Hyaluronan (HA) is suggested to play a role in the pathophysiology of multiple myeloma. To further investigate the role of HA in this disease, we examined hyaluronan synthase (Has) gene expression and HA production in bone marrow mesenchymal progenitor cells (bmMPCs) derived from multiple myeloma patients. The relative abundance of mRNA for each HAS gene was determined using competitive reverse transcription–polymerase chain reaction (cRT-PCR), whereas HA production was detected by fluorophore-assisted carbohydrate electrophoresis (FACE). We determined the basal expression of Has isoforms in myeloma bmMPCs and then compared this expression with expression in healthy donor bmMPCs. Of the 3 Has isoforms, Has1 mRNA was expressed predominantly in myeloma bmMPCs, with expression 7.6-fold greater than Has2. Compared with normal bmMPCs, Has1 mRNA expression was 20-fold greater in myeloma bmMPCs. Normal bmMPCs predominantly expressed Has2 mRNA (8.2-fold greater than myeloma bmMPCs). Upon coculture of myeloma bmMPCs with plasma cells, Has1 transcript was strongly attenuated. FACE results show that myeloma bmMPCs synthesize 5.7-fold more HA than those from healthy donors. These data suggest that myeloma bmMPCs could be an important component of the myeloma pathophysiology in vivo by their increased expression of extracellular matrix (ECM) components relevant to plasma cell growth and survival.
Introduction
Hyaluronan, a major constituent of the extracellular matrix (ECM), is a large nonprotein glycosaminoglycan composed of repeating units of glucuronic acid andN-acetylglucosamine. Hyaluronan has been shown to play an important role in matrix assembly, cell proliferation, cell migration, and embryonic/tissue development.1-4 There is increasing evidence that hyaluronan plays an important role in tumorigenesis and in the process of tumor angiogenesis.5 In particular, hyaluronan production is up-regulated in a variety of malignant tissues compared with normal cells, and increased hyaluronan levels have been shown to correlate with increased tumor cell invasion, migration, and proliferation.6-8
A role for hyaluronan in multiple myeloma pathophysiology has recently been suggested, where abnormally high or low serum hyaluronan has been correlated with poor prognosis,9,10 conveying the prognostic importance of this molecule. Multiple myeloma is a malignancy characterized by the accumulation of malignant plasma cells within the bone marrow, wherein soluble factors and cell-cell contact likely provide optimal conditions for proliferation and survival. In vitro coculture systems have shown that in addition to the bone marrow environment providing cell-cell contact and the elaboration of growth factors/interleukins, interactions with bone marrow ECM likely affect the proliferation and survival of the myeloma cells and their response to chemotherapeutic agents.11-13 The bone marrow ECM consists primarily of fibronectin, collagen types I and IV, laminin, and the glycosaminoglycans heparan sulfate, chondroitin sulfate, and hyaluronan.14 Myeloma cells express multiple adhesion receptors, including the hyaluronan receptors CD44 and RHAMM (receptor for hyaluronan-mediated motility).15-17Interaction of hyaluronan with hyaluronan receptors initiates intracellular signal–mediated events, including cell adhesion, migration, proliferation, and protection of myeloma cells from apoptosis.18-20 Clinically, the expression of the v9 isoform of CD44 by myeloma plasma cells is related to decreased overall patient survival.21 22
The overall production of hyaluronan is determined by the enzymatic activities of hyaluronan synthase and hyaluronidase. Three hyaluronan synthase (Has) genes have been described, each encoding a plasma membrane protein responsible for hyaluronan synthesis.23-25 Has genes are differentially expressed during embryonic development, and Has1, Has2, and Has3 have been identified in various cell types, among them adult peripheral blood lymphocytes. Although expression of any one HAS gene is sufficient for hyaluronan synthesis, the contribution of the 3 HAS gene products to hyaluronan production and expression is not yet clear.
As a first step toward discerning the role of hyaluronan as an integral component of the bone marrow extracellular matrix in multiple myeloma patients, we have characterized Has gene expression and hyaluronan production in bone marrow mesenchymal progenitor cells (bmMPCs) derived from multiple myeloma and healthy donors. We show that myeloma bmMPCs predominantly express message for Has1 and furthermore synthesize more hyaluronan than do healthy donors. Because myeloma plasma cells reside within the bone marrow in close proximity with cellular and extracellular components of the bone marrow microenvironment, these data support the idea that further characterization of the bone marrow compartment is required to assess its role in the pathophysiology of multiple myeloma.
Materials and methods
Cell lines and myeloma plasma cells
The myeloma cell line ARH77 was obtained from American Type Culture Collection (ATCC) (Manassas, VA) and was grown in RPMI supplemented with 5% fetal bovine serum (FBS), streptomycin, andl-glutamine. The cell lines ANBL-6, RPMI 8226, and U266 (gifts from Dr B. Van Ness, Department of Genetics, Cell Biology and Development, University of Minnesota, Minneapolis) were grown in 10% FBS, streptomycin, and l-glutamine. The 8226 and U266 cell lines grow in the absence of interleukin-6 (IL-6), whereas ANBL-6 requires IL-6 for growth and survival. Mononuclear cells were isolated by Ficoll-Hypaque gradient from multiple myeloma patient bone marrow aspirates obtained upon informed consent. Plasma cells were sorted as CD38hi large cells and confirmed for plasma cell content by morphology (Giemsa-Wright stain). Approval was obtained from the institutional review board of the Park-Nicollet Institute, Minneapolis, MN, for these studies. Informed consent was provided according to the Declaration of Helsinki.
Reagents
Trizol reagent was obtained from Gibco (Grand Island, NY), dexamethasone (Dex) was obtained from Sigma (St Louis, MO), and 2-aminoacridone HCl (AMAC) was purchased from Molecular Probes (Eugene, OR). Glacial acetic acid (> 99.99%), dimethyl sulfoxide (DMSO) (99.9%), sodium cyanoborohydride (95%), glycerol (99.5%), and d-glucose were from Aldrich-Sigma. Phenol red (0.5% wt/vol) and proteinase K were purchased from Gibco. MONO composition gels and MONO gel running buffer were from Glyko. Chondroitinase ABC, hyaluronidase SD, glucoamylase, maltooligosaccharides (maltose, maltotriose, and maltotetraose), and unsaturated hyaluronan (ΔDiHA) and chondroitin sulfate (including ΔDi0S) disaccharide standards were purchased from Seikagaku America (Falmouth, MA). Monoclonal antibody (mAb) against CD45, conjugated to fluorescein isothiocyanate (FITC), was obtained from Becton Dickinson (San Jose, CA).
Isolation and growth of bmMPCs
Fresh bone marrow aspirate samples were obtained from newly diagnosed myeloma patients (n = 13) upon receiving informed consent. Fresh bone marrow aspirate samples were obtained from age-matched healthy donors (n = 14) with no prior history of malignancy, after informed consent was obtained. Bone marrow aspirates were harvested into preservative-free heparin.
BmMPCs were isolated and expanded by incubating isolated bone marrow mononuclear cells at a concentration of 1 × 106/mL in 5 mL α-minimal essential medium (α-MEM) supplemented with 10% FBS (Hyclone, Logan, UT) in T-25 cm2 tissue culture flasks (Corning, NY). Of 4 Hyclone FBS lots examined, lot AGM 7470 was found to best promote growth of bmMPCs. Therefore, this FBS lot was used for all experiments. Medium was replaced on day 4 and every 7 days thereafter. For passage, cells were detached with 0.25% trypsin and split 1:3 in α-MEM supplemented as indicated above. Normal and myeloma bmMPC cultures were examined between passage 1 and 7, with a normal and patient bmMPC culture observed between passages 1 and 2 and between passages 2 and 5. The bmMPC cultures were morphologically and phenotypically absent of hematopoietic cells as previously described26 but express the mesenchymal progenitor cell marker Stro-1. BmMPCs grown in this manner do not express osteogenic, adipogenic, or endothelial cell–specific markers but are capable of differentiation to adipogenic and osteogenic lineages under appropriate growth conditions (A. Panaskoltsis-Mortari et al, manuscript submitted, May 2002). It has been shown that increasing cell density results in a marked decrease in hyaluronan synthesis.27Therefore, subconfluent bmMPC cultures, at no more than 80% confluency (3.5 × 105 cells), were examined in these studies. BmMPC cultures were given a complete medium change and incubated for 48 hours prior to reverse transcription–polymerase chain reaction (RT-PCR) or fluorophore-assisted carbohydrate electrophoretic (FACE) analyses. To assess the impact of dexamethasone, bone marrow MPC cultures were grown in medium supplemented with dexamethasone (10−6/L), which was added at the initiation of plating MPC cultures.
Oligonucleotide primers
Sense and antisense primers were prepared by Integrated DNA Technologies (IDT, Coralville, IA). Has primers were derived outside the conserved coding sequences to ensure specific amplification of Has1, Has2, and Has3. β-Actin was used for normalization of cDNA as well as for evaluation of RNA integrity. Bands obtained by PCR amplification with each primer set were excised, purified from the agarose gel, sequenced, and the resultant sequence was compared with each HAS gene sequence. Each primer set was found to specifically amplify Has1, Has2, and Has3 only. The primers used are indicated in Table 1.
Primers used in RT-PCR
| . | Forward primer . | Reverse primer . | Product size, base pairs . |
|---|---|---|---|
| Primers for RT-PCR | |||
| HAS1 | 5′-GTGAGTGGCTGTACAACGCG-3′ | 5′-AGAGGGACGTAGTTAGCGGC-3′ | 354 |
| HAS2 | 5′-TGGCATCACACCTCATCATC-3′ | 5′-ACCAATTGCGTTACGTGTTG-3′ | 368 |
| HAS3 | 5′-TTGGCTGTGTGCAGTGTATTAGT-3′ | 5′-GGTCTCTGTGAGGCACTTGG-3′ | 200 |
| β-actin | 5′-TCCCTGGAGAAGAGCTACGA-3′ | 5′-AGCACTGTGTTGGCGTACAG-3′ | 194 |
| Insert primers for competitive RT-PCR | |||
| HAS1 forward | 5′-TAATACGACTCACTATAGGGTGAGTGGCTGTACAACGCGTCCCTGGAGAAGAGCTACGA-3′ | ||
| HAS1 reverse | 5′-TTTTTTTTTTTTTTTTTTAGAGGGACGTAGTTAGCGGCAGCACTGTGTTGGCGTACCG-3′ | 194 | |
| HAS2 forward | 5′-TAATACGACTCACTATAGGTGGCATCACACCTCATCATCTCCCTGGAGAAGAGCTACGA-3′ | ||
| HAS2 reverse | 5′-TTTTTTTTTTTTTTTTTTACCAATTGCGTTACGTGTTGAGCACTGTGTTGGCGTACCG-3′ | 194 | |
| . | Forward primer . | Reverse primer . | Product size, base pairs . |
|---|---|---|---|
| Primers for RT-PCR | |||
| HAS1 | 5′-GTGAGTGGCTGTACAACGCG-3′ | 5′-AGAGGGACGTAGTTAGCGGC-3′ | 354 |
| HAS2 | 5′-TGGCATCACACCTCATCATC-3′ | 5′-ACCAATTGCGTTACGTGTTG-3′ | 368 |
| HAS3 | 5′-TTGGCTGTGTGCAGTGTATTAGT-3′ | 5′-GGTCTCTGTGAGGCACTTGG-3′ | 200 |
| β-actin | 5′-TCCCTGGAGAAGAGCTACGA-3′ | 5′-AGCACTGTGTTGGCGTACAG-3′ | 194 |
| Insert primers for competitive RT-PCR | |||
| HAS1 forward | 5′-TAATACGACTCACTATAGGGTGAGTGGCTGTACAACGCGTCCCTGGAGAAGAGCTACGA-3′ | ||
| HAS1 reverse | 5′-TTTTTTTTTTTTTTTTTTAGAGGGACGTAGTTAGCGGCAGCACTGTGTTGGCGTACCG-3′ | 194 | |
| HAS2 forward | 5′-TAATACGACTCACTATAGGTGGCATCACACCTCATCATCTCCCTGGAGAAGAGCTACGA-3′ | ||
| HAS2 reverse | 5′-TTTTTTTTTTTTTTTTTTACCAATTGCGTTACGTGTTGAGCACTGTGTTGGCGTACCG-3′ | 194 | |
Reverse transcription–polymerase chain reaction
Total RNA was extracted using Trizol reagent (Gibco) from bmMPC cultures at no greater than 80% confluence. Extraction was performed following the manufacturer's protocol. The RNA pellet was resuspended in 20 μL DEPC-treated water and the RNA concentration was determined. RNA was stored at −70°C until ready for use.
Reverse transcription was performed using total RNA under the following conditions: 1.0 μg total RNA, 1 × reaction buffer, 5 mM MgCl2, 1 mmol deoxyribonucleoside triphosphates (dNTPs), 1.6 μg oligo-p(dT)18 primer, 40 units RNase inhibitor, and 20 units avian myeloblastosis virus (AMV) reverse transcriptase (Seikagaku America) to a total volume of 20 μL. The samples were incubated at 25°C for 10 minutes, followed by 42°C for 60 minutes. PCR was used to detect the presence of Has1, Has2, and Has3. β-Actin amplification was used for normalization as well as for detection of DNA contamination. The PCR was performed under the following conditions in a 30 μL total volume: 1 μL cDNA template, 1 × PCR buffer, 2 mM MgCl2, 0.3 mmol dNTPs, 60 pmol of each forward and reverse primer, and 5 units Taq polymerase (Qiagen, Valencia, CA). The cycling conditions used were 94°C for 5 minutes; followed by 30 cycles at 30 seconds at 94°C, 30 seconds at 60°C, and 30 seconds at 68°C; and extension at 68°C for 7 minutes. The PCR products were visualized on a 1.5% agarose gel stained with ethidium bromide.
Competitive reverse transcription coupled with polymerase chain reaction
Construction of the recombinant Has DNA to be used as the internal standard (Has competitor) used primers containing sequences for a portion of β-actin mRNA.28 The 3′ end of the forward primer contained 20 bases complementary to the β-actin gene, with the remaining 39 bases containing Has 5′ sequence and T7 promoter sequence. The reverse primer contains reverse β-actin complementary sequence at the 3′ end as well as the reverse Has sequence and a poly(dT)18 sequence. These primers were used to amplify the recombinant internal standard (Has competitor) from normal bone marrow cDNA. The PCR reaction was performed as follows: 1 μL cDNA, 1 × PCR buffer, 2 mM MgCL2, 0.5 μM dNTPs, 0.8 μM Has competitive RT-PCR (cRT-PCR) 5′ insert primer, 0.8 μM Has cRT-PCR 3′ insert primer, and 2.5 units Taq polymerase (Qiagen). Cycling conditions were 1 cycle at 95°C for 5 minutes and 49°C for 45 seconds; 72°C for 1 minute; 2 cycles at 95°C for 1 minute, 49°C for 45 seconds, and 72°C for 1 minute; 30 cycles at 95°C for 1 minute, 58°C for 45 seconds, and 72°C for 1 minute; and 1 cycle at 95°C for 1 minute, 58°C for 45 seconds, and 72°C for 10 minutes. The internal standard PCR fragment was gel purified using the QIAquick gel extraction kit (Qiagen), eluted in 20 μL Tris (tris(hydroxymethyl)aminomethane)–HCl (pH 8.5), and used directly for in vitro transcription. The transcription reaction was performed using the Riboquant kit (Pharmingen). The insert was purified by phenol-chloroform extraction and ethanol precipitation and resuspended in DEPC-treated water. The recombinant RNA concentrations were determined by spectrophotometry. Serial dilutions were made at 10 pg/μL, 1 pg/μL, 0.1 pg/μL, 0.01 pg/μL, 0.001 pg/μL, 0.0001 pg/μL, 0.000 01 pg/μL, 0.000 001 pg/μL, and 0.000 0001 pg/μL and used in cRT-PCR.
The cRT-PCR was performed with 1 μg total RNA and recombinant RNA at concentrations indicated above. RT was performed in a total volume of 5 μL containing 0.4 μg oligo-p(dT)18 primer, 1 × reaction buffer, 1.25 mmol MgCl2, 0.25 mmol dNTPs, 10 units RNase inhibitor, and 5 units AMV and incubated at 25°C for 10 minutes followed by 42°C for 60 minutes. The entire volume of generated cDNA was then used immediately for PCR. PCR was performed in a 25 μL total volume containing 1 × PCR buffer, 2 mmol MgCl2, 0.3 mmol dNTPs, 60 pmol primer, and 1.25 units Taq polymerase; with 30 cycles at 94°C for 5 minutes, 30 seconds at 94°C, 30 seconds at 60°C, and 30 seconds at 68°C; followed by 7 minutes 68°C. The PCR products were visualized on a 1.5% agarose gel stained with ethidium bromide.
To determine copy number, the cDNA prepared from 1.0 μg total RNA was kept constant and the competitor concentration was decreased as indicated above in log dilutions. The PCR products were scanned by densitometry, and the scanning units were normalized to the size of the competitor. The densitometry units were plotted against the number of copies of the competitor template. The copy numbers for Has transcripts in myeloma and normal bmMPCs are the crossover points at which the competitor and the bmMPC cDNA gave equal amounts of PCR product.
Coculture experiments
BmMPCs were established, as indicated above, in 24-well plates. When cultures reached 80% confluency, the medium of each was aspirated, and 1 × 105 plasma cells in α-MEM, 10% FBS were added to each well. BmMPCs and plasma cells were in coculture for 1, 6, and 12 hours at 37°C, 5% CO2, followed by treatment with 0.01% trypsin for 2 minutes to remove any adherent plasma cells. After trypsin treatment, bmMPC monolayers were rinsed 3 times in 2.0 mL PBS, followed by lysis in Trizol reagent, and used for RT-PCR as indicated above. Removal of the plasma cells and purity of the bmMPCs after trypsinization were determined by flow cytometry. Purity of bmMPCs and removal of plasma cells were determined by the presence of cells expressing CD45 as measured by flow cytometry.
Immunofluorescence
Analysis of surface marker expression on bmMPCs was performed by single-color immunofluorescence. Briefly, cells were stained with an antibody conjugated to FITC for 60 minutes at 4°C, followed by 2 washes in PBS containing 10% FBS and 0.02% azide. Samples were analyzed on a FACSCalibur (Becton Dickinson). Files of 10 000 events were collected. Staining with a specific mAb is compared with staining with the appropriate isotype-matched control in all cases.
Proteinase K digestion of culture media and cell layer fractions
At the end of the culture period (48 hours after medium change), each medium fraction (about 5 mL) was transferred to a pretared tube and stored at −20°C. Culture flasks containing the cell layers were stored at −20°C. A 2.5 mg/mL stock solution of proteinase K (PK) was made fresh in 0.0005% phenol red, 100 mmol ammonium acetate, pH 7.0 (digest buffer); a 250 μL aliquot of the stock solution added to each medium fraction; and the samples digested for 2 hours at 60°C with mixing every 30 minutes. The PK stock solution was diluted 1:10 (250 μg/mL final concentration) with digest buffer, 3 mL added to each cell layer, and the samples digested for 2 hours at 60°C with mixing every 30 minutes. A second 250 μL aliquot of a fresh PK stock solution was added to each medium sample prior to digestion for an additional 2 hours at 60°C with mixing every 30 minutes. A second 3 mL aliquot of a 1:10 dilution of fresh PK stock solution was added to each cell layer sample prior to digestion for an additional 2 hours at 60°C with mixing every 30 minutes. The cell layer PK digests were transferred from each culture flask to a pretared tube, and each flask was rinsed twice with 2 mL digest buffer. Rinses were combined with their corresponding PK digests for each cell layer sample. Both the medium and cell layer samples were then heated at 90°C for 10 minutes to inactivate the PK. The medium fractions were adjusted to 5.5 mL by addition of digest buffer; 5 mL culture medium was processed as described above and served as a control for any preexisting hyaluronan and/or chondroitin sulfate present in the FBS supplement.
Double ethanol precipitation
All of each PK-digested cell layer sample (about 10 mL) and one fifth of each PK-digested medium sample (1.1 mL) were transferred to pretared tubes and concentrated on a vacuum concentrator to 300 μL as determined by weight (1 μL = 1 mg). For the cell/matrix fractions this required multiple transfer and concentration steps. A total of 1.0 mL of −20°C absolute ethanol was added to the sample concentrate (about 77% final ethanol concentration). The samples were mixed thoroughly and incubated overnight at −20°C. The samples were centrifuged at 10 000g for 15 minutes at 4°C to pellet macromolecular material including hyaluronan. The supernatant fractions were aspirated to waste. Each pellet was washed with 1 mL of −20°C absolute ethanol and centrifuged at 10 000g for 15 minutes at 4°C. The pellet wash was aspirated to waste. The precipitate was dried and resuspended in 300 μL digest buffer. A total of 1.0 mL of −20°C absolute ethanol was added to each resuspended pellet and the samples were precipitated a second time as described above. The supernatant and pellet wash from each sample were aspirated to waste. The precipitate fractions were resuspended in 100 or 200 μL digest buffer prior to enzymatic digestion.
Enzymatic digestion
A total of 100 μL of each medium and precipitate fraction was treated as follows: 1 aliquot was digested for 1 hour at 37°C, with 100 mU/mL hyaluronidase SD, followed by 1 hour at 37°C with 100 mU/mL chondroitinase ABC and 2 hours at 37°C with 0.5 U/mL glucoamylase. Where applicable, an aliquot was left untreated with any enzyme activity. The samples were frozen on dry ice and lyophilized prior to derivatization as described below.
Fluorescent derivatization with 2-aminoacridone, fluorophore-assisted carbohydrate electrophoresis, gel imaging, and data analysis
All samples were derivatized as previously described29 by addition of 40 μL of 12.5 mmol AMAC (500 nmol) in 85% DMSO/15% acetic acid followed by incubation for 15 minutes at room temperature. A total of 40 μL of 1.25 M sodium cyanoborohydride (50 000 nmol) in ultrapure water was added followed by incubation for 16 hours at 37°C. After derivatization, 20 μL glycerol (20% final concentration) was added to each sample prior to electrophoresis.
A total of 5 μL (1:20) of each derivatization reaction was electrophoresed using MONO composition gels with MONO gel running buffer as previously described.29 The gels were illuminated with UV light (365 nm) from an Ultra Lum Transilluminator and imaged with a Quantix cooled CCD camera from Roper Scientific/Photometrics with the specifications previously described.30 The images were analyzed by Gel-Pro Analyzer 3.0 (Media Cybernetics). Digital images for each gel were taken at 2 exposures: one with oversaturated pixel intensity to allow visualization of less abundant derivatized structures and a second exposure with pixels within a linear 12-bit depth range that was used for quantitation. Baseline was determined using the “join valleys” method set at 1%.
Statistical analysis of HAS expression
For each HAS isoform, we made 2 comparisons. Among normal and myeloma bmMPCs, we compared the fold difference in relative transcript levels of Has1 versus Has2, respectively. We also compared the fold difference in relative transcript levels of Has1 versus Has2 between normal and myeloma bmMPCs. Within each patient population, relative amounts of Has1 and Has2 were transformed on the log scale and then compared using a paired t test. The difference in the relative amounts of Has1 and Has2, also calculated on the log scale, was then compared between patient populations using the Studentt test.
Results
Differential expression of Has mRNA in bone marrow mesenchymal progenitor cells
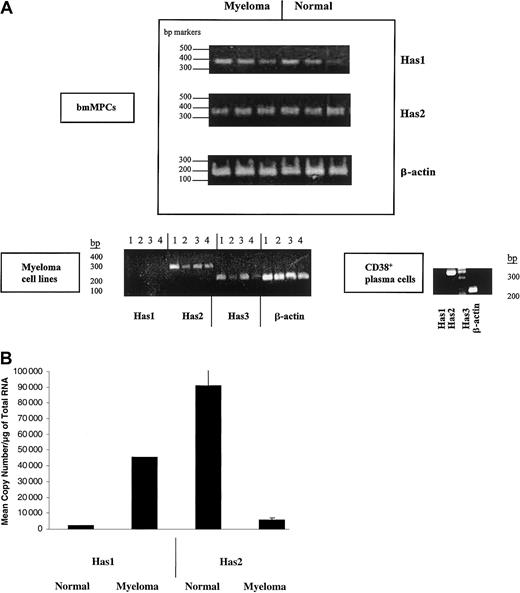
We examined Has isoform expression in bmMPC cultures derived from normal and multiple myeloma donors by RT-PCR. The primers used to detect each HAS isoform are indicated in Table 1 and were derived from the coding sequences of Has1, Has2, and Has3. PCR products for Has1 and Has2 were detected in all normal and myeloma bmMPCs examined (Figure1A). However, using 2 separate sets of RT-PCR primers, Has3 product was only variably detected in normal donor bmMPCs and detected in only 1 of 7 multiple myeloma patient bmMPC cultures (data not shown). These same primer sets were able to amplify Has2 and Has3 from 4 myeloma plasma cell lines. Similarly, Has2 but not Has1 was expressed in CD38+ bone marrow plasma cells from a myeloma patient; however, Has3 was not detectable (Figure 1A). The lack of detectable Has3 in CD38+ bone marrow plasma cells was further confirmed in CD38+ bone marrow plasma cells from 3 additional patients (data not shown). Based on the weak and/or no expression of Has3 in both myeloma bmMPCs and bone marrow CD38+ plasma cells, we focused our further analysis of HAS expression to Has1 and Has2.
HAS gene expression in healthy donor bmMPCs and myeloma cells.
RT-PCR products were amplified by competitive RT-PCR and resolved on a 1.5% agarose gel. Band intensity was determined by densitometry as indicated in “Materials and methods.” (A) Expression of Has mRNA in bmMPCs: RT-PCR for Has1, Has2, and β-actin in 3 healthy donors and 3 multiple myeloma patients and done as described in “Materials and methods” using primers indicated in Table 1. A representative result of 3 experiments is shown. Expression of HAS mRNA in myeloma plasma cell lines and ex vivo bone marrow plasma cells: (lane 1) ANBL-6, (lane 2) U266, (lane 3) ARH77, (lane 4) RPMI 8226. RT-PCR for Has1, Has2, Has3, and β-actin was done as described in “Materials and methods” and using primers indicated in Table 1. β-Actin was amplified to determine that all samples contained equivalent amounts of cDNA. (B) Competitive RT-PCR for Has1 and Has2 in 3 multiple myeloma patients and 3 healthy donors. One microgram of total RNA was reverse transcribed as indicated in “Materials and methods”; 2.0 μL cDNA was subjected to competitive PCR using primers for Has1 and Has2 as indicated in Table 1. RT-PCR was carried out in duplicate. The graph in panel B shows the results from duplicate quantifications of Has mRNA from 3 myeloma and 3 healthy donors.
HAS gene expression in healthy donor bmMPCs and myeloma cells.
RT-PCR products were amplified by competitive RT-PCR and resolved on a 1.5% agarose gel. Band intensity was determined by densitometry as indicated in “Materials and methods.” (A) Expression of Has mRNA in bmMPCs: RT-PCR for Has1, Has2, and β-actin in 3 healthy donors and 3 multiple myeloma patients and done as described in “Materials and methods” using primers indicated in Table 1. A representative result of 3 experiments is shown. Expression of HAS mRNA in myeloma plasma cell lines and ex vivo bone marrow plasma cells: (lane 1) ANBL-6, (lane 2) U266, (lane 3) ARH77, (lane 4) RPMI 8226. RT-PCR for Has1, Has2, Has3, and β-actin was done as described in “Materials and methods” and using primers indicated in Table 1. β-Actin was amplified to determine that all samples contained equivalent amounts of cDNA. (B) Competitive RT-PCR for Has1 and Has2 in 3 multiple myeloma patients and 3 healthy donors. One microgram of total RNA was reverse transcribed as indicated in “Materials and methods”; 2.0 μL cDNA was subjected to competitive PCR using primers for Has1 and Has2 as indicated in Table 1. RT-PCR was carried out in duplicate. The graph in panel B shows the results from duplicate quantifications of Has mRNA from 3 myeloma and 3 healthy donors.
For quantification of Has1 and Has2 mRNA expression, competitive quantitative RT-PCR was performed. Figure 1B indicates the amplification of Has1 and Has2 mRNA in 3 individual healthy donor and multiple myeloma–derived bmMPC cultures. The amount of competitor in the PCR ranged from 0.1 × 102 to 1 × 10−6 for Has1 and Has2. Within myeloma-derived bmMPCs, the Has1 mRNA was at highest-level expression of 4.5 × 104 copies per microgram RNA compared with 6.1 × 103 copies per microgram RNA of Has2 mRNA (7.6-fold higher Has1 than Has2) (Figure 1B; Table2). Expression of Has1 mRNA was also higher in myeloma bmMPCs than in normal donor bmMPCs, where expression was 20-fold higher for myeloma compared with normal donor bmMPCs (4.5 × 104 copies per microgram RNA in myeloma bmMPCs compared with 2.3 × 103/μg RNA in normal bmMPCs) (Figure 1B; Table 2).
Fold difference and P for Has expression in myeloma and normal MPC cultures
| Comparisons . | Copy no., × 104(variation) . | Fold difference . | P . | |
|---|---|---|---|---|
| Has1 . | Has2 . | |||
| Has1 vs Has2 | ||||
| Normal | 0.2 (0) | 9.1 (0.1) | 21.5 | .0044 |
| Multiple myeloma | 4.5 (0) | 0.6 (6.8) | 7.6 | .0572 |
| Multiple myeloma vs normal | ||||
| Has1 | 4.5 (0) | 0.6 (6.8) | 20 | N/A (no variation) |
| Has2 | 0.2 (0) | 9.1 (0.1) | 8.2 | .0539 |
| Comparisons . | Copy no., × 104(variation) . | Fold difference . | P . | |
|---|---|---|---|---|
| Has1 . | Has2 . | |||
| Has1 vs Has2 | ||||
| Normal | 0.2 (0) | 9.1 (0.1) | 21.5 | .0044 |
| Multiple myeloma | 4.5 (0) | 0.6 (6.8) | 7.6 | .0572 |
| Multiple myeloma vs normal | ||||
| Has1 | 4.5 (0) | 0.6 (6.8) | 20 | N/A (no variation) |
| Has2 | 0.2 (0) | 9.1 (0.1) | 8.2 | .0539 |
Fold difference in Has1 and Has2 expression in multiple myeloma and normal donor MPCs was calculated on the log scale and compared using a paired or unpaired t test as appropriate.
Normal donor bmMPCs predominantly expressed Has2 mRNA, with expression 21.5-fold greater than Has1 (9.1 × 104 copies per microgram RNA Has2 and 2.3 × 103 copies per microgram RNA Has1). Has2 mRNA in normal bmMPCs was 8.2-fold greater than Has2 mRNA in myeloma bmMPCs (9.1 × 104 copies per microgram RNA in normal bmMPCs and 6.1 × 103 copies per microgram RNA in myeloma MPCs) (Figure 1B, Table 2).
Coculture of myeloma plasma cells modulates bmMPC Has expression
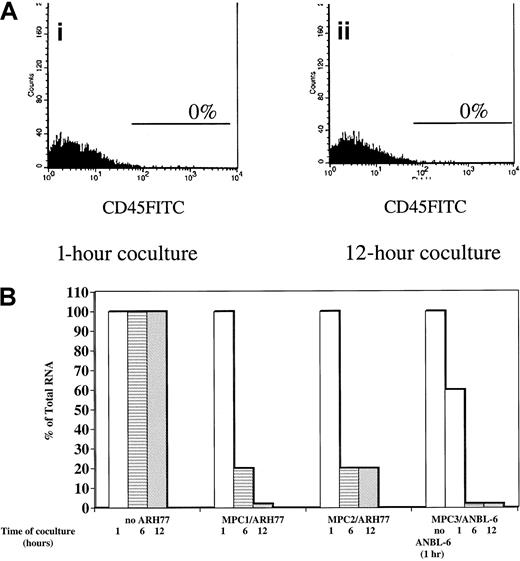
Because myeloma plasma cells reside within the bone marrow in juxtaposition with stromal cells, we examined the consequence of plasma cell contact on HAS expression in myeloma bmMPCs. By RT-PCR we noted that of 4 plasma cell lines examined, all expressed mRNA for Has2 and Has3 but not for Has1 (Figure 1A). The presence of Has2 and lack of Has1 mRNA observed in the plasma cell lines is identical to the expression pattern observed in ex vivo bone marrow plasma cells (Figure1A). Due to the difficulty in obtaining sufficient numbers of purified bone marrow plasma cells to carry out the coculture experiments, we chose ANBL-6 (IL-6 dependent) and ARH77 (IL-6 independent) myeloma plasma cell lines to coculture with myeloma bmMPCs for 1, 6, and 12 hours. Both ANBL-6 and ARH77 cells were in direct contact with bmMPCs during the time course assay. A total of 35% ± 8% of ARH77 cells and 30% ± 5% of ANBL-6 cells were found to adhere to bmMPC monolayers by 1 hour, and this percentage did not change over the time course of coculture (data not shown). After the coculture period, nonadherent and adherent cells were removed as described in “Materials and methods,” and Has expression was determined. Removal of adherent plasma cells was determined by assessing the presence of CD45+ cells, which is expressed by bone marrow plasma cells but not by bmMPCs. As shown in Figure 2, no CD45+ cells remained in the bmMPC monolayer after trypsinization at either the 1-hour or 12-hour time points.
Has1 expression in myeloma bmMPCs after coculture with the plasma cell lines ARH77 and ANBL-6.
Multiple myeloma bone marrow bmMPCs were established as indicated in “Materials and methods.” When cultures reached 80% confluency, the medium was aspirated and 1 × 105 ANBL-6 and ARH77 cells were added to each well. Cocultures were incubated for 1, 6, and 12 hours, followed by treatment with 0.01% trypsin. RNA was isolated as described in “Materials and methods.” Coculture time course assays were done in quadruplicate, and RNA was pooled for each time point. Values indicated are the expression of Has1 relative to that observed in myeloma bmMPCs in the absence of coculture (100%). There were no significant differences in the total RNA values at the 1-, 6-, and 12-hour ARH77/ANBL-6 time points. Total RNA values at 1, 6, and 12 hours are shown for ARH77, and total RNA value at 1 hour is shown for ANBL-6. As such, and for consistency, all values were normalized to the 1-hour ARH77/ANBL-6 time point. SEM was less than 5% for all samples. (A) shows a representative histogram plot (representative of 3 experiments with 3 individual myeloma bmMPCs) for CD45-FITC–positive cells in ANBL-6:bmMPC cocultures after treatment with 0.01% trypsin at the (i) 1-hour and (ii) 12-hour time points. Staining above isotype-matched control mAb is represented by the marker in each histogram plot. BmMPCs do not express CD45 either as determined by flow cytometry (0% CD45-FITC–positive cells, data not shown) or as determined by immunohistochemistry.25 To assure that treatment with 0.01% trypsin did not result in the removal of CD45, U937 cells (CD45+) were treated with trypsin as indicated in “Materials and methods” and CD45-FITC expression was determined by flow cytometry. Prior to trypsin treatment, 70% U937 cells expressed CD45-FITC at a median channel of 147, whereas 75% of U937 cells expressed CD45 at a median channel of 151 after trypsin treatment.
Has1 expression in myeloma bmMPCs after coculture with the plasma cell lines ARH77 and ANBL-6.
Multiple myeloma bone marrow bmMPCs were established as indicated in “Materials and methods.” When cultures reached 80% confluency, the medium was aspirated and 1 × 105 ANBL-6 and ARH77 cells were added to each well. Cocultures were incubated for 1, 6, and 12 hours, followed by treatment with 0.01% trypsin. RNA was isolated as described in “Materials and methods.” Coculture time course assays were done in quadruplicate, and RNA was pooled for each time point. Values indicated are the expression of Has1 relative to that observed in myeloma bmMPCs in the absence of coculture (100%). There were no significant differences in the total RNA values at the 1-, 6-, and 12-hour ARH77/ANBL-6 time points. Total RNA values at 1, 6, and 12 hours are shown for ARH77, and total RNA value at 1 hour is shown for ANBL-6. As such, and for consistency, all values were normalized to the 1-hour ARH77/ANBL-6 time point. SEM was less than 5% for all samples. (A) shows a representative histogram plot (representative of 3 experiments with 3 individual myeloma bmMPCs) for CD45-FITC–positive cells in ANBL-6:bmMPC cocultures after treatment with 0.01% trypsin at the (i) 1-hour and (ii) 12-hour time points. Staining above isotype-matched control mAb is represented by the marker in each histogram plot. BmMPCs do not express CD45 either as determined by flow cytometry (0% CD45-FITC–positive cells, data not shown) or as determined by immunohistochemistry.25 To assure that treatment with 0.01% trypsin did not result in the removal of CD45, U937 cells (CD45+) were treated with trypsin as indicated in “Materials and methods” and CD45-FITC expression was determined by flow cytometry. Prior to trypsin treatment, 70% U937 cells expressed CD45-FITC at a median channel of 147, whereas 75% of U937 cells expressed CD45 at a median channel of 151 after trypsin treatment.
Upon coculture with ARH77 cells, Has1 mRNA expression was reduced by 80% ± 5% in bmMPCs derived from 2 individual myeloma donors within the first 6 hours of coculture and remained at this level after 12 hours of coculture with ARH77 cells (Figure 2). In the second myeloma MPC culture, Has1 expression was reduced to less than 5% by 12 hours of coculture with ARH77 (Figure 2). In contrast to Has1 expression, Has2 mRNA expression in myeloma bmMPCs was unaltered after 12 hours of coculture (data not shown). Down-regulation of Has1 was also observed upon coculture of myeloma bmMPCs with the ANBL-6 plasma cell line, where Has1 expression was reduced by 40% as early as 1 hour after coculture, to less than 5% by 6 hours, and remained at this level at 12 hours of coculture (Figure 2). As observed with ARH77 coculture, Has2 mRNA expression in bmMPCs remained unaltered (data not shown).
Fluorophore-assisted carbohydrate electrophoretic analysis of hyaluronan in bmMPC cultures
FACE was used to measure the relative amounts of HA present in normal and myeloma bmMPC cultures. BmMPCs were also cultured with a corticosteroid (Dex), which has been shown to reduce HA expression in other cell culture models.31 In this study, HA synthesis is determined through measurement of AMAC-derivatized ΔDiHA, the smallest characteristic repeat saccharide for HA generated by hyaluronidase SD digestion (unsaturated glucuronic acid residue at the nonreducing terminal linked to N-acetylglucosamine).
Most of the HA synthesized by bmMPCs was secreted into the culture medium with all of the HA partitioning into the medium ethanol precipitate fraction (Figure 3). No HA was detected in fresh control medium, which was never used to culture cells (data not shown); therefore, all of the HA detected as AMAC-derivatized ΔDiHA in Figure 3 was synthesized by the bmMPCs during the 48-hour culture period. Figure 3 shows that bmMPCs derived from the myeloma patients synthesized 9.6-fold more HA in the absence of Dex than bmMPC cultures from the healthy donor. In the presence of Dex, the amounts of HA synthesized by bmMPCs derived from healthy donors and myeloma patients (37%) were significantly decreased, but the HA synthesized by the bmMPCs from the myeloma patient remained 10-fold higher than for the bmMPCs from healthy donors. The chondroitin sulfate detected was approximately the same in all 3 cultures (as evident from the ΔDi0S band) as well as in the control medium, indicating that it was derived from medium serum and was not synthesized by the bmMPCs. Collectively, the bmMPCs derived from all 4 myeloma patients synthesized 5.7-fold more HA in the absence of Dex than the bmMPCs derived from the 4 healthy donors (data not shown).
Relative amounts of hyaluronan in medium from healthy donor and multiple myeloma patient–derived bmMPC cultures.
FACE analysis of HA in the medium ethanol precipitate fractions of bmMPC cultures derived from a healthy donor (lanes 1-4) and a multiple myeloma patient (lanes 5-8). Cultures were incubated as indicated in the presence (+; lanes 3, 4, 7, and 8) or absence (−; lanes 1, 2, 5, and 6) of dexamethasone (DEX) as described in “Materials and methods.” Samples were digested as indicated with (+; lanes 2, 4, 6, and 8) or without (−; lanes 1, 3, 5, and 7) hyaluronidase SD, chondroitinase ABC, and glucoamylase (EZ) as described in “Materials and methods.” The gel image was exposed such that all pixels were within the linear 12-bit depth range required for quantitation. The percent HA detected as AMAC-derivatized ΔDiHA in each culture is listed above each lane and is normalized to the (−) DEX multiple myeloma value in each image. Note the absence of AMAC-derivatized ΔDiHA or ΔDi0S in the absence of enzyme digestion and the similar intensity of the AMAC-derivatized ΔDi0S (the unsulfated disaccharide of chondroitin sulfate) in each sample. Each lane represents 0.5% (1:200) of the total medium sample. Data presented are representative of 4 healthy donors and 4 myeloma patients. Similar results were observed in all donors.
Relative amounts of hyaluronan in medium from healthy donor and multiple myeloma patient–derived bmMPC cultures.
FACE analysis of HA in the medium ethanol precipitate fractions of bmMPC cultures derived from a healthy donor (lanes 1-4) and a multiple myeloma patient (lanes 5-8). Cultures were incubated as indicated in the presence (+; lanes 3, 4, 7, and 8) or absence (−; lanes 1, 2, 5, and 6) of dexamethasone (DEX) as described in “Materials and methods.” Samples were digested as indicated with (+; lanes 2, 4, 6, and 8) or without (−; lanes 1, 3, 5, and 7) hyaluronidase SD, chondroitinase ABC, and glucoamylase (EZ) as described in “Materials and methods.” The gel image was exposed such that all pixels were within the linear 12-bit depth range required for quantitation. The percent HA detected as AMAC-derivatized ΔDiHA in each culture is listed above each lane and is normalized to the (−) DEX multiple myeloma value in each image. Note the absence of AMAC-derivatized ΔDiHA or ΔDi0S in the absence of enzyme digestion and the similar intensity of the AMAC-derivatized ΔDi0S (the unsulfated disaccharide of chondroitin sulfate) in each sample. Each lane represents 0.5% (1:200) of the total medium sample. Data presented are representative of 4 healthy donors and 4 myeloma patients. Similar results were observed in all donors.
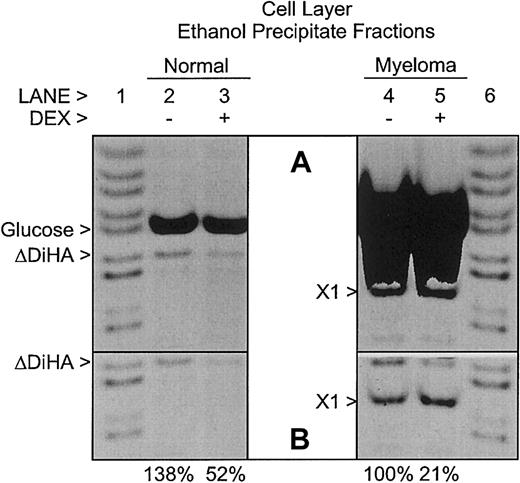
Although most of the HA synthesized by the bmMPCs in culture was secreted into the culture medium (Figure 3), some HA was retained in the cell layer fraction. As shown in Figure4, the cell layer ethanol precipitate fractions contain all of the HA present in the cell layers. Unlike in the medium samples in Figure 3, in the absence of Dex, the amount of HA present in the cell layer from the bmMPCs of myeloma patients was slightly lower than that in healthy donors. However, similar to the medium samples in Figure 3, the amounts of HA in the cell layers were reduced in the presence of Dex. In the absence of enzyme digestion, no AMAC-derivatized glucose or glycosaminoglycan-related bands were observed (data not shown) similar to those seen in Figure 3. An unknown AMAC-derivatized saccharide (X1) is present in only the myeloma patient–derived bmMPCs. The AMAC-derivatized glucose in the samples in Figure 4 is derived from cellular macromolecular glycogen recovered in the precipitate fraction and depolymerized by the action of glucoamylase. There is approximately 10-fold more glycogen in the myeloma cultures than in the normal cultures. As shown in Figure 4B, the AMAC-derivatized glucose bands in lanes 6 and 7 were covered to allow imaging of the AMAC-derivatized ΔDiHA bands.
Relative amounts of hyaluronan in cell layers from healthy donor and multiple myeloma patient–derived bmMPC cultures.
FACE analysis of HA in the cell layer ethanol precipitate fractions of bmMPC cultures derived from a healthy donor (lanes 2 and 3) and a myeloma patient (lanes 4 and 5). Cultures were incubated as indicated in the presence (+; lanes 3 and 5) or absence (−; lanes 2 and 4) of dexamethasone (DEX) as described in “Materials and methods.” All samples were digested with hyaluronidase SD, chondroitinase ABC, and glucoamylase as described in “Materials and methods.” No AMAC-derivatized bands were detected in the absence of enzyme digestion (data not shown), similar to the results shown in Figure 3. The gel image in panel A was overexposed to show minor bands, whereas the gel image in panel B was exposed such that all pixels were within the linear 12-bit depth range required for quantitation. In panel B, the AMAC-derivatized glucose bands in lanes 4 and 5 were covered to allow imaging of the AMAC-derivatized ΔDiHA bands. The percent HA detected as AMAC-derivatized ΔDiHA in each culture is listed below each lane in panel B and is normalized to the (−) DEX myeloma value. Note the presence of a prominent unknown band in the samples from the multiple myeloma patient (X1). AMAC-derivatized standards (lanes 1 and 6) include, from top to bottom,N-acetylgalactosamine, maltotetraose, maltotriose, maltose, glucose, ΔDiHA, ΔDi0S, 6-sulfatedN-acetylgalactosamine, and 4-sulfatedN-acetylgalactosamine. Each lane represents either 2.5% (healthy donor) or 5% (myeloma patients) of the total cell layer samples. Data presented are representative of 4 healthy donors and 4 myeloma patients. Similar results were observed in all donors.
Relative amounts of hyaluronan in cell layers from healthy donor and multiple myeloma patient–derived bmMPC cultures.
FACE analysis of HA in the cell layer ethanol precipitate fractions of bmMPC cultures derived from a healthy donor (lanes 2 and 3) and a myeloma patient (lanes 4 and 5). Cultures were incubated as indicated in the presence (+; lanes 3 and 5) or absence (−; lanes 2 and 4) of dexamethasone (DEX) as described in “Materials and methods.” All samples were digested with hyaluronidase SD, chondroitinase ABC, and glucoamylase as described in “Materials and methods.” No AMAC-derivatized bands were detected in the absence of enzyme digestion (data not shown), similar to the results shown in Figure 3. The gel image in panel A was overexposed to show minor bands, whereas the gel image in panel B was exposed such that all pixels were within the linear 12-bit depth range required for quantitation. In panel B, the AMAC-derivatized glucose bands in lanes 4 and 5 were covered to allow imaging of the AMAC-derivatized ΔDiHA bands. The percent HA detected as AMAC-derivatized ΔDiHA in each culture is listed below each lane in panel B and is normalized to the (−) DEX myeloma value. Note the presence of a prominent unknown band in the samples from the multiple myeloma patient (X1). AMAC-derivatized standards (lanes 1 and 6) include, from top to bottom,N-acetylgalactosamine, maltotetraose, maltotriose, maltose, glucose, ΔDiHA, ΔDi0S, 6-sulfatedN-acetylgalactosamine, and 4-sulfatedN-acetylgalactosamine. Each lane represents either 2.5% (healthy donor) or 5% (myeloma patients) of the total cell layer samples. Data presented are representative of 4 healthy donors and 4 myeloma patients. Similar results were observed in all donors.
Discussion
HA, a ubiquitous component of the ECM, is synthesized by a class of membrane-bound HAS proteins.23-25 There is increasing evidence that HA production is up-regulated in tumors and may play a significant role in tumor progression.5-8 Results from this study suggest that HA expression within bone marrow mesenchymal progenitor cells derived from myeloma patients is increased. To examine HA synthesis in bmMPCs from multiple myeloma patients, we have examined the expression of HAS mRNA, specifically Has1 and Has2 in bmMPCs by quantitative RT-PCR and HA synthesis by FACE analysis. We have found that myeloma bmMPCs express predominantly Has 1 and synthesize 5- to 10-fold greater HA than do normal bmMPCs.
Within the bone marrow microenvironment, cell-cell contact and elaboration of growth factors and cytokines are important in the maintenance of normal homeostasis and hematopoiesis. In multiple myeloma, the bone marrow microenvironment is likewise believed to play an important role, providing favorable signals for plasma cell growth and survival. In particular, IL-6 produced by the bone marrow stromal cells is a major survival and proliferation factor for myeloma cells.32-34 We have shown that IL-6 and other cytokines and growth factors are up-regulated in myeloma bone marrow mesenchymal progenitor cells in the absence of plasma cell contact, indicating an abnormality within the myeloma bone marrow environment that may impact plasma cell growth and survival. It has been previously shown that myeloma bone marrow stromal cells exhibit a simpler deposition of ECM proteins such as fibronectin, laminin, and collagen type IV compared with that observed on normal donor bone marrow stromal cells.35 The data presented in this paper provide further and novel evidence of differences within the HA component of bone marrow ECM in myeloma compared with healthy donors. The clinical implications of altered bone marrow ECM on myeloma disease progression are not yet clear.
FACE analysis of the HA synthesized by bmMPCs in culture showed that bmMPCs derived from myeloma patients synthesized 5- to 10-fold more HA in the absence of dexamethasone than bmMPCs from healthy donors. In the presence of dexamethasone, the amount of HA synthesized by bmMPCs derived from healthy donors and myeloma patients was reduced to approximately a third of initial values, but the HA synthesized by the bmMPCs from the multiple myeloma patients remained 10-fold higher than that for the bmMPCs from healthy donors. There was a differential retention of HA in the cell layers of bmMPC cultures from healthy donors and multiple myeloma patients, with less retained when cultured with dexamethasone.
Because most of the HA is secreted into the medium, the cell layer results may reflect differential contamination of the cell layers with their respective culture medium. However, if this were true, the ratio of ΔDiHA to ΔDi0S in the cell layers would be the same as in their respective medium samples, but it is not. In fact, little chondroitin sulfate as indicated by ΔDi0S was measured in any of the cell layer samples. Therefore, the difference in the relative amounts of HA retained in the cell layers of the bmMPCs derived from healthy donors and multiple myeloma patients probably reflects a differential ability of the various cells to retain HA in a pericellular matrix.
Among the 3 HAS isoforms examined, Has1 transcript levels were up-regulated to the greatest extent in myeloma bmMPCs compared with normal donor bmMPCs. Although our studies did not determine the molecular weight of HA produced in bmMPCs, the increase in Has1 transcript did correspond to an observed increase in HA production. Each HAS protein can independently catalyze HA synthesis, although each HAS protein has different enzymatic properties. Has3 has been shown to be more active and produce a shorter HA chain than either Has1 or Has2, whereas Has1 and Has2 produce HA chain lengths of similar molecular weight. The various HA chain lengths may in turn impact different cell functions such as proliferation and migration,4 although to date differences in biologic function have been observed in vitro comparing small oligosaccharide chains of HA (30-50 kDa in size) with large HA chains of 1 million to 3 million kDa. Our observations indicate a switch between Has2 and Has1 in myeloma bmMPCs that likely will not reflect a difference in overall chain size but may reflect differences in overall function or deposition of HA within the bone marrow extracellular matrix and may have important implications for tissue remodeling.
Cytokine and growth factors have been shown to modulate both HAS message and HA synthesis, where differential regulation of HAS transcripts and cell type–specific regulation in response to factors have been observed. Transforming growth factor-β (TGF-β), insulinlike growth factor-1 (IGF-1), and basic fibroblast growth factor (bFGF) have been shown to up-regulate Has1, Has2, and Has3 gene expression in skin fibroblasts, with an associated increase in HA production.36,37 While bFGF did enhance all 3 HAS genes in skin fibroblasts, there was a greater enhancement of Has2 compared with Has1 or Has3. Similarly, IL-1β and tumor necrosis factor-α (TNF-α) have been shown to up-regulate Has 1 and Has 3 transcripts and HA expression in fibroblasts; however, TNF-α effects were observed in fetal fibroblasts but not adult fibroblasts.38 These data are suggestive of independent regulation of the Has genes at the transcription level. Coculture of myeloma plasma cells with myeloma bmMPCs resulted in down-regulation of Has 1 mRNA with no concomitant modulation of Has2, which supports the concept of independent regulation of Has 1 and Has 2 in myeloma bmMPCs as well. We have previously shown that cytokine and growth factor expression are up-regulated in myeloma bmMPCs.26 We have also recently identified constitutive expression of TGF-β, IGF-1, and bFGF in myeloma bmMPCs, where expression levels for all 3 cytokines were similar to levels observed in normal bmMPCs (A, Panaskoltsis-Mortari et al, manuscript submitted, May 2002). Although bmMPCs express cytokines known to modulate HAS and HA expression in other cell types, it is yet to be determined whether bmMPCs are able to respond to these cytokines in a paracrine fashion. The relevance of HAS expression and increased HA synthesis by myeloma bone marrow mesenchymal progenitor cells is not yet determined; however, differential expression of Has1 compared with normal donor mesenchymal progenitor cells is suggestive of a distinct functional role for Has1-directed HA synthesis in these cells.
We are grateful to the staff of the Biomedical Imaging and Processing Laboratory and Flow Cytometry Core Facility (University of Minnesota) and to Robin Bliss (BioStatistics Core, University of Minnesota Cancer Center) for expert technical assistance and advice. We thank Aniq Darr, Hyunjin Rho, Alicia Felthauser, Stephanie Wallace, and Nancy Gin for their expert technical assistance. We thank the staff of the TwinCities Spine Center (Minneapolis, MN) and the University of Minnesota Cancer Center Tissue Procurement Facility for provision of normal donor bone marrow.
Prepublished online as Blood First Edition Paper, June 7, 2002; DOI 10.1182/blood-2002-01-0030.
Supported in part by the Cleveland Clinic Foundation and by grant support from the Allina Medical Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anna M. Masellis, Virginia Piper Cancer Institute-39419, Abbott Northwestern Hospital, 800 E 28th St, Minneapolis, MN 55407; e-mail:anna.masellis@allina.com.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal