The inhibitor of the apoptosis protein (IAP) survivin is expressed in proliferating cells such as fetal tissues and cancers. We previously reported that survivin is expressed and growth factor regulated in normal adult CD34+ cells. Herein, we examined survivin expression in CD34+ cells before and after cell cycle entry and demonstrate a role for survivin in cell cycle regulation and proliferation. Analysis of known human IAPs revealed that only survivin is cytokine regulated in CD34+ cells. Survivin expression is coincident with cell cycle progression. Up-regulation of survivin by thrombopoietin (Tpo), Flt3 ligand (FL), and stem cell factor (SCF) occurred in underphosphorylated-retinoblastoma protein (Rb)positive, Ki-67negative, and cyclin DnegativeCD34+ cells. Quantitative real-time reverse transcription–polymerase chain reaction (RT-PCR) and multivariate flow cytometry demonstrated that Tpo, SCF, and FL increase survivin mRNA and protein in quiescent G0 CD34+cells without increasing Ki-67 expression, indicating that cytokine-stimulated up-regulation of survivin in CD34+cells occurs during G0, before cells enter G1. Selective inhibition of the PI3-kinase/AKT and mitogen-activated protein kinase (MAPKp42/44) pathways blocked survivin up-regulation by growth factors before arresting cell cycle. Retrovirus transduction of survivin-internal ribosome entry site–enhanced green fluorescent protein (survivin-IRES-EGFP) in primary mouse marrow cells increased granulocyte macrophage–colony-forming units (CFU-GM) by 1.7- to 6.2-fold and the proportion of CFU-GM in S phase, compared to vector control. An antisense survivin construct decreased total and S-phase CFU-GM. These studies provide further evidence that survivin up-regulation by growth factors is not a consequence of cell cycle progression and strongly suggest that survivin is an important early event for cell cycle entry by CD34+cells.
Introduction
Apoptosis and cell cycle regulation are tightly orchestrated processes involving multiple effector molecules.1-3 Pathways that regulate cell cycle and cell survival overlap, but distinct mechanisms are involved.4,5The inhibitor of apoptosis protein (IAP) family proteins inhibit apoptosis by inactivating several caspases. There are 7 known IAP family proteins: NAIP,6,7 XIAP,7c-IAP1,7-9 c-IAP2,7,8survivin,10,11 livin,12 and murine Bruce13 and its human homolog, Apollon.14Survivin and livin are frequently overexpressed in cancer cells and fetal tissues but are barely detectable in adult quiescent tissues.10-12,15 While most IAPs block apoptosis,3,8,12,16 17 their roles in cell cycle regulation are unclear.
We recently reported that survivin is expressed and cytokine regulated in normal adult marrow CD34+ cells, umbilical cord blood (UCB) CD34+ cells, and adult peripheral blood T cells.18 Survivin blocks caspase-3 activity and inhibits apoptosis in cancer cells,11,15,16,19-21 and an inverse correlation between survivin and active caspase-3 expression was observed in CD34+ cells.18 Survivin expression in cytokine-stimulated CD34+ cells was associated with cell cycle progression, being highest in G2/M. However, in contrast to expression occurring only during G2/M in cancer cells,15 survivin is expressed in all phases of the cell cycle in cytokine-stimulated CD34+cells.18 These studies indicate that survivin is not a cancer-specific protein and suggest that survivin plays a role in the proliferation and survival of normal hematopoietic cells.
Since most proliferating cells express survivin, including cancer cells,10,11,15 normal T cells,18 and normal CD34+ cells,18 it is unclear whether survivin is expressed simply because cells are dividing or whether survivin expression directly affects cell cycle progression and proliferation. Since survivin interacts with the cdk4/cyclin D complex and enhances Rb phosphorylation4,22,23 and overexpression of survivin enhances cell cycle progression in hepatoma cells, survivin may be involved in cell cycle regulation, at least in cancer cells.24 Our previous findings that survivin is found in G0 CD34+ cells after growth factor stimulation for 48 hours18 raised the question of whether up-regulation of survivin by growth factors occurs in quiescent G0 CD34+ cells before they enter G1.
In this study, we examined whether IAPs in general, and survivin in particular, are expressed and growth factor regulated in normal CD34+ cells. Using real-time reverse transcription–polymerase chain reaction (RT-PCR) and multivariate flow cytometry, we now demonstrate that up-regulation of survivin expression by growth factors occurs in quiescent (G0) CD34+ cells before entering G1and that survivin expression is not a consequence of cell cycle progression. Overexpression of survivin in normal primary mouse bone marrow cells enhanced granulocyte macrophage–colony-forming unit (CFU-GM) production and cell cycle, suggesting that survivin plays a regulatory role in cell cycle entry and proliferation of normal hematopoietic cells.
Materials and methods
Animals
Specific pathogen-free female C57Bl/6 mice, 8 to 12 weeks of age, were purchased from Harlan Sprague-Dawley, Indianapolis, IN. Mice were provided continuous access to rodent chow and acidified water. The Institutional Animal Care and Use Committee of Indiana University School of Medicine approved all experimental procedures.
Growth factors, antibodies, and reagents
Recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF), Flt3 ligand (FL), and thrombopoietin (Tpo) were provided by Immunex, Seattle, WA. Recombinant human and mouse stem cell factor (SCF) were a gift from Dr Karl Nocka, UCB Research (Cambridge, MA). Recombinant murine GM-CSF was purchased from BioVision (Palo Alto, CA). Affinity purified antihuman survivin polyclonal antibody (AF886) and mouse IgG1 were purchased from R&D Systems (Minneapolis, MN). We previously described the specificity of the AF886 survivin antibody for intracellular staining in CD34+ cells.18 Monoclonal antihuman survivin antibody (6E4) was purchased from Cell Signaling (Beverly, MA). Antihuman retinoblastoma protein (Rb) monoclonal antibody (mAb) (G3-245), fluorescein isothiocyanate (FITC)–conjugated anti–underphosphorylated-Rb mAb (clone G99-549), FITC–antihuman cyclin D1/D2/D3 mAb (clone G124-259), FITC–anti-Ki-67 mAb (clone B56), FITC–antimouse Ig, FITC-mouse IgG1 and 7-AAD (Via Probe) were obtained from BD Pharmingen (San Diego, CA). Normal rabbit IgG was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). FITC- and phycoerythrin (PE)-goat anti–rabbit IgG was from Caltag Laboratories (Burlingame, CA). FITC- and Cy-chrome anti-CD34 antibodies (BIRMA-K3) were from Dako (Carpinteria, CA). Affinity purified polyclonal antihuman phosphorylated- Rb (Ser780, Ser795, and Ser807/811) antibodies were obtained from Cell Signaling. LY294002 and PD98059 were from BioMol (Plymouth Meeting, PA) and Calbiochem (San Diego, CA), respectively.
Isolation of cord blood CD34+ cells and cell culture
Normal UCB was obtained with institutional review board approval. Low-density mononuclear cells were separated on Ficoll-Paque (1.077 g/mL) (Amersham Pharmacia Biotech, Piscataway, NJ) and CD34+ cells isolated with antihuman CD34 mAb (QBEND/10) and 2 sequential positive selections with immunomagnetic beads (Miltenyi Biotech, Auburn, CA) as previously described.18 The purity of CD34+ cells routinely exceeded 95%. Fresh CD34+ cells or cells incubated with 100 ng/mL each of thrombopoietin (Tpo), Flt3 ligand (FL), and stem cell factor (SCF) in 10% heat-inactivated fetal bovine serum (HI-FBS) (Hyclone Laboratories, Logan, UT), 2 mM glutamine, and Iscove modified Dulbecco medium (IMDM) for up to 48 hours were fixed in 4% paraformaldehyde and frozen. For cell cycle fractionation, fresh or cultured CD34+ cells were stained with Hoechst 33342 (Hst) (Molecular Probes, Eugene, OR) and pyronin Y (PY) (Polysciences, Warrington, PA) as described.18,25,26 G0 and G1 cells were defined as Hstlow/PYlow and Hstlow/PYhigh, respectively. The 20% dimmest cells in the pyronin Y gate having 2N DNA were collected as G0 cells in all experiments.27 In some experiments, fresh G0 CD34+ cells were sorted based on Hst/PY staining and then stained with 1 μM of 5- (and 6-) carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes) in Hanks balanced salt solution (HBSS) (Gibco-BRL/Invitrogen, Carlsbad, CA) for 10 minutes at 37°C. The reaction was stopped with 500 μL of ice-cold HBSS with 10% FBS, then cells were washed 3 times and resuspended at 1-2 × 105cells/mL in IMDM with 10% HI-FBS, 2 mM glutamine, plus 100 ng/mL each of Tpo, FL, and SCF.28 29 After culture, CD34+ cells were sorted and reanalyzed for cell cycle and cell division based on CFSE and Hst/PY staining.
RNA isolation and RT-PCR
Total RNA isolation and RT-PCR were performed as previously described.18 PCR primers for amplification of IAPs were: survivin: 5′-GAGCTGCAGGTTCCTTATC-3′ and 5′-ACAGCATCGAGCCAAGTCAT-3′18; livin: 5′-TGAGGTGCTTCTTCTGCTAT -3′ and 5′-TTTCAGACTGGACCTCTCTC-3′; XIAP: 5′-GAAGACCCTTGGGAACAACA-3′ and 5′-GTCCTTGAAACTGAACCCCA-3′; c-IAP1: 5′-GCCTTTCTCCAAACCCTCTT-3′ and 5′-CATTCGAGCTGCATGTGTCT-3′; c-IAP2: 5′-CAGTGGATATTTCCGTGGCT-3′ and 5′-ATTTTCCACCACAGGCAAAG-3′; NAIP: 5′-CCGAACAGGAACTGCTTCTC-3′ and 5′-AAATTTGGCAAACTGGCAAC-3′; Apollon: 5′-AAGTGGCACCCGTAAATCTG-3′ and 5′-CCTGCCTCAAAGAAGCAAAC-3′. Amplification of all IAPs was carried out for 30 to 40 cycles of denaturation at 95°C for 1 minute, annealing at 55°C for 1 minute, and elongation for 1 minute at 72°C, followed by final extension for 7 minutes at 72°C. For NAIP, annealing was carried out at 60°C. PCR products were visualized in 2% agarose gels stained with ethidium bromide.
Quantitative real-time RT-PCR
Primers and probes for human survivin, Ki-67, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were designed using Primer Express Software (Applied Biosystems, Foster City, CA) and purchased from Applied Biosystems. For survivin, the forward and reverse primers were 5′-TGAACTTCAGGTGGATGAGGAGA-3′ and 5′-GTCTAATCACACAGCAGTGGCAA-3′, and the TaqMan probe was 6 FAM-AATAGAGTGATAGGAAGCGTCTGGCAGATACTC-TAMRA. For Ki-67, the forward and the reverse primers were 5′-CCACACTGTGTCGTCGTTTG-3′ and 5′-CCGTGCGCTTATCCATTCA-3′, and the TaqMan probe was 6 FAM-CCTATGTTCTCCAGGGCACGGTGG-TAMRA. Total RNA was treated with DNase (Promega, Madison, WI) for 30 minutes at 37°C, followed by real-time RT-PCR. The RT-PCR cycle parameters were 48°C for 30 minutes and 95°C for 10 minutes, followed by 50 cycles at 95°C for 15 seconds and 60°C for 1 minute. Survivin, Ki-67, and GAPDH PCR reactions were performed in separate tubes in triplicate, and the average threshold cycle (CT), representing the cycle at which a significant increase in fluorescence occurs, was used in subsequent calculations. The relative differences for survivin and Ki-67, before and after cytokine stimulation, were determined using the CT method as outlined in the Applied Biosystems protocol for RT-PCR. Briefly, a CT value for GAPDH was subtracted from the CT values for survivin and Ki-67 for each sample. The CT values were then converted to fold differences compared to unstimulated cells by raising 2 to the −CT power.
Intracellular staining and flow cytometry
Multivariate intracellular staining of CD34+ cells with anti–total-Rb, phosphorylated-Rb, underphosphorylated-Rb, cyclin-D, Ki-67, and survivin antibodies in combination with DNA staining were performed as previously described18 with minor modifications. CD34+ cells were fixed with 4% paraformaldehyde, washed with 0.1% bovine serum albumin/phosphate-buffered saline (BSA/PBS), resuspended in ice-cold 80% ethanol, and incubated for 24 hours at −20°C. Cells were washed with PBS containing 0.25% Triton X-100 and 1% BSA and stained with either FITC–anti-underphosphorylated-Rb, FITC–anti-Ki-67, or FITC–anti-cyclin D mAbs and antihuman survivin antibody. DNA staining was performed using 7-AAD. Stained cells were analyzed using a FACScan and ModFIT (for cell cycle) and CellQuest software (Becton Dickinson, San Jose, CA).
Retrovirus production and infection of mouse bone marrow progenitor cells
Human survivin cDNA was obtained from Dr Hari Nakshatri (Indiana University School of Medicine, Indianapolis, IN). Mouse survivin cDNA was amplified by RT-PCR from total RNA harvested from NIH3T3 cells using random hexamers and the primers 5′-GTTTGAGTCGTCTTGGCGGAGGTTGTGGTGACGCCATC-3′ and 5′-CTCAGGTCCAAGTTATCTCAGCAAAGGCTCAGCA-3′. The bicistronic retrovirus plasmid MIEG3 containing internal ribosome entry site–enhanced green fluorescent protein (IRES-EGFP)30 was obtained from Dr David Williams (Indiana University). Full-length human and mouse survivin cDNAs were cloned into the MIEG3 plasmid. The orientation and sequence of every construct were confirmed before transfection. A clone showing a reverse direction was used as an antisense construct. Ecotropic retrovirus containing human, mouse, and antisense-mouse survivin and MIEG3 backbone were produced using Phoenix eco cells (ATTC, Manassas, VA) as described.30Briefly, 5 × 106 Phoenix eco cells were seeded onto 100-mm dishes and transfected with 8 μg plasmid using Lipofectamine and Plus Reagent (Gibco BRL/Invitrogen) 16 hours later. Transfected cells were incubated in serum-free media for 3 hours, followed by readdition of serum. After 24 hours, cells were exposed to 50 mM sodium butyrate for 8 hours, followed by sequential washing with PBS. Cells were fed with IMDM containing 10% HI-FBS and 2 mM glutamine, incubated at 32°C for 24 hours, and the supernatant was collected, filtered, and stored at −70°C. Virus titer was usually 1-2 × 105 plaque-forming units (pfu)/mL. Mouse bone marrow cells were harvested, and mononuclear cells were isolated on Lympholyte-M (Cedarlane Laboratories, ON, Canada) and stimulated with 100 ng/mL each human Tpo, murine SCF, and human G-CSF for 48 hours.30 Media were replaced with freshly thawed retrovirus supernatant containing the identical cytokine cocktail, and the cells were cultured on wells precoated with recombinant fibronectin fragment CH296 (Takara Shuzo, Otsu, Japan) for 48 hours.
CFU-GM and thymidine suicide assay
Mouse bone marrow cells cultured with retrovirus supernatant were collected and FACS sorted based on green fluorescence protein (GFP) expression. Ten thousand GFP+ cells were plated in 0.3% agar (Difco Laboratories, Detroit, MI) containing supplemented McCoy 5a medium with 15% HI-FBS, 10 ng/mL rmGM-CSF, and 50 ng/mL rmSCF.31 CFU-GMs were scored after 7 days' incubation at 37°C in a humidified 5% CO2, 5% O2 air atmosphere. S-phase CFU-GMs were quantitated by thymidine suicide as previously described.32 Briefly, GFP+ cells were incubated with either 5 mg/mL thymidine (Sigma Chemical, St Louis, MO) or 50 μCi (1.85 MBq) [methyl-3H]thymidine (20 Ci/mmol [740 GBq/mmol], New England Nuclear, Boston, MA) at 37°C for 30 minutes. Reactions were terminated by adding 100-fold excessive thymidine (300 μg/mL). Cells were washed twice with IMDM and CFU-GM/1 × 104 cells quantitated as described above. In some experiments, growth factor addition was delayed for up to 48 hours and colonies scored after 10 days.
Western blot analyses
Western blot analyses were performed on lysates from 5 × 105 GFP+ cells from each transduced group using the rabbit polyclonal antihuman survivin (AF886) antibody. Preliminary flow cytometry and Western blot analyses experiments demonstrated the cross-reactivity of this antibody for murine survivin.
Results
Expression of mRNA for inhibitor of apoptosis proteins in cord blood CD34+ cells
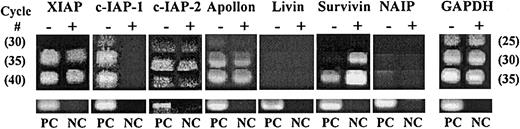
Since we previously demonstrated that survivin is expressed and cytokine regulated in normal CD34+ cells, we examined expression and growth factor regulation of all other human IAPs. Comparison of mRNA before and after culture with Tpo, SCF, and FL by RT-PCR for 48 hours demonstrated that in addition to survivin, XIAP, c-IAP1, c-IAP2, and Apollon are expressed in fresh CD34+cells (Figure 1). Livin and NAIP were not detectable. Only survivin mRNA expression was up-regulated after cytokine stimulation, whereas expression of c-IAP1 and c-IAP2 decreased. Identical results were observed in CD34+ cells from 5 UCB samples, although in 2 of 5 samples, c-IAP1 remained unchanged after culture with Tpo, SCF, and FL, while decreasing in 3 of 5 samples.
Semiquantitative RT-PCR for IAPs in UCB CD34+ cells before and after growth factor stimulation.
Total RNA from CD34+ cells before (−) and after (+) Tpo, SCF, and FL stimulation was subjected to 30, 35, and 40 cycles of RT-PCR for all 7 known human IAPs and 25, 30, and 35 cycles for GAPDH. The number of PCR cycles is shown in parentheses. Total RNA derived from human melanoma G361 cells was used as positive control (PC). NC represents reactions without RNA template. Reaction products were visualized in 2% agarose gels stained with ethidium bromide.
Semiquantitative RT-PCR for IAPs in UCB CD34+ cells before and after growth factor stimulation.
Total RNA from CD34+ cells before (−) and after (+) Tpo, SCF, and FL stimulation was subjected to 30, 35, and 40 cycles of RT-PCR for all 7 known human IAPs and 25, 30, and 35 cycles for GAPDH. The number of PCR cycles is shown in parentheses. Total RNA derived from human melanoma G361 cells was used as positive control (PC). NC represents reactions without RNA template. Reaction products were visualized in 2% agarose gels stained with ethidium bromide.
Survivin, Rb protein, cyclin D, and Ki-67 expression in CD34+ cells
A number of proteins have been linked to cell cycle entry. D cyclins are induced upon mitogenic stimulation in quiescent cells,33-35 and Ki-67 is a nuclear antigen found exclusively in proliferating cells.33,34,36,37 Upon mitogenic stimulation, Rb becomes phosphorylated, allowing cells to transit from G1 to S phase.5,35,38-40 In unstimulated lymphocytes, underphosphorylated Rb predominates, and the proportion of cells with underphosphorylated Rb decreases within 3 to 8 hours of mitogenic stimulation.39 To address whether survivin up-regulation by growth factors in CD34+ cells occurs before or after cell cycle entry, survivin, total Rb, underphosphorylated Rb, phosphorylated Rb (Ser780, Ser795, and Ser807/811), D cyclins, Ki-67, and cell cycle status were measured by multivariate intracellular flow cytometry following cytokine stimulation. Survivin mRNA was also quantitated by real-time RT-PCR. Within 2 hours, cytokine-stimulated up-regulation of survivin mRNA and protein was observed coincident with up-regulation of D cyclins and Ki-67 (Table 1). Total Rb protein gradually increased, whereas phosphorylated Rb increased dramatically. Underphosphorylated-Rb protein remained constant or marginally decreased (not shown). The ratio of phosphorylated Rb to total Rb protein was dramatically elevated following growth factor addition, whereas the ratio of underphosphorylated Rb to total Rb gradually declined (Table 1). The increase in the percentage of S + G2M phase cells correlated with up-regulation of survivin, Ki-67, D cyclins, and phosphorylated Rb. Because cells that express underphosphorylated Rb and are negative for Ki-67 and D cyclins are believed to be quiescent, expression of survivin in underphosphorylated-Rbpositive, Ki-67negative, and cyclin Dnegative CD34+ cells was examined before and after growth factor stimulation (Table2). Survivin protein was up-regulated by growth factors in underphosphorylated-Rbpositive, Ki-67negative, and cyclin DnegativeCD34+ cells as well as in underphosphorylated-Rbnegative, Ki-67positive, and cyclin Dpositive CD34+ cells, suggesting that survivin expression is up-regulated in quiescent cells before cell cycle entry.
Survivin, Ki-67, cyclin D, phosphorylated-Rb, underphosphorylated-Rb, and % S + G2/M cells in Tpo-, SCF-, and FL-stimulated CD34+ cells
| Cell population . | No. of experiments . | Fold increase . | |||
|---|---|---|---|---|---|
| 2 hours after growth factor addition* . | 4 hours after growth factor addition* . | 6 hours after growth factor addition* . | 18 hours after growth factor addition* . | ||
| Survivin protein | 4 | 1.4 ± 0.2 | 1.8 ± 0.2‡ | 2.4 ± 0.5† | 6.2 ± 2.4† |
| Survivin mRNA | 3 | 1.1 ± 0.1 | 1.2 ± 0.1† | 2.1 ± 0.5† | 10.3 ± 3.8† |
| %S + G2M phase | 4 | 7.5 ± 1.2 | 6.9 ± 1.0 | 8.5 ± 1.2† | 19.9 ± 3.1† |
| Ki-67 | 2 | 1.3 ± 0.011-153 | 1.3 ± 0.021-153 | 1.2 ± 0.011-153 | 1.5 ± 0.21-153 |
| Cyclin D | 5 | 1.1 ± 0.1† | 1.2 ± 0.1† | 1.2 ± 0.1‡ | 1.5 ± 0.2† |
| Phosphorylated Rb/Total Rb | 2 | 1.7 ± 0.6† | 1.5 ± 0.5† | 1.8 ± 0.7† | 2.3 ± 0.6† |
| Underphosphorylated Rb/Total Rb | 3 | 0.8 ± 0.1† | 0.7 ± 0.1‡ | 0.7 ± 0.1† | 0.5 ± 0.11-153 |
| Cell population . | No. of experiments . | Fold increase . | |||
|---|---|---|---|---|---|
| 2 hours after growth factor addition* . | 4 hours after growth factor addition* . | 6 hours after growth factor addition* . | 18 hours after growth factor addition* . | ||
| Survivin protein | 4 | 1.4 ± 0.2 | 1.8 ± 0.2‡ | 2.4 ± 0.5† | 6.2 ± 2.4† |
| Survivin mRNA | 3 | 1.1 ± 0.1 | 1.2 ± 0.1† | 2.1 ± 0.5† | 10.3 ± 3.8† |
| %S + G2M phase | 4 | 7.5 ± 1.2 | 6.9 ± 1.0 | 8.5 ± 1.2† | 19.9 ± 3.1† |
| Ki-67 | 2 | 1.3 ± 0.011-153 | 1.3 ± 0.021-153 | 1.2 ± 0.011-153 | 1.5 ± 0.21-153 |
| Cyclin D | 5 | 1.1 ± 0.1† | 1.2 ± 0.1† | 1.2 ± 0.1‡ | 1.5 ± 0.2† |
| Phosphorylated Rb/Total Rb | 2 | 1.7 ± 0.6† | 1.5 ± 0.5† | 1.8 ± 0.7† | 2.3 ± 0.6† |
| Underphosphorylated Rb/Total Rb | 3 | 0.8 ± 0.1† | 0.7 ± 0.1‡ | 0.7 ± 0.1† | 0.5 ± 0.11-153 |
Survivin, cyclin D, Ki-67, phosphorylated-Rb (Ser 807/811), underphosphorylated-Rb, and total-Rb protein levels were determined by intracellular flow cytometry in the CD34+ cell population from UCB. The fold increases in mean channel fluorescence for each protein and the ratio of fold changes of phosphorylated Rb/total Rb and underphosphorylated Rb/total Rb are shown as means ± SEM of the number of experiments indicated. Survivin mRNA was determined by real-time RT-PCR. The percentage of CD34+ cells in S + G2/M phase was measured by DNA staining with 7-AAD and analyzed using ModFit software. Data are presented as means ± SEM of 4 experiments.
100 ng/mL each rhTpo, rhFL, rhSCF.
P < .05.
P < .01.
P < .005 compared with time 0.
Survivin expression in CD34+ cell populations following growth factor addition
| Cell population . | Fold increase in survivin protein . | ||
|---|---|---|---|
| 2 hours after growth factor addition* . | 4 hours after growth factor addition* . | 6 hours after growth factor addition* . | |
| CD34+, cyclin Dnegative | 1.8 ± 0.3† | 2.2 ± 0.4‡ | 2.4 ± 0.4‡ |
| CD34+, cyclin Dpositive | 1.7 ± 0.2‡ | 2.3 ± 0.5‡ | 2.7 ± 0.5‡ |
| CD34+, Ki-67negative | 2.4 ± 0.22-153 | 3.2 ± 0.22-153 | 4.2 ± 0.52-153 |
| CD34+, Ki-67positive | 3.9 ± 1.0‡ | 5.4 ± 1.8† | 6.5 ± 2.0‡ |
| CD34+, underphosphorylated Rbpositive | 2.2 ± 0.22-153 | 3.2 ± 0.32-153 | 3.7 ± 0.62-153 |
| CD34+, underphosphorylated Rbnegative | 1.9 ± 0.3‡ | 3.2 ± 0.52-153 | 3.5 ± 0.62-153 |
| Cell population . | Fold increase in survivin protein . | ||
|---|---|---|---|
| 2 hours after growth factor addition* . | 4 hours after growth factor addition* . | 6 hours after growth factor addition* . | |
| CD34+, cyclin Dnegative | 1.8 ± 0.3† | 2.2 ± 0.4‡ | 2.4 ± 0.4‡ |
| CD34+, cyclin Dpositive | 1.7 ± 0.2‡ | 2.3 ± 0.5‡ | 2.7 ± 0.5‡ |
| CD34+, Ki-67negative | 2.4 ± 0.22-153 | 3.2 ± 0.22-153 | 4.2 ± 0.52-153 |
| CD34+, Ki-67positive | 3.9 ± 1.0‡ | 5.4 ± 1.8† | 6.5 ± 2.0‡ |
| CD34+, underphosphorylated Rbpositive | 2.2 ± 0.22-153 | 3.2 ± 0.32-153 | 3.7 ± 0.62-153 |
| CD34+, underphosphorylated Rbnegative | 1.9 ± 0.3‡ | 3.2 ± 0.52-153 | 3.5 ± 0.62-153 |
Survivin, cyclin D, Ki-67, and underphosphorylated-Rb protein levels were determined by intracellular flow cytometry in the CD34+ cell population from UCB. Data are expressed as means ± SEM from 3 experiments.
100 ng/mL each rhTpo, rhFL, rhSCF.
P < .05.
P < .01.
P < .005 compared with time 0.
Survivin expression in G0 CD34+ cells before and after growth factor stimulation
We previously demonstrated that survivin expression is elevated in G0 cells after incubation of unseparated CD34+cells with cytokines.18 However, these studies did not address the issue of whether these cells had yet to enter cell cycle or had already completed mitosis and returned to G0. We therefore investigated whether survivin up-regulation by growth factors observed in G0 cells is specifically regulated before cells enter cell cycle. Fresh G0 CD34+ cells were isolated based upon Hoechst 33342/pyronin Y (Hst/PY) staining (Figure2A, gate R1) and incubated for 12 hours with 100 ng/mL each of Tpo, SCF, and FL. After culture, cells were restained with Hst/PY, and Hstlow, PYlow cells (gate R2), representing cells in G0, were collected by FACS sorting. Replicate freshly isolated G0 cells were stained with CFSE before culture to monitor cell division. No cell division occurred during the 12-hour culture period as defined by CFSE analysis before sorting (Figure 2A). The purity of the G0 population (Figure 2A, gate R2) assessed by Hst/PY resorting was 98% (Figure 2B, gate R2) and was 97% ± 2% in 5 experiments. No cell division in the G0CD34+ population (gate R2) was observed based on CFSE analysis. Because Ki-67 is undetectable in G0cells41 but expressed in cells entering G1, Ki-67, and 7-AAD, staining was used to validate the Hst/PY sort for cell cycle status of CD34+ cells. Approximately 99% of cells in the R2 gate, that is, G0 CD34+ cells, did not express Ki-67 after culture with cytokines (Figure 2C). In 3 experiments, 89.9% ± 9.1% of fresh G0 cells (R1) and 95.2% ± 4.3% of cultured G0 cells (R2) were Ki-67negative, whereas only 65.9% ± 18.3% of fresh G1 cells were Ki-67negative (not shown). Survivin protein increased 2.1 ± 0.3 fold in G0CD34+ cells (R2 gate) following culture with growth factors, and the percentage of survivin-positive cells increased from 4.7% ± 1.4% (R1) to 32.5% ± 9.3% (R2) (Figure 2D; Table3). Because the G0 cells isolated based on Hst/PY staining were not 100% negative for Ki-67 expression (98.9%, Figure 2C, gate R2), survivin protein expression was examined in Ki-67negative cells in the R1 and R2 gates (Figure 2E; Table 3). Survivin protein increased 1.9 ± 0.1-fold in G0 CD34+ cells (R2 gate) after culture, and the percentage of survivin-positive cells increased from 1.6% ± 0.7% to 40.0% ± 18.5%. Up-regulation of survivin in G0 cells after growth factor incubation was verified at the mRNA level using quantitative real-time RT-PCR (Figure 2F; Table 3). Cells in the R1 and R2 gates were harvested and total RNA analyzed for survivin and Ki-67 expression. Ki-67 expression was essentially negative both before and after growth factor stimulation, confirming the quiescent nature of these cells. The threshold CT values for survivin were 34.7 and 37.8 in R2 and R1 cells, respectively, representing a 2.4 ± 0.7-fold increase in survivin mRNA in R2 cells compared to R1 cells (2 experiments). No increase in Ki-67 mRNA was observed in R2 cells compared with cells in R1 (0.8 ± 0.3-fold, 2 experiments), indicating that survivin is up-regulated in quiescent G0 cells after cytokine stimulation. Real-time RT-PCR analysis of freshly isolated G0 cells and CFSEbright G0 cells cultured for 48 hours with growth factors produced similar results (not shown).
Expression of survivin protein and mRNA in G0 CD34+ cells isolated by Hoechst 33342/pyronin Y staining before and after 12 hours' growth factor stimulation.
(A) Hoechst 33342/pyronin Y staining of UCB CD34+cells before and after incubation for 12 hours with 100 ng/mL each Tpo, SCF, and FL. The left panel represents the gate for fresh G0 CD34+ cells (R1). Fresh G0 cells (R1) were incubated with Tpo, SCF, and FL for 12 hours, restained with Hoechst 33342/pyronin Y, and G0 cells isolated by FACS sorting (middle panel; gate R2). In replicate cultures, R1 cells were prestained with CFSE before incubation, and cell division in the unseparated cell population was analyzed after 12 hours' culture (right panel). Data represent 1 of 5 identical experiments. (B) Postsort analysis of Hoechst 33342/pyronin Y and CFSE staining on cells from the R1 and R2 gates in Figure 2A. Data represent 1 of 5 identical experiments. The R1 and R2 gates were set so that the 20% dimmest pyronin Y cells were collected for fresh and cultured CD34+cells. The gates for the postsort analysis were adjusted for the characteristic progressive loss of pyronin Y fluorescence with time.27 33 (C) Intracellular Ki-67 protein expression and DNA staining of fresh G0 cells (R1) and G0 cells isolated after culture for 12 hours with growth factors (R2) from Figure 2A. The percentage of cells negative for Ki-67 expression, that is, below isotype staining (horizontal bar), is shown. Data represent 1 of 3 independent experiments. (D) Intracellular survivin protein expression in fresh G0 CD34+cells (R1) and in G0 cells (R2) from Figure 2A, harvested after 12 hours' incubation with growth factors. Mean channel fluorescence (MCF) of survivin and the percentage of cells staining positive (above isotype control [horizontal bar]) for survivin are shown below each blot. Data represent 1 of 3 identical experiments. (E) Survivin expression was quantitated in the Ki-67–negative fraction of fresh or cultured G0 cells from Figure 2C. The MCF for survivin and the percentage of survivin-positive cells (above isotype control [horizontal bar]) are shown beneath the blot. Data represent 1 of 3 identical experiments. (F) Total RNA from G0 cells before (R1) and after (R2) growth factor incubation was subjected to real-time RT-PCR to quantify survivin and Ki-67 mRNA expression. GAPDH was used as the internal control. Because Ki-67 expression of both samples was extremely low, G1CD34+ cells were used to verify the PCR reaction of Ki-67. The y-axis represents ΔRn, which indicates the magnitude of the signal generated at each cycle. The x-axis shows the reaction cycle. Data represent 1 of 2 identical experiments.
Expression of survivin protein and mRNA in G0 CD34+ cells isolated by Hoechst 33342/pyronin Y staining before and after 12 hours' growth factor stimulation.
(A) Hoechst 33342/pyronin Y staining of UCB CD34+cells before and after incubation for 12 hours with 100 ng/mL each Tpo, SCF, and FL. The left panel represents the gate for fresh G0 CD34+ cells (R1). Fresh G0 cells (R1) were incubated with Tpo, SCF, and FL for 12 hours, restained with Hoechst 33342/pyronin Y, and G0 cells isolated by FACS sorting (middle panel; gate R2). In replicate cultures, R1 cells were prestained with CFSE before incubation, and cell division in the unseparated cell population was analyzed after 12 hours' culture (right panel). Data represent 1 of 5 identical experiments. (B) Postsort analysis of Hoechst 33342/pyronin Y and CFSE staining on cells from the R1 and R2 gates in Figure 2A. Data represent 1 of 5 identical experiments. The R1 and R2 gates were set so that the 20% dimmest pyronin Y cells were collected for fresh and cultured CD34+cells. The gates for the postsort analysis were adjusted for the characteristic progressive loss of pyronin Y fluorescence with time.27 33 (C) Intracellular Ki-67 protein expression and DNA staining of fresh G0 cells (R1) and G0 cells isolated after culture for 12 hours with growth factors (R2) from Figure 2A. The percentage of cells negative for Ki-67 expression, that is, below isotype staining (horizontal bar), is shown. Data represent 1 of 3 independent experiments. (D) Intracellular survivin protein expression in fresh G0 CD34+cells (R1) and in G0 cells (R2) from Figure 2A, harvested after 12 hours' incubation with growth factors. Mean channel fluorescence (MCF) of survivin and the percentage of cells staining positive (above isotype control [horizontal bar]) for survivin are shown below each blot. Data represent 1 of 3 identical experiments. (E) Survivin expression was quantitated in the Ki-67–negative fraction of fresh or cultured G0 cells from Figure 2C. The MCF for survivin and the percentage of survivin-positive cells (above isotype control [horizontal bar]) are shown beneath the blot. Data represent 1 of 3 identical experiments. (F) Total RNA from G0 cells before (R1) and after (R2) growth factor incubation was subjected to real-time RT-PCR to quantify survivin and Ki-67 mRNA expression. GAPDH was used as the internal control. Because Ki-67 expression of both samples was extremely low, G1CD34+ cells were used to verify the PCR reaction of Ki-67. The y-axis represents ΔRn, which indicates the magnitude of the signal generated at each cycle. The x-axis shows the reaction cycle. Data represent 1 of 2 identical experiments.
Fold increase in survivin and Ki-67 in G0CD34+ cells after growth factor addition
| . | 0 hours after growth factor addition3-150 . | 12 hours after growth factor addition3-150 . |
|---|---|---|
| Fold increase survivin protein | – | 2.1 ± 0.33-151(1.9 ± 0.1)3-151 |
| % survivin-positive cells | 4.7 ± 1.4 (1.6 ± 0.7) | 32.5 ± 9.33-151(40.0 ± 18.5)3-151 |
| Fold increase survivin mRNA | – | 2.4 ± 0.73-152 |
| Fold increase Ki-67 protein | – | 0.9 ± 0.1 |
| % Ki-67negativecells | 89.9 ± 9.1 | 95.2 ± 4.3 |
| Fold increase Ki-67 mRNA | – | 0.8 ± 0.3 |
| . | 0 hours after growth factor addition3-150 . | 12 hours after growth factor addition3-150 . |
|---|---|---|
| Fold increase survivin protein | – | 2.1 ± 0.33-151(1.9 ± 0.1)3-151 |
| % survivin-positive cells | 4.7 ± 1.4 (1.6 ± 0.7) | 32.5 ± 9.33-151(40.0 ± 18.5)3-151 |
| Fold increase survivin mRNA | – | 2.4 ± 0.73-152 |
| Fold increase Ki-67 protein | – | 0.9 ± 0.1 |
| % Ki-67negativecells | 89.9 ± 9.1 | 95.2 ± 4.3 |
| Fold increase Ki-67 mRNA | – | 0.8 ± 0.3 |
Survivin and Ki-67 protein expression (means ± SEM from 3 experiments) was determined by intracellular flow cytometry; mRNA levels were determined by real-time RT-PCR (means ± SD from 2 experiments). Data are expressed as the fold increase in survivin and Ki-67 protein or percentage survivin-positive or Ki-67negative cells in G0 CD34+cells or in the G0 CD34+Ki-67negative cell population (shown in parentheses).
100 ng/mL each rhTpo, rhFL, and rhSCF.
P < .005.
Range, 1.42-3.27-fold; n = 2 experiments.
PI3-kinase/AKT and MAP kinase pathway inhibitors block survivin up-regulation by growth factors before cell cycle arrest
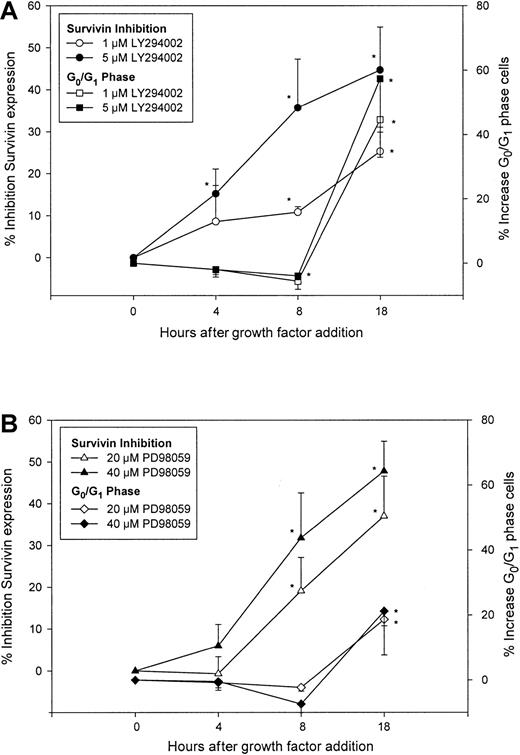
Activation of the PI3-kinase and MAPKp42/44pathways correlates with survival of CD34+ cells stimulated by hematopoietic cytokines,42,43 and inhibition of these pathways results in reduction of cytokine action44 and decreased proliferation of normal CD34+cells.45 Because inhibition of PI3-kinase/AKT and MAPKp42/44 pathways by LY294002 and PD98059, respectively, resulted in down-regulation of survivin accompanied by cell cycle arrest in cytokine-stimulated AML cells,46 we used these selective inhibitors to determine whether changes in survivin expression preceded cell cycle arrest induced by these inhibitors. CD34+ cells were treated with 1 and 5 μM LY294002, 20 and 40 μM PD98059 or dimethyl sulfoxide (DMSO) for 1 hour, followed by incubation with 100 ng/mL each Tpo, SCF, and FL. Survivin expression and cell cycle status were analyzed over time by flow cytometry. In control cultures, multivariate staining of survivin and DNA in CD34+ cells demonstrated that survivin expression was up-regulated in a time-dependent manner coincident with cell cycle progression (not shown). The PI3-kinase/AKT pathway inhibitor LY294002 (5 μM) reduced survivin expression by 15.2% ± 5.9% and 35.7% ± 11.7% (means ± SEM, 3 experiments) at 4 and 8 hours after cytokine stimulation, respectively, with no effect on cell cycle progression. At 18 hours, survivin expression was reduced by 44.7% ± 10.3% with a concomitant 57.3% ± 15.5% increase in the number of CD34+ cells in G0/G1 phase (Figure3A). Similar results were observed with 1 μM LY294002. CD34+ cell viability was greater than 90% in all groups up to 8 hours after cytokine addition, but was ∼85% in cells treated with 1 or 5 μM LY294002 at 18 hours. Inhibition of survivin was observed in both G0/G1 and S + G2/M cells (data not shown). The MAPKp42/44 inhibitor PD98059 at 20 μM reduced survivin expression in CD34+ cells by 19.1% ± 8.0% at 8 hours, without any effect on the ability of CD34+ cells to progress through cell cycle. However, at 18 hours, survivin expression was reduced by 37.0% ± 9.5%, concomitant with an 18.6% ± 11% increase in G0/G1 cells (Figure 3B). Similar results were obtained using 40 μM PD98059. Forward and side scatter analysis indicated that greater than 90% of cells in all groups remained viable. Incubation of CD34+ cells with 20 and 40 μM PD98059 for longer than 24 hours or at concentrations greater than or equal to 60 μM for up to 20 hours inhibited survivin up-regulation and reduced cell cycle (not shown). Similar to LY294002, inhibition of survivin up-regulation by PD98059 was observed in S + G2/M and G0/G1 cells (not shown).
Effects of PI3-kinase and MAPKp42/44inhibitors on survivin expression and cell cycle in CD34+cells.
CD34+ cells were pretreated with either DMSO, 1 and 5 μM LY294002 (A), or 20 and 40 μM PD98059 (B), followed by incubation with 100 ng/mL each Tpo, SCF, and FL. Cells were fixed at 4, 8, and 18 hours after growth factor stimulation and analyzed for survivin protein and cell cycle. Percent inhibition of survivin expression was calculated as percent reduction of mean channel fluorescence of survivin compared to DMSO control at each time point. Percent increase in G0/G1 phase cells was calculated based on the increase in G0/G1 population over DMSO control at each time. Data are shown as means ± SEM of 3 independent experiments. *P < .05.
Effects of PI3-kinase and MAPKp42/44inhibitors on survivin expression and cell cycle in CD34+cells.
CD34+ cells were pretreated with either DMSO, 1 and 5 μM LY294002 (A), or 20 and 40 μM PD98059 (B), followed by incubation with 100 ng/mL each Tpo, SCF, and FL. Cells were fixed at 4, 8, and 18 hours after growth factor stimulation and analyzed for survivin protein and cell cycle. Percent inhibition of survivin expression was calculated as percent reduction of mean channel fluorescence of survivin compared to DMSO control at each time point. Percent increase in G0/G1 phase cells was calculated based on the increase in G0/G1 population over DMSO control at each time. Data are shown as means ± SEM of 3 independent experiments. *P < .05.
Overexpression of wild-type and antisense survivin cDNA in primary mouse bone marrow cells
Mouse bone marrow mononuclear cells were infected with the retrovirus MIEG3 vector, human, mouse, or antisense-mouse survivin. GFP+ cells were sorted, collected, and assayed for CFU-GM. Approximately 13% of GFP+ cells from each group were c-kit+, and 0.5% were c-kit+, Sca-1+. Intracellular staining of GFP+ cells for survivin indicated that 72.3% ± 3.0% and 54.9% ± 16.3% of human and mouse transduced cells, respectively, were survivin positive compared with 33.9% ± 1.3% of vector control cells, whereas 14.5% ± 1.2% of antisense-mouse survivin–transduced cells were positive for survivin (Figure 4A). Western analysis of human or mouse survivin-transduced marrow cells confirmed elevated expression of survivin, while reduced survivin was observed in cells transduced with an antisense-mouse survivin (Figure4A, insert). The number of proliferating CFU-GM was increased 1.7 to 6.2-fold in marrow cells transduced with human (301% ± 66% increase, P < .005, 4 experiments) or mouse (285% ± 79% increase, P < .01, 4 experiments) survivin, compared with cells transduced with vector (Figure 4B). Transduction of mouse marrow cells with an antisense-mouse survivin construct decreased total CFU-GM by 69% ± 5% (P < .05, 4 experiments). Analysis of the proportion of CFU-GM in S phase indicated that 78.9% ± 1.8% (P < .05; 3 experiments) and 77.6% ± 3.0% (P < .01, 3 experiments) of CFU-GM transduced with human or mouse survivin, respectively, were in S phase, compared with vector control (55.0% ± 5.0%) (Figure 4C). Transduction of marrow cells with antisense-mouse survivin reduced the proportion of S-phase CFU-GM to 42.1% ± 4.7% (P < .05, 3 experiments). Delayed addition of growth factors for 24 and 48 hours to vector or human (h)-survivin–transduced marrow cells resulted in progressive apoptosis of CFU-GM. However, after normalization for CFU-GM enhancement at time 0, 16% to 77% and 61% to 244% more CFU-GM (2 experiments) were observed in h-survivin–transduced cells after 24 and 48 hours of delayed growth factor addition, respectively, than in vector control cultures, suggesting that survivin overexpression blocked CFU-GM apoptosis caused by cytokine starvation.
Retrovirus transduction of IRES-EGFP control (control), human survivin IRES-EGFP (h-survivin), mouse survivin IRES-EGFP (m-survivin), and antisense survivin-IRES-EGFP (AS-m-survivin) into primary mouse bone marrow cells.
(A) GFP-positive bone marrow mononuclear cells in each transduced group were FACS sorted and the percentage of survivin-positive cells was determined by flow cytometry after staining with PE-antisurvivin antibody. Data are expressed as means ± SEM from 2 experiments. Western analysis for survivin in GFP+mononuclear cells from each transduced group is shown in the insert. The same filter was stripped and reprobed with antihuman actin antibody as a loading control. Cross-reactivity of this antibody to mouse has been validated by the manufacturer. (B) CFU-GM production in transduced mouse bone marrow cells. Ten thousand GFP-positive marrow mononuclear cells were cultured in soft agar with 10 ng/mL rmGM-CSF and 50 ng/mL rmSCF and CFU-GM quantitated after 7 to 10 days at 37°C, 5% CO2, 5% O2 in air. The average number and SEM of CFU-GM from triplicate plates of 4 individual experiments are shown. Combined data from all 4 experiments are shown in the insert. Retrovirus harboring human survivin was used in experiments 1, 3, and 4. Mouse survivin was used in experiments 2, 3, and 4. Vector backbone and antisense-mouse survivin were used in all 4 experiments. *P < .005; **P < .001. (C) The proportion of CFU-GM in S phase of the cell cycle was determined by thymidine suicide with high specific activity [3H]thymidine. Data are the averages ± SEM of 3 independent experiments.
Retrovirus transduction of IRES-EGFP control (control), human survivin IRES-EGFP (h-survivin), mouse survivin IRES-EGFP (m-survivin), and antisense survivin-IRES-EGFP (AS-m-survivin) into primary mouse bone marrow cells.
(A) GFP-positive bone marrow mononuclear cells in each transduced group were FACS sorted and the percentage of survivin-positive cells was determined by flow cytometry after staining with PE-antisurvivin antibody. Data are expressed as means ± SEM from 2 experiments. Western analysis for survivin in GFP+mononuclear cells from each transduced group is shown in the insert. The same filter was stripped and reprobed with antihuman actin antibody as a loading control. Cross-reactivity of this antibody to mouse has been validated by the manufacturer. (B) CFU-GM production in transduced mouse bone marrow cells. Ten thousand GFP-positive marrow mononuclear cells were cultured in soft agar with 10 ng/mL rmGM-CSF and 50 ng/mL rmSCF and CFU-GM quantitated after 7 to 10 days at 37°C, 5% CO2, 5% O2 in air. The average number and SEM of CFU-GM from triplicate plates of 4 individual experiments are shown. Combined data from all 4 experiments are shown in the insert. Retrovirus harboring human survivin was used in experiments 1, 3, and 4. Mouse survivin was used in experiments 2, 3, and 4. Vector backbone and antisense-mouse survivin were used in all 4 experiments. *P < .005; **P < .001. (C) The proportion of CFU-GM in S phase of the cell cycle was determined by thymidine suicide with high specific activity [3H]thymidine. Data are the averages ± SEM of 3 independent experiments.
Discussion
The IAP proteins are the only known endogenous caspase inhibitors that suppress apoptosis.47 The IAPs XIAP, c-IAP1, and c-IAP2 directly bind to and inhibit caspases 3, 7, and 9 via their BIR domains.48 In this report, we demonstrate that in addition to survivin, XIAP, c-IAP1, and c-IAP2 are expressed in CD34+ cells; however, survivin is the only cytokine-regulated IAP in these cells and therefore is the only likely IAP mediating suppression of apoptosis by hematopoietic cytokines. Similarly, survivin is the only IAP up-regulated by CD40 ligation in B-CLL cells.49 The relationship between survivin and other antiapoptotic molecules that play a role in hematopoietic cells, such as Bcl2,50,51 is not known. Both proteins are up-regulated in breast cancer cells,52 and their expression can be regulated by the tumor suppressor gene p53.52 53 Whether there is a direct link between the Bcl2 family members and survivin or, given the importance of apoptosis, that their effects are independent, remains to be determined.
Since the original reports that cancer cells and embryonic tissues, but not normal adult tissues, express survivin,10,15,54 others and we have described survivin expression in normal adult cells. Survivin expression is not observed in resting endothelial cells but is up-regulated in a cell cycle–dependent manner55 by vascular endothelial growth factor55,56 or angiopoietin-1.57 The murine homolog of survivin,Tiap, is induced in T lymphocytes activated by mitogens.11 We previously reported that survivin is expressed and growth factor regulated in normal adult CD34+cells and T lymphocytes in all phases of cell cycle.18Because it is apparent that survivin is expressed in proliferating cells, the questions of whether survivin expression in CD34+ cells is cytokine regulated or simply reflects cell division and cell cycle progression and whether survivin expression affects proliferation and cell cycle of normal hematopoietic cells are raised. To address these questions, we examined survivin expression in CD34+ cells relative to the cell cycle markers Ki-67, cyclin D, and underphosphorylated-Rb and in Hstlow, PYlow, CFSEbright G0CD34+ cells relative to Ki-67 expression. We also investigated the effects of PI3-kinase/AKT and MAPKp42/p44pathway inhibitors on survivin expression and cell cycle in CD34+ cells. Finally, we introduced survivin cDNA into primary mouse bone marrow cells and examined the effects of modulating survivin expression on cell cycle and proliferation of myeloid progenitor cells. Up-regulation of survivin expression in CD34+ cells by hematopoietic growth factors was coincident with up-regulation of phosphorylated Rb, D cyclins, and Ki-67, indicating that survivin expression parallels cell cycle progression. Multivariate intracellular staining of survivin, Rb, Ki-67, cyclin D, and DNA in CD34+ cells upon growth factor stimulation demonstrated that survivin is up-regulated not only in underphosphorylated-Rbnegative, cyclin Dpositive, and KI-67positive cells, but also in the underphosphorylated-Rbpositive,5,38-40,58-60cyclin Dnegative,33-35 and Ki-67negative33,34,36 37 cells. Isolation of G0 CD34+ cells sorted based on Hoechst 33342/Pyronin Y and staining with CFSE before incubation with growth factors indicated that survivin mRNA and protein are up-regulated in cytokine-stimulated G0 CD34+ cells that had not yet up-regulated Ki-67 and had not yet divided. These data demonstrate that survivin is up-regulated in CD34+ cells by growth factors during G0 before cells enter G1 and that survivin expression is specifically regulated by growth factors in CD34+ cells and not merely a consequence of cell cycle progression.
We have previously shown that like cancer cells, survivin expression in CD34+ cells is highest during G2/M.18 Our present study clearly demonstrates that unlike cancer cells, survivin expression is up-regulated in quiescent CD34+ cells following growth factor stimulation before cell cycle entry. This raises the question of whether survivin expression in quiescent CD34+ cells is unique to normal hematopoietic cells. Murine survivin (TIAP) mRNA has been demonstrated in quiescent T cells, though its expression was low.11 In addition, survivin up-regulation in synchronized NIH3T3 cells following 12 hours' serum stimulation was observed, where greater than 90% of the cells still remained in G0/G1 without any increase in S + G2/M cells.11 These studies indicate that survivin expression is observed before cell cycle entry in nonhematopoietic cells. Li et al, who reported the specific expression of survivin during G2/M in HeLa cells, used cell cycle synchronization and Northern and Western analyses15to demonstrate that treatment of HeLa cells with mimosine reduced survivin expression coincident with G1 arrest. Failure to detect survivin expression in G1-arrested cells might be due to the methodology employed. We used real-time RT-PCR and intracellular flow cytometry to demonstrate G0 expression of survivin mRNA and protein, which are more sensitive. Furthermore, survivin inhibition by mimosine could be due to a direct effect on survivin expression rather than an effect on cell cycle arrest.
The selective PI3-kinase/AKT pathway inhibitor LY294002 and MAPKp42/p44 pathway inhibitor PD98059 significantly blocked up-regulation of survivin expression by growth factors in CD34+ cells. Survivin protein expression decreased in the absence of any effect on cell cycle progression or cell viability, at least during the initial 8 hours, although there was reduction of cell cycle progression after longer exposure. Most importantly, these compounds inhibited up-regulation of survivin expression before an arresting effect on cell cycle was observed. If survivin expression were a result of cell cycle progression, reduction of survivin would be accompanied by cell cycle arrest. Although we did not analyze changes in the proportion of G0 and G1 cells, these data strongly suggest that survivin expression in CD34+cells is regulated by growth factors and is not merely a consequence of cell cycle progression. These findings also indicate that both the PI3-kinase/AKT and MAPKp42/p44 pathways are involved in cytokine regulation of survivin expression in normal CD34+cells, which is consistent with the involvement of these pathways in regulating survivin expression in AML cells.46
Overexpression of human or mouse survivin cDNA in primary mouse bone marrow cells dramatically enhanced CFU-GM proliferation and the proportion of CFU-GM in S phase of the cell cycle. An antisense-mouse survivin construct had the opposite effect. These findings indicate that modulating survivin expression modulates proliferation and cell cycle of primary hematopoietic progenitor cells. Although we did not quantitate survivin protein levels in survivin or antisense survivin-transduced CFU-GM due to technical limitations, an increase in survivin-positive marrow cells could be quantitated by flow cytometry following transduction with human or mouse survivin. A decrease in survivin-positive cells was observed following transduction with antisense mouse survivin. A similar trend in survivin protein expression in transduced primary marrow cells was observed by Western analysis. In addition, following transduction of these same MIEG3 constructs into BaF/3 cells, elevated protein levels of h-survivin (237%-466% increase, 3 experiments), mouse (m)-survivin (118%-287% increase, 3 experiments), and antisense-m–survivin (9%-25% decrease in survivin protein levels, 3 experiments) were observed. Survivin has been shown to interact with cdk4, and overexpression of survivin enhances Rb phosphorylation in hepatoma cells by releasing p16INK4a and p21WAF1/CIP1from the cdk4/p16INK4a and cdk4/p21WAF1/CIP1complexes, respectively.4 22-24 This suggests that survivin promotes cell cycle progression by inactivating the p16INK4a/Rb pathway, a finding consistent with our observation that overexpression of survivin enhances the cell cycle rate of primary mouse CFU-GM. Because survivin is an antiapoptotic protein, it is possible that the enhanced CFU-GM proliferation observed following survivin transduction results from increased CFU-GM survival. This is consistent with reduced apoptosis of survivin-transduced CFU-GM observed in cultures in which growth factor addition was delayed. However, this does not adequately explain the increase in the proportion of S-phase CFU-GMs observed following survivin transduction. There is no reason to assume that enhanced CFU-GM survival means that the additional surviving cells are in S phase.
In conclusion, we have provided evidence demonstrating that survivin expression in CD34+ cells is growth factor regulated and not a consequence of cell cycle progression. First, survivin is up-regulated in CD34+ cells that are underphosphorylated Rbpositive, Ki-67negative, and cyclin-Dnegative, that is, quiescent cells. Second, survivin up-regulation by growth factors occurs in G0CD34+ cells before these cells enter G1. Third, inhibition of survivin by selective PI3-kinase/AKT and MAPKp42/44 pathway inhibitors occurs before cell cycle arrest. Finally, survivin overexpression enhances and antisense survivin reduces cell cycle rate and proliferation of CFU-GM. Taken together, these data demonstrate that survivin expression is specifically regulated by growth factors in quiescent CD34+cells and strongly suggest that up-regulation of survivin is an early and important event for cell cycle entry and proliferation of normal hematopoietic stem and progenitor cells.
The authors thank Suzan Rice for sorting cells by flow cytometry, Charlie Mantel for helpful suggestions, and Hui-min Bian, Jonathan Pelus, and Jessie Mindel for technical assistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Louis M. Pelus, Walther Oncology Center, Indiana University School of Medicine, 1044 W Walnut St, Indianapolis, IN 46202; e-mail: lpelus@iupui.edu.

![Fig. 2. Expression of survivin protein and mRNA in G0 CD34+ cells isolated by Hoechst 33342/pyronin Y staining before and after 12 hours' growth factor stimulation. / (A) Hoechst 33342/pyronin Y staining of UCB CD34+cells before and after incubation for 12 hours with 100 ng/mL each Tpo, SCF, and FL. The left panel represents the gate for fresh G0 CD34+ cells (R1). Fresh G0 cells (R1) were incubated with Tpo, SCF, and FL for 12 hours, restained with Hoechst 33342/pyronin Y, and G0 cells isolated by FACS sorting (middle panel; gate R2). In replicate cultures, R1 cells were prestained with CFSE before incubation, and cell division in the unseparated cell population was analyzed after 12 hours' culture (right panel). Data represent 1 of 5 identical experiments. (B) Postsort analysis of Hoechst 33342/pyronin Y and CFSE staining on cells from the R1 and R2 gates in Figure 2A. Data represent 1 of 5 identical experiments. The R1 and R2 gates were set so that the 20% dimmest pyronin Y cells were collected for fresh and cultured CD34+cells. The gates for the postsort analysis were adjusted for the characteristic progressive loss of pyronin Y fluorescence with time.2733 (C) Intracellular Ki-67 protein expression and DNA staining of fresh G0 cells (R1) and G0 cells isolated after culture for 12 hours with growth factors (R2) from Figure 2A. The percentage of cells negative for Ki-67 expression, that is, below isotype staining (horizontal bar), is shown. Data represent 1 of 3 independent experiments. (D) Intracellular survivin protein expression in fresh G0 CD34+cells (R1) and in G0 cells (R2) from Figure 2A, harvested after 12 hours' incubation with growth factors. Mean channel fluorescence (MCF) of survivin and the percentage of cells staining positive (above isotype control [horizontal bar]) for survivin are shown below each blot. Data represent 1 of 3 identical experiments. (E) Survivin expression was quantitated in the Ki-67–negative fraction of fresh or cultured G0 cells from Figure 2C. The MCF for survivin and the percentage of survivin-positive cells (above isotype control [horizontal bar]) are shown beneath the blot. Data represent 1 of 3 identical experiments. (F) Total RNA from G0 cells before (R1) and after (R2) growth factor incubation was subjected to real-time RT-PCR to quantify survivin and Ki-67 mRNA expression. GAPDH was used as the internal control. Because Ki-67 expression of both samples was extremely low, G1CD34+ cells were used to verify the PCR reaction of Ki-67. The y-axis represents ΔRn, which indicates the magnitude of the signal generated at each cycle. The x-axis shows the reaction cycle. Data represent 1 of 2 identical experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/7/10.1182_blood.v100.7.2463/3/m_h81923215002.jpeg?Expires=1767840979&Signature=JLYqF6iryvYQ0HzqWBwy0gJs6BA2UpHnL2RKVqwdvAzww4GSvO8-Q~BYraBp5kp9wU8u66egAGq~2WnJYzABK7X18CfA-oQ6pP1STK-CEOm~P6oZHHa-vxKpwUMTomk9OhRd2SVE8PlxKY9mb-dichjuzYOcUDVlcR8hnBxPslV6MZP3z~ZJAA9tE1VnZ2Ez7Y4l5IpOoD4gb5wStU9DeTs8Xuc0W~q4GGQ3jS-uQQBqWFhwAmrCaA18ikG9E7aBK~Qek3iuWtlj56Pr4z5N1mur2LQMGoF19KKUwA2InOs0V0INC8LJYWir8pumFi3fFJUOm5lrmZF0IFpHknR4Yw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Retrovirus transduction of IRES-EGFP control (control), human survivin IRES-EGFP (h-survivin), mouse survivin IRES-EGFP (m-survivin), and antisense survivin-IRES-EGFP (AS-m-survivin) into primary mouse bone marrow cells. / (A) GFP-positive bone marrow mononuclear cells in each transduced group were FACS sorted and the percentage of survivin-positive cells was determined by flow cytometry after staining with PE-antisurvivin antibody. Data are expressed as means ± SEM from 2 experiments. Western analysis for survivin in GFP+mononuclear cells from each transduced group is shown in the insert. The same filter was stripped and reprobed with antihuman actin antibody as a loading control. Cross-reactivity of this antibody to mouse has been validated by the manufacturer. (B) CFU-GM production in transduced mouse bone marrow cells. Ten thousand GFP-positive marrow mononuclear cells were cultured in soft agar with 10 ng/mL rmGM-CSF and 50 ng/mL rmSCF and CFU-GM quantitated after 7 to 10 days at 37°C, 5% CO2, 5% O2 in air. The average number and SEM of CFU-GM from triplicate plates of 4 individual experiments are shown. Combined data from all 4 experiments are shown in the insert. Retrovirus harboring human survivin was used in experiments 1, 3, and 4. Mouse survivin was used in experiments 2, 3, and 4. Vector backbone and antisense-mouse survivin were used in all 4 experiments. *P < .005; **P < .001. (C) The proportion of CFU-GM in S phase of the cell cycle was determined by thymidine suicide with high specific activity [3H]thymidine. Data are the averages ± SEM of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/7/10.1182_blood.v100.7.2463/3/m_h81923215004.jpeg?Expires=1767840979&Signature=OG~VgbmKDM0kjrtquZuRmFrDOIldoT-8fBZoXRO5MeQPU87Res-k-EZ5RuQf5PIqdCaMLiVTNDRZ-gHAe6YfOKEZEFMopLxiOJoDUbh0rjim-9Cq~T5fLNeMkwz2iBumTQnmu5F6jcUmiow9eoidlX5HXrGRdg4jHeWfZMk5RlKv61j31g6cfjkUbnpDddDKE9bJxgh9t8m87dIFZzgqM-maVCgnHjwbIQeDiumk~txEIcI4AmkGN-hG4GeBJsAqof3x5oFJvCEd3niVgw2KBvYYzeM5c7aQddVvspx5hnbeJSdOET3Q8rI3mGkkCEJDag-4Q5~~L-ynzwYTQVYftg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal