUVECs) that had been pretreated with SDF-1β (5 μg/mL). After the nonadherent cells had been discarded, the cells that adhered to the HUVECs were lysed, and fluorescence was measured by using a spectrofluorometer as described.35
MMP/TIMP expression
To evaluate MMP and TIMP activities, RMS cells were incubated for 24 hours in serum-free media in the absence (control) or presence of SDF-1β (300 ng/mL), and zymography and reverse zymography were carried out as previously described by us.36 To evaluate gene expression for MMPs and TIMPs in these cells, total RNA was extracted (as described previously).36,37 The conversion of mRNA to cDNA was carried out using avian myeloblastosis virus reverse transcriptase (AMVRT) (Seigaku America, Ijamsville, MD), and PCRs were carried out following the “primer dropping” method. Sequences for human MMP-2, MMP-9, TIMP-1, and TIMP-2 were obtained from Genbank (Los Alamos, NM) and were used to design primer pairs, as described by us previously.37
Chemoinvasion assay
The ability of various malignant cells to invade the reconstituted basement membrane Matrigel is regarded as an important measure of their metastatic potential. Two ARMS cell lines, RH30 and RH28, were evaluated in a chemoinvasion assay as originally described38 and modified by us.39 Briefly, cells that were preincubated either with the MMP inhibitoro-phenanthroline (0.5 mM, 30 minutes) or in control conditions (media) were loaded onto the upper compartments of Boyden chambers (105 cells/chamber; BD Biosciences, Bedford, MA) and incubated for 3 hours. Cells that invaded the Matrigel barrier toward media alone or toward an SDF-1β gradient (300 ng/mL) were counted on the undersides of filters after fixation and staining with crystal violet. A chemoinvasion index was calculated as the ratio of the number of cells invading the Matrigel toward an SDF-1 gradient to the number of cells invading toward media alone.
Blockade of CXCR4–SDF-1 axis using T140
Some of the adhesion and directional migration experiments were performed on cells preincubated for 30 minutes at 37°C in the presence of 1 μM T140-truncated polyphemusin analog (a gift from Dr N. Fuji, Kyoto University, Japan) or preincubated with anti-CXCR4 (10 mg/mL). In the chemotaxis experiments, cultured bone marrow stroma cells were pretreated with anti–SDF-1 (100 μg/mL; R&D Systems) as described.28
Statistical analysis
All results are presented as mean ± SEM. Statistical analysis of the data was performed using the nonparametric Mann-Whitney test, with P < .05 considered significant.
Results
CXCR4 is highly expressed in ARMS but not ERMS cell lines
First, we phenotyped several human breast and lung cancer as well as melanoma cell lines for expression of CXCR4 and found it to be expressed at a low level on only 1 (HTB 22) of 6 breast cancer cell lines (Table 1). In this cell line less than 15% of cells stained positive for CXCR4. All 5 lung cancer cell lines investigated in this study as well as 8 melanoma cell lines were negative for CXCR4 expression as determined by FACS. In contrast, CXCR4 was expressed on 7 of 7 human RMS cell lines tested (Table 1; Figure 1).
Expression of CXCR4 as determined by FACS
| Cell lines . | Positive for CXCR4 . | Negative for CXCR4 . |
|---|---|---|
| Breast cancer | HTB 22 | HTB-26, T47D, BT-549, BT20, MDA-175 |
| Lung cancer | HTB-177, HTB-183, CRL 2066, CRL 2062, A549 | |
| Melanomas | WM-39, WM-164, WM-1205, WM-451, WM-115, WM-278, HTB-140, SBCL2 | |
| Rhabdomyosarcomas | RH28, RH30, CW9019, RHRKMP-4, RH5, SMS-CTR, RD | |
| Other sarcomas | A204, A673 |
| Cell lines . | Positive for CXCR4 . | Negative for CXCR4 . |
|---|---|---|
| Breast cancer | HTB 22 | HTB-26, T47D, BT-549, BT20, MDA-175 |
| Lung cancer | HTB-177, HTB-183, CRL 2066, CRL 2062, A549 | |
| Melanomas | WM-39, WM-164, WM-1205, WM-451, WM-115, WM-278, HTB-140, SBCL2 | |
| Rhabdomyosarcomas | RH28, RH30, CW9019, RHRKMP-4, RH5, SMS-CTR, RD | |
| Other sarcomas | A204, A673 |
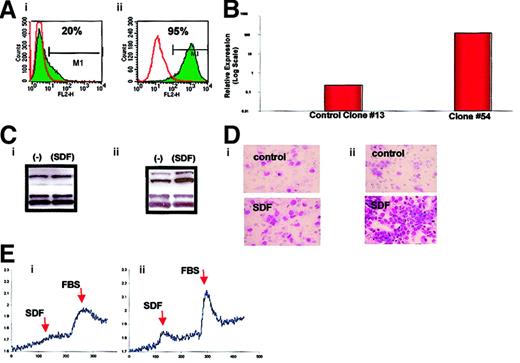
Expression of CXCR4 on human ARMS and ERMS cell lines.
(A) ARMS cell lines: RhRKMP-4 (i), RH28 (ii), CW9019 (iii), RH5 (iv), and RH30 (v). (B) ERMS cell lines: SMS-CTR (i) and RD (ii). The experiment was repeated 3 times with similar results. A representative study is shown.
Expression of CXCR4 on human ARMS and ERMS cell lines.
(A) ARMS cell lines: RhRKMP-4 (i), RH28 (ii), CW9019 (iii), RH5 (iv), and RH30 (v). (B) ERMS cell lines: SMS-CTR (i) and RD (ii). The experiment was repeated 3 times with similar results. A representative study is shown.
Because the RMS cell lines we investigated included both ARMS and ERMS cell lines, we attempted to correlate the expression of CXCR4 on these cell lines with their histologic phenotype (ARMS versus ERMS). We observed that all 5 ARMS (RH5, RH28, RH30, RHRKMP-4, and CW9019) cell lines stained highly positive for CXCR4 (> 90% of cells). CXCR4 was expressed at lower levels in about 20% of SMS-CTR and RD ERMS cells, and 2 other non-RMS sarcoma lines, A673 and A204, were negative for its expression as determined by FACS (Figure 1).
CXCR4 expression increases after transfection of the ERMS cell line with PAX3-FKHR
All ARMS cell lines used in our studies displayed translocations typical of ARMS: t(2;13) (RH5, RH28, RH30, and RHRKMP-4) and t(1;13) (CW9019), known to be associated with the fusion protein products PAX3-FKHR and PAX7-FKHR, respectively.11-20
Thus, having found a strong correlation between the ARMS phenotype and the expression of CXCR4, we sought to determine whether thePAX3-FKHR fusion gene regulates the expression of CXCR4. To address this issue, the ERMS cell line RD, which constitutively expresses a low level of CXCR4, was stably transfected with an expression vector containing the PAX3-FKHR fusion gene cDNA. We found that RD cells transfected with this fusion gene (Figure2) expressed substantially more CXCR4, as determined by FACS, with expression increasing from 20% to 95%, and as determined by RNase protection, with CXCR4 mRNA expression increasing by 3 orders of magnitude. In addition, in agreement with the studies shown below for ARMS and ERMS cell lines, RD2/PAX3-FKHR cells demonstrated increased response to SDF-1 by (1) MAPK p42/44 phosphorylation, (2) calcium flux, and (3) directional chemotaxis (Figure 2).
Analysis of wild-type RD cells (left panels) and RD cells transfected with the
PAX3-FKHR fusion gene (right panels). (A) Expression of CXCR4 by FACS. (B) Expression of CXCR4 mRNA by RNase protection. (C) Phosphorylation of MAPK p42/44 by SDF-1β. (D) Directional chemotaxis to SDF-1β. Original magnification, × 400. (E) Calcium flux in response to SDF-1β or FBS. The experiments were repeated 3 times with similar results.
Analysis of wild-type RD cells (left panels) and RD cells transfected with the
PAX3-FKHR fusion gene (right panels). (A) Expression of CXCR4 by FACS. (B) Expression of CXCR4 mRNA by RNase protection. (C) Phosphorylation of MAPK p42/44 by SDF-1β. (D) Directional chemotaxis to SDF-1β. Original magnification, × 400. (E) Calcium flux in response to SDF-1β or FBS. The experiments were repeated 3 times with similar results.
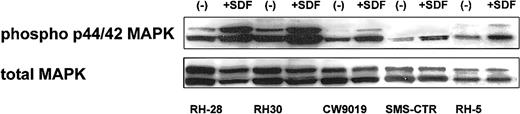
SDF-1 induces phosphorylation of MAPK p42/44
Next, we turned our attention to the putative role of the CXCR4–SDF-1 axis in regulating the biology of RMS cells. We determined whether stimulation of CXCR4 by SDF-1 induces phosphorylation of MAPK p42/44 and serine-threonine kinase AKT, which had been reported by others and by us to be involved in signaling from activated CXCR4.7,23,40 41 We found that 4 ARMS cell lines (RH28, RH30, CW9019, RH5) and one ERMS cell line (SMS-CTR) responded to SDF-1 by phosphorylation of MAPK p42/44, and an example of this is shown in Figure 3. In contrast, SDF-1 did not stimulate phosphorylation of either serine threonine kinase AKT or STAT-1 to -6 proteins (data not shown).
Phosphorylation of MAPK p42/44 in human RMS stimulated by SDF-1β (500 ng/mL for 5 minutes).
The experiment was repeated twice with similar results. A representative study is shown.
Phosphorylation of MAPK p42/44 in human RMS stimulated by SDF-1β (500 ng/mL for 5 minutes).
The experiment was repeated twice with similar results. A representative study is shown.
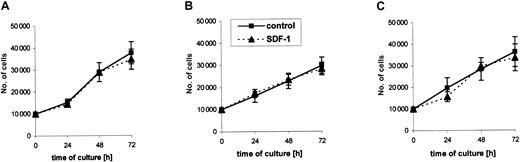
SDF-1 does not influence proliferation of RMS cell lines
Further, we selected 2 ARMS cell lines (RH30, CW9019) and one ERMS cell line (SMS-CTR) that responded to SDF-1 stimulation by phosphorylation of MAPK p42/44 to determine whether their proliferation is affected by SDF-1. We stimulated them with this chemokine or not (control) under serum-free conditions or in media supplemented with 2% BCS. We found that during 72 hours the RMS cell lines proliferated intensively in both types of media (Figure4). The kinetics of their proliferation were similar and were not affected by the presence of SDF-1 in the culture, even if the cells were cultured up to 7 days (data not shown).
Kinetics of growth of RMS cell lines in the absence (control) or presence of SDF-1β (300 ng/mL).
(A) CW9019, (B) RH30, and (C) SMS-CTR. Data from 4 independent experiments are demonstrated.
Kinetics of growth of RMS cell lines in the absence (control) or presence of SDF-1β (300 ng/mL).
(A) CW9019, (B) RH30, and (C) SMS-CTR. Data from 4 independent experiments are demonstrated.
Because the biology of various tumors may be regulated by autocrine/paracrine axes, we asked whether RMS cells express the CXCR4 ligand, the α-chemokine SDF-1. We investigated SDF-1 expression both by RT-PCR and Western blotting. We observed that 3 of 5 cell lines investigated in this study expressed mRNA for SDF-1 (CW9019, RH28, SMS-CTR). The presence of SDF-1 at the protein level was subsequently confirmed by Western blotting for CW9019 cells (data not shown). Blocking the putative CXCR4–DF-1 autocrine regulatory axis by adding anti–SDF-1 blocking MoAb did not affect, however, the proliferation kinetics of CW9019 cells (data not shown).
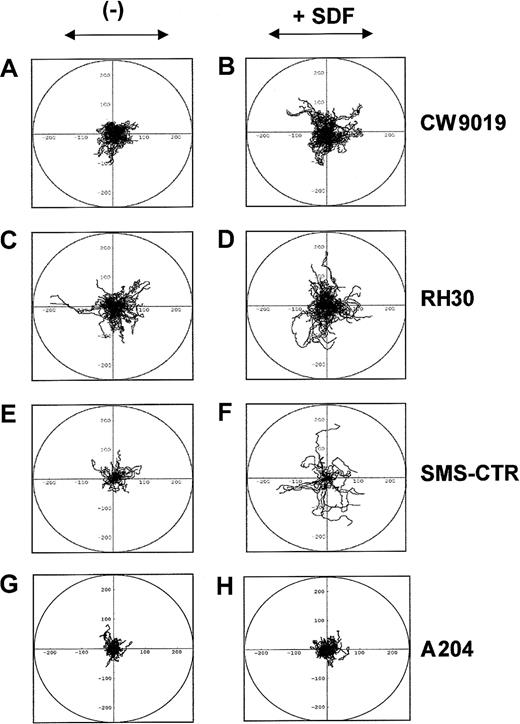
SDF-1 induces locomotion of individual RMS cells
Next, we studied whether SDF-1 influences the locomotion on plastic dishes of 2 selected human ARMS cell lines (CW9019 and RH30), one ERMS (SMS-CTR), and, as a control, a non-RMS sarcoma cell line A204 (not expressing CXCR4), using time-lapse monitoring of movement of individual cells. Figure 5 shows the trajectories of RMS cell migration in the absence (left panel) or presence (right panel) of SDF-1β in the culture medium. Analysis of these trajectories and mean values and standard errors for the parameters of cell locomotion are summarized for CW9019, RH30, SMS-CTR, and A204 cells in Table 2.
SDF-1β induces locomotion of RMS cells.
Trajectories of 100 CW9019, 50 SMS-CTR, 100 RH30, and 100 A204 cells locomoting in RPMI-1640 (control conditions; A,C,E,G, respectively) and in the presence of SDF-1β (300 ng/mL; B,D,F,H, respectively) displayed in circular diagrams drawn with the initial point of each trajectory placed at the origin of the plot. The images were recorded for 200 minutes, and the positions of cell centroids at 5-minute time intervals were determined.
SDF-1β induces locomotion of RMS cells.
Trajectories of 100 CW9019, 50 SMS-CTR, 100 RH30, and 100 A204 cells locomoting in RPMI-1640 (control conditions; A,C,E,G, respectively) and in the presence of SDF-1β (300 ng/mL; B,D,F,H, respectively) displayed in circular diagrams drawn with the initial point of each trajectory placed at the origin of the plot. The images were recorded for 200 minutes, and the positions of cell centroids at 5-minute time intervals were determined.
Parameters characterizing the movement of CW9019, RH30 ARMS, and SMS-CTR ERMS cells and A204 sarcoma cells in control medium (—) and in the presence of SDF-1β
| Cell line . | TLcT . | AScM . | TLcD . | AvcD . | CME . |
|---|---|---|---|---|---|
| CW9019 | |||||
| (—) | 130.21 ± 2.33 | 0.65 ± 0.01 | 39.58 ± 2.44 | 0.20 ± 0.01 | 0.31 ± 0.02 |
| SDF-1 | 175.46 ± 4.51* | 0.88 ± 0.02* | 55.26 ± 3.44* | 0.28 ± 0.02* | 0.32 ± 0.02 |
| RH30 | |||||
| (—) | 167.00 ± 4.27 | 0.83 ± 0.02 | 46.75 ± 3.91 | 0.23 ± 0.02 | 0.28 ± 0.02 |
| SDF-1 | 184.50 ± 5.73* | 0.92 ± 0.03* | 64.72 ± 3.87 | 0.32 ± 0.02 | 0.35 ± 0.02 |
| SMS-CTR | |||||
| (—) | 134.80 ± 7.46 | 0.67 ± 0.04 | 45.98 ± 3.12 | 0.23 ± 0.02 | 0.37 ± 0.02 |
| SDF-1 | 184.65 ± 6.73* | 0.92 ± 0.03* | 94.08 ± 7.43* | 0.47 ± 0.04* | 0.50 ± 0.03* |
| A204 | |||||
| (—) | 95.45 ± 2.35 | 0.48 ± 0.01 | 16.60 ± 1.42 | 0.08 ± 0.01 | 0.17 ± 0.01 |
| SDF-1 | 110.99 ± 3.05 | 0.55 ± 0.02 | 19.28 ± 1.49 | 0.09 ± 0.01 | 0.17 ± 0.01 |
| Cell line . | TLcT . | AScM . | TLcD . | AvcD . | CME . |
|---|---|---|---|---|---|
| CW9019 | |||||
| (—) | 130.21 ± 2.33 | 0.65 ± 0.01 | 39.58 ± 2.44 | 0.20 ± 0.01 | 0.31 ± 0.02 |
| SDF-1 | 175.46 ± 4.51* | 0.88 ± 0.02* | 55.26 ± 3.44* | 0.28 ± 0.02* | 0.32 ± 0.02 |
| RH30 | |||||
| (—) | 167.00 ± 4.27 | 0.83 ± 0.02 | 46.75 ± 3.91 | 0.23 ± 0.02 | 0.28 ± 0.02 |
| SDF-1 | 184.50 ± 5.73* | 0.92 ± 0.03* | 64.72 ± 3.87 | 0.32 ± 0.02 | 0.35 ± 0.02 |
| SMS-CTR | |||||
| (—) | 134.80 ± 7.46 | 0.67 ± 0.04 | 45.98 ± 3.12 | 0.23 ± 0.02 | 0.37 ± 0.02 |
| SDF-1 | 184.65 ± 6.73* | 0.92 ± 0.03* | 94.08 ± 7.43* | 0.47 ± 0.04* | 0.50 ± 0.03* |
| A204 | |||||
| (—) | 95.45 ± 2.35 | 0.48 ± 0.01 | 16.60 ± 1.42 | 0.08 ± 0.01 | 0.17 ± 0.01 |
| SDF-1 | 110.99 ± 3.05 | 0.55 ± 0.02 | 19.28 ± 1.49 | 0.09 ± 0.01 | 0.17 ± 0.01 |
Values are given as the mean ± SEM.
Statistically significant at P < .05.
TLcT indicates total length of cell trajectory (μm); AScM, average speed of cell movement (μm/min)—the average speed of cell; TLcD, total length of cell displacement (μm); AvcD, average velocity of cell displacement (μm/min)—the average velocity of cell displacement is defined as length of final cell displacement/time of recording; CME, coefficient of movement efficiency—the ratio of cell displacement to cell trajectory length. CME would equal 1 for cells moving persistently along one straight line in one direction and 0 for random movement.
Analysis of the time-lapse recording showed that the RMS cells migrated extensively in the presence of SDF-1, and SDF-1 increased the final cell displacement and the average velocity of cell displacement (Figure 5). The trajectories of CW9019 cells were significantly changed in medium containing SDF-1 (Figure 5B), and the distance of the cell displacement increased almost 1.5 times (Table 2). The trajectories of RH30 cells migrating in the control medium and in the presence of SDF-1β are shown in Figure 5C,D, respectively. SDF-1β increased the final cell displacement almost 1.4 times compared with the control condition (Table 2). The trajectories of SMS-CTR cells were also changed in the presence of SDF-1 (Figure 5F). The final cell displacement increased by almost double, in keeping with other parameters of cell locomotion (Table 2). In contrast the A204 sarcoma cell line did not show locomotion in response to SDF-1 (Figure5H and Table 2). It is worth nothing that, because the migrating RMS cells made tortuous tracks, their final displacement may be much smaller than the total length of their trajectories (Table 2).
SDF-1 alters the actin cytoskeleton
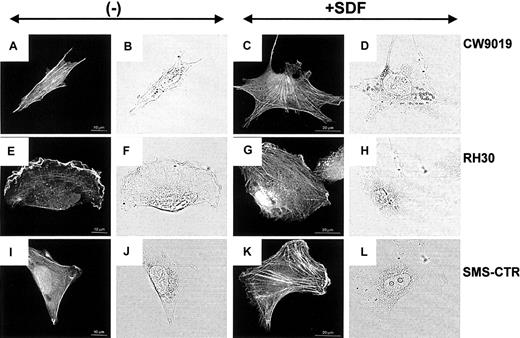
Examination of the actin cytoskeleton organization by confocal microscopy revealed striking differences between the chemokine-treated and the untreated cells. RMS cells grown in control medium displayed well-developed bundles of F-actin arranged in parallel to the long axis of the cell (Figure6A,E,I). Incubation of all RMS cells in the SDF-1–containing medium for 12 hours induced a change in the organization of actin filaments and significantly increased both the number and thickness of F-actin bundles (Figure 6C,G,K).
Stress fiber formation in human RMS cells.
Upper panel, CW 9019 cells; middle panel, RH30 cells; and lower panel, SMS-CTR cells. Left half of the figure shows cells not exposed to SDF-1β; right half of the figure shows cells exposed to SDF-1β (300 ng/mL). Dark images are immunofluorescence pictures; light images are normal light pictures. Representative cells were selected.
Stress fiber formation in human RMS cells.
Upper panel, CW 9019 cells; middle panel, RH30 cells; and lower panel, SMS-CTR cells. Left half of the figure shows cells not exposed to SDF-1β; right half of the figure shows cells exposed to SDF-1β (300 ng/mL). Dark images are immunofluorescence pictures; light images are normal light pictures. Representative cells were selected.
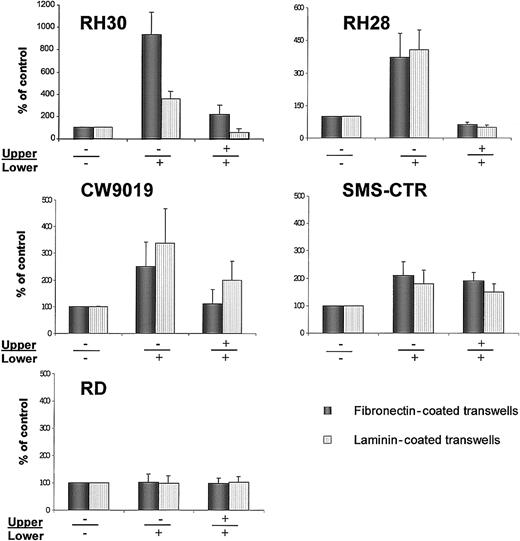
SDF-1 increases migration through fibronectin- or laminin-covered transwell membranes
Next, we investigated directed migration of RMS cells through transmembranes covered with fibronectin or laminin (Figure7). We selected for this study all the cell lines that had responded to SDF-1 stimulation by phosphorylation of MAPK p42/44 (Figure 3). The CXCR4-null A204 cell line was used as a negative control.
Chemotaxis of RMS cells across transwell membranes covered with fibronectin (black bars) or laminin (dashed bars).
(A) RH30, (B) RH28, (C) CW9019, (D) SMS-CTR, and (E) RD cells. Left part of each panel shows chemotaxis to control medium (no SDF-1β in upper (−) and lower (−) chambers; middle part of each panel, chemotaxis to SDF-1β present (+) in lower chamber; right part of each panel, chemotaxis in the presence of SDF-1 in both upper (+) and lower (+) chambers. Data from 5 separate experiments are pooled together.
Chemotaxis of RMS cells across transwell membranes covered with fibronectin (black bars) or laminin (dashed bars).
(A) RH30, (B) RH28, (C) CW9019, (D) SMS-CTR, and (E) RD cells. Left part of each panel shows chemotaxis to control medium (no SDF-1β in upper (−) and lower (−) chambers; middle part of each panel, chemotaxis to SDF-1β present (+) in lower chamber; right part of each panel, chemotaxis in the presence of SDF-1 in both upper (+) and lower (+) chambers. Data from 5 separate experiments are pooled together.
We observed that SDF-1 statistically increased the chemotactic activity of the ARMS cell lines RH30, RH28, and CW9019. Interestingly, RH30, which showed the highest spontaneous locomotion on plastic dishes in medium without SDF-1 (Figure 7) and Table 2), owed the strongest chemotaxis to SDF-1, especially through fibronectin-covered membranes. In contrast, the ERMS cell line SMS-CTR, in which locomotion activity on plastic dishes was strongly induced after the addition of SDF-1, showed relatively weak chemotaxis to SDF-1 across transwell membranes covered by fibronectin or laminin. In addition, the RD (ERMS) cell line did not show chemotaxis to SDF-1 (Figure 7E).
SDF-1 increases adhesion to fibronectin and laminin
We subsequently decided to investigate whether SDF-1 regulates expression/activation of integrins on human RMS. By using FACS analysis, we did not find any change in the level of expression of VLA-4, VLA-5, PECAM-1, or ICAM-1 on RMS cells after incubation with SDF-1 for 24 hours (data not shown). However, we observed that SDF-1 affected the adhesion of RH30, RH28, and CW9019 to fibronectin and laminin (Figure 8A) and that this adhesion correlated with their migration through fibronectin- or laminin-covered transwell membranes (Figure 7).
Adhesion of human RSM cells and chemoinvasion of RH28 and RH30 cells.
(A) Adhesion of human RMS cells to fibronectin or laminin; (i) adhesion to laminin, (ii) adhesion to fibronectin, and (iii) adhesion to BSA. Cells not stimulated with SDF-1 are shown as 100% (0), cells stimulated with SDF-1β are RH30 (1), RH28 (2), CW9019 (3), SMS-CTR (4), and RD (5). Data from 4 separate experiments are pooled together. *Indicates P < .01. (B) Chemoinvasion of RH28 and RH30 cells across the Matrigel barrier. Cells invading the Matrigel were counted on the undersides of the filters as described in “Materials and methods.” Results are expressed as a chemoinvasion index. Left part of the panel shows the number of cells crossing the Matrigel in the absence of an SDF-1 gradient (−/−), and middle panel the number crossing in the presence of an SDF-1 gradient (−/+). Right part of panel shows reduced chemoinvasion toward an SDF-1 gradient after preincubation of cells with the MMP inhibitoro-phenanthroline (phen/+). Data from 3 separate experiments are pooled together and means ± SD are shown.
Adhesion of human RSM cells and chemoinvasion of RH28 and RH30 cells.
(A) Adhesion of human RMS cells to fibronectin or laminin; (i) adhesion to laminin, (ii) adhesion to fibronectin, and (iii) adhesion to BSA. Cells not stimulated with SDF-1 are shown as 100% (0), cells stimulated with SDF-1β are RH30 (1), RH28 (2), CW9019 (3), SMS-CTR (4), and RD (5). Data from 4 separate experiments are pooled together. *Indicates P < .01. (B) Chemoinvasion of RH28 and RH30 cells across the Matrigel barrier. Cells invading the Matrigel were counted on the undersides of the filters as described in “Materials and methods.” Results are expressed as a chemoinvasion index. Left part of the panel shows the number of cells crossing the Matrigel in the absence of an SDF-1 gradient (−/−), and middle panel the number crossing in the presence of an SDF-1 gradient (−/+). Right part of panel shows reduced chemoinvasion toward an SDF-1 gradient after preincubation of cells with the MMP inhibitoro-phenanthroline (phen/+). Data from 3 separate experiments are pooled together and means ± SD are shown.
SDF-1 stimulates MMP-2 secretion in selected ARMS cell lines
Extensive experimental data show that MMPs actively contribute to cancer progression and metastasis and that relatively benign cells acquire malignant properties when MMP activity is increased or TIMP activity is diminished.42 Also, it was previously demonstrated by us that SDF-1 stimulates MMP-9 and MMP-2 in normal hematopoietic CD34+ cells, and we postulated that, through this action, SDF-1 modulates homing of hematopoietic progenitors.6-9 In this study we evaluated whether RMS cells express MMPs and whether SDF-1 stimulates their secretion and/or has any effect on TIMP production. We observed MMP-2 as well as TIMP-1 and TIMP-2 transcripts in all the RMS cell lines tested (Table3). After SDF-1 stimulation pro–MMP-2 activity (as measured by zymography) increased in RH5 and RH28 but not in the other cell lines tested, and pro–MMP-9 activity was not affected (Table 3). However, by using reverse zymography we found that SDF-1 stimulation diminished TIMP-1 and TIMP-2 protein secretion in all RMS lines, with the exception of SMS-CTR (Table 3).
MMP/TIMP gene and protein expression in RMS cell lines without (control, c) and after (SDF) SDF-1 stimulation
| Cell line . | MMP-9 . | MMP-2 . | TIMP-1 . | TIMP-2 . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene . | Protein . | Gene . | Protein . | Protein . | Protein . | |||||||
| c . | SDF . | c . | SDF . | c . | SDF . | c . | SDF . | c . | SDF . | c . | SDF . | |
| RH5 | − | − | − | − | + | ++ | + | ++ | + | < | + | < |
| RH28 | − | ND | − | − | + | + | + | ++ | + | < | + | < |
| RH30 | − | − | − | − | + | + | + | + | + | + | + | < |
| RHRKMP4 | + | + | + | + | + | + | + | + | + | < | + | < |
| CW9019 | + | + | + | + | + | + | + | + | + | < | + | < |
| RD | + | + | + | + | + | + | + | + | + | < | + | < |
| SMS-CTR | + | + | + | + | + | + | + | + | + | ++ | + | + |
| Cell line . | MMP-9 . | MMP-2 . | TIMP-1 . | TIMP-2 . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene . | Protein . | Gene . | Protein . | Protein . | Protein . | |||||||
| c . | SDF . | c . | SDF . | c . | SDF . | c . | SDF . | c . | SDF . | c . | SDF . | |
| RH5 | − | − | − | − | + | ++ | + | ++ | + | < | + | < |
| RH28 | − | ND | − | − | + | + | + | ++ | + | < | + | < |
| RH30 | − | − | − | − | + | + | + | + | + | + | + | < |
| RHRKMP4 | + | + | + | + | + | + | + | + | + | < | + | < |
| CW9019 | + | + | + | + | + | + | + | + | + | < | + | < |
| RD | + | + | + | + | + | + | + | + | + | < | + | < |
| SMS-CTR | + | + | + | + | + | + | + | + | + | ++ | + | + |
− indicates lack of transcripts/protein; +, presence of transcripts/protein; ++, 1.5-fold increase and greater; <, 1.5-fold decrease and greater; ND, not done.
MMP inhibitor diminishes SDF-1–directed chemoinvasion
Because tumor cell invasion is one of the characteristics of highly metastatic cells, we evaluated this feature by using a chemoinvasion Matrigel assay. We found that the invasive capability of RH30 and RH28 cell lines increases 2- to 3.5-fold in the presence of an SDF-1 gradient (Figure 8B). Furthermore, we present evidence that a synthetic inhibitor of MMPs, o-phenanthroline, inhibits invasion of these cell lines by approximately 50%, further suggesting that MMPs play a role in the aggressive behavior of ARMS cells (Figure 8B).
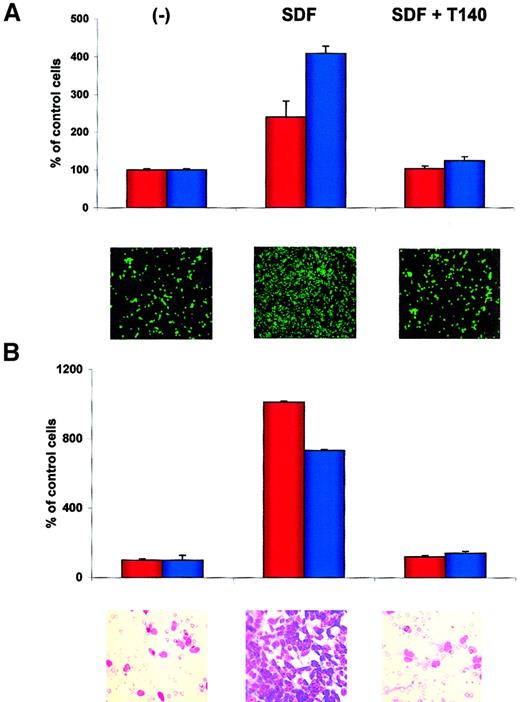
T140 inhibits adhesion of RMS cells to SDF-1–pretreated HUVECs and directional chemotaxis to bone marrow stroma
Finally, we used T140, which is a recently described specific blocking agent for CXCR4,43-46 in an experiment to see whether this molecule affects the metastatic behavior of RMS cells. In one set of experiments we found that T140 inhibited SDF-1–directed adhesion of RH30 and CW 9019 cells to HUVECs (Figure9A) and chemotaxis of RH30 and RH28 cells toward established cultures of bone marrow stroma fibroblasts (Figure9B). Similarly, the chemotaxis of RMS cells toward bone marrow stromal cultures was inhibited significantly by preincubating RMS cells with anti-CXCR4 or bone marrow stroma cultures with anti–SDF-1 (data not shown), suggesting that SDF-1 is a major stromal-derived factor that attracts RMS cells.
T140 inhibits the SDF-1–directed metastatic behavior of RMS cells.
(A) Effect of T140 on adhesion of RH30 (red bars) and CW9019 (blue bars) cells to HUVECs. Upper panel, relative number of the cells; lower panel, calcein-am–labeled RH30 cells present on HUVECs, as detected by immunofluorescence microscopy. (B) Effect of T140 on chemotaxis of RH30 (red bars) and RH28 (blue bars) to the cultured bone marrow stromal cells. Upper panel, relative number of cells; lower panel, RH30 cells present on the lower side of pore membrane stained by hematoxylin-eosin. The experiments were repeated 4 times with similar results. Original magnifications: panel A, × 100; panel B, × 400.
T140 inhibits the SDF-1–directed metastatic behavior of RMS cells.
(A) Effect of T140 on adhesion of RH30 (red bars) and CW9019 (blue bars) cells to HUVECs. Upper panel, relative number of the cells; lower panel, calcein-am–labeled RH30 cells present on HUVECs, as detected by immunofluorescence microscopy. (B) Effect of T140 on chemotaxis of RH30 (red bars) and RH28 (blue bars) to the cultured bone marrow stromal cells. Upper panel, relative number of cells; lower panel, RH30 cells present on the lower side of pore membrane stained by hematoxylin-eosin. The experiments were repeated 4 times with similar results. Original magnifications: panel A, × 100; panel B, × 400.
Discussion
RMS is the most common soft-tissue sarcoma of adolescence and childhood and accounts for 5% of all malignant tumors in patients younger than 15 years.11-20 Most of these tumors originate in the head and neck region, the urogenital tract, and the extremities. The 2 major subtypes of RMS (ARMS and ERMS) share a common myogenic differentiation but are not histogenetically related.12,15Accumulated clinical evidence suggests that ARMS is more aggressive than ERMS. Almost all the reported cases associated with marrow involvement, including those presenting as possible acute leukemia, have been of the ARMS type.13
Recently, the involvement of the CXCR4–SDF-1 axis in metastatic cancer has been postulated.47 In this study we did not, however, observe the CXCR4 protein in most lung and breast cancer and melanoma cell lines, but we found it to be highly expressed on the surface of 7 of 7 human RMS cell lines. Hence, we hypothesized that the CXCR4–SDF-1 axis could potentially have an important effect on the metastasis of RMS cells into bone marrow. Bone marrow involvement at diagnosis of RMS in children and adolescents is a poor prognostic sign and represents a continuing challenge to current treatment modalities.21Moreover, patients with RMS frequently undergo treatment with high-dose chemotherapy combined with autologous bone marrow/peripheral blood stem cell transplantation, and contamination of the transplanted grafts with tumor cells can affect patients' survival rates.21
In this study we found that CXCR4 was expressed by both ARMS and ERMS cells, but its expression was much higher (> 90%) on the former. Thus, CXCR4 expression correlated with the more aggressive ARMS subtype that is associated with poor clinical outcome and a propensity for bone marrow metastasis. Moreover, after stable transfection of the ERMS cell line RD with a PAX3-FKHR gene expression construct, these modified RD cells expressed significantly more CXCR4 mRNA and protein than control transfectants. Examination of the CXCR4 promoter sequence reveals several potential PAX3 binding sites; thus, we hypothesize that the CXCR4 gene may be a direct transcriptional target of wild-type PAX3 (which is expressed by embryonal RMS cells) and the alveolar RMS-specific PAX3-FKHR fusion protein. Furthermore, PAX3-FKHR+ RD cells responded to SDF-1 by phosphorylation of MAPK p42/44, calcium flux, and directional chemotaxis. The regulation of CXCR4 expression in RMS cells may be even more complex. For example, one report indicates that CXCR4 expression in T lymphocytes is regulated by the protooncogene c-myc.48 Because RMS cells often display aberrant expression of N-myc and c-myc, and as N-myc and c-myc expression has been reported to correlate with the metastatic potential of ARMS cells,49 the role of myc protooncogenes in coregulating, together with PAX3 and PAX7, the expression of CXCR4 in human RMS cells needs further investigation.
In our studies we focused on those RMS cell lines that responded to stimulation by SDF-1 by phosphorylation of MAPK p42/44. However, despite the fact that MAPK p42/44 has been shown to be involved in regulating cell proliferation and survival,50 we did not find any effect of SDF-1 on proliferation of RMS cell lines, although SDF-1 stimulated phosphorylation of MAPK p42/44 in these cells.
Although we did not observe any effect of SDF-1 on proliferation or survival of RMS cells, we found that SDF-1 stimulated processes related to cell metastatic/invasive behavior, which is an integrated sequence of events, including induction of cell polarity, migration, matrix degradation, and adhesion. Accordingly, we demonstrated that SDF-1 may induce motility of RMS cells, their polarity (appearance of the leading edge), and cytoskeletal rearrangements (formation of stress fibers) as well as enhance production of certain metalloproteinases. We believe that together all these processes may contribute to the egress of RMS cells from the tumor. In further support of this idea, we present evidence that all RMS cell lines tested express MMP-2 and that SDF-1 stimulates its secretion in some of them. We also observed in most of the RMS cell lines that SDF-1 down-regulated the secretion of the natural MMP tissue inhibitors TIMP-1 and TIMP-2. Thus, simultaneous up-regulation of MMP-2 and/or down-regulation of TIMP-1 and TIMP-2 further support the role of the CXCR4–SDF-1 axis in the metastatic behavior of RMS. This concept is further supported by our findings, which show that the synthetic MMP inhibitor o-phenanthroline significantly reduced the chemoinvasion of these cells in Matrigel invasion assays. The ability of various malignant cells to penetrate Matrigel (which consists of collagen type IV, laminin, and heparin sulfate proteoglycans in a structure representative of most basement membranes, including those of blood and lymphatic vessels) is an indicator of their metastatic potential in vivo.37,38,51Further, this assay allows discrimination between invasive and noninvasive cell populations.51 Moreover, we also demonstrated that SDF-1 increases adhesion of RMS cells to endothelium and that they follow a directional chemotaxis toward SDF-1 secreted by bone marrow stroma. Thus, all these SDF-1–induced processes such as adhesion to endothelium, migration though basement membrane, and directional chemotaxis toward bone marrow stroma may influence the invasiveness of circulating RMS cells and their “homing” from the peripheral blood into the SDF-1–rich environment of the bone marrow or lymph nodes. Moreover, we found that the responsiveness of RMS cells to SDF-1 correlated with the level of CXCR4 surface expression, the presence of the PAX3-FKHR fusion gene, and the ARMS phenotype.
Generally, the biologic effects of SDF-1 on RMS cells observed here are consistent with those reported by us and others for human hematopoietic cells6-8,22,23 and involve primarily cell interactions with the microenvironment and their migration rather than cell proliferation and/or survival.6-9,22,23 However, the possibility that SDF-1 influences the metastatic potential of RMS in synergy/cooperation with other factors cannot be excluded. A potential candidate is, for example, the scatter factor. Both the scatter factor/hepatocyte growth factor and SDF-1 are highly expressed by bone marrow and lymph node stromal cells, and their respective receptors c-met52 and CXCR4, as shown here, are highly expressed by RMS cells. Furthermore, c-met has also been shown to be a direct transcriptional target of wild-type PAX3 and PAX3-FKHR fusion protein.52 Hence, we postulate that, although SDF-1 primarily directs/attracts RMS cells to bone marrow and lymph nodes, the scatter factor, alone or in combination with other factors, stimulates proliferation and survival of metastasizing cells.
We also demonstrated that blocking of the CXCR4 receptor in RMS cells by the specific small-molecule inhibitor T140 perturbed adhesion of these cells to the HUVECs and inhibited directional chemotaxis toward bone marrow fibroblast cultures. These findings indicate the CXCR4–SDF-1 axis plays a significant role in the metastasis of the cells to the bone marrow. Because T140 is well tolerated in vivo,53 we suggest that it could be used as an inhibitor of metastasis of CXCR4+ cells. We are currently testing this hypothesis in an in vivo murine model.
On the basis of our observations we conclude that the CXCR4–SDF-1 axis very likely plays an important role in tumor dissemination and metastasis to bone marrow, particularly of RMS. Hence, molecular strategies aimed at inhibiting this axis, eg, using small-molecule inhibitors,54-56 may lead to therapies that complement conventional radiotherapy or chemotherapy in preventing dissemination of RMS cells into hematopoietic organs.
Acknowledgments
We thank Lisa Ross, Ryan Reca, and Jacek Kijowski for their technical assistance, and Patsy Cotterill for editorial assistance.
J.L. was on leave of absence from SMM, School of Molecular Medicine, Warsaw, Poland.
Prepublished online as Blood First Edition Paper, June 7, 2002; DOI 10.1182/blood-2002-01-0031.
Supported by grant R01 HL61796-01 and KBN 3P05E10122 (M.Z.R.) and R01 CA64202 (F.G.B.) from the National Institutes of Health and by Canadian Blood Services (CBS) R&D grant (A.J.-W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Mariusz Z. Ratajczak, Stem Cell Biology Program at James Graham Brown Cancer Center, University of Louisville, 529 S Jackson St, Louisville, KY 40202; e-mail: mzrata01@louisville.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal