Monocyte chemoattractant protein (MCP)–3 is inactivated upon cleavage by the matrix metalloproteinase (MMP) gelatinase A (MMP-2). We investigated the susceptibility to proteolytic processing of the 4 human MCPs by 8 recombinant MMPs to determine whether MCP-3 is an isolated example or represents a general susceptibility of chemokines to proteolytic inactivation by these important inflammatory proteases. In addition to MMP-2, MCP-3 is efficiently cleaved by membrane type 1 (MT1)–MMP, the cellular activator of MMP-2, and by collagenase-1 and collagenase-3 (MMP-1, MMP-13) and stromelysin-1 (MMP-3). Specificity was shown by absence of cleavage by matrilysin (MMP-7) and the leukocytic MMPs neutrophil collagenase (MMP-8) and gelatinase B (MMP-9). The closely related chemokines MCP-1, MCP-2, and MCP-4 were not cleaved by MMP-2 or MT1-MMP, but were cleaved by MMP-1 and MMP-3 with varying efficiency. MCPs were typically cleaved between residues 4 and 5, but MCP-4 was further processed at Val7-Pro8. Synthetic MCP analogs corresponding to the MMP-cleaved forms bound CC chemokine receptor (CCR)–2 and CCR-3, but lacked chemoattractant activity in pre-B cells transfected with CCR-2 and CCR-3 or in THP-1 monocytic cells, a transformed leukemic cell line. Moreover, the truncated products of MCP-2 and MCP-4, like MCP-3, were potent antagonists of their cognate CC chemokine receptors in transwell cell migration assays in vitro. When they were injected 24 hours after the initiation of carrageenan-induced inflammation in rat paws, their in vivo antagonist activities were revealed by a greater than 66% reduction in inflammatory edema progression after 12 hours. We propose that MMPs have an important role in modulating inflammatory and immune responses by processing chemokines in wound healing and in disease.
Introduction
Chemokines are potent chemoattractant cytokines that are produced locally in tissues and direct the migration and homing of leukocytes. Tissue gradients of inflammatory chemokines attract and maintain inflammatory cells at sites of host challenge in infection, inflammation, and cancer.1 Chemokines can be divided into families according to the position and spacing of N-terminal cysteine residues. Presently, the C, CC, CXC, and CX3C families are recognized,2 with more than 54 human chemokines currently identified. The monocyte chemoattractant proteins (MCPs) of the CC family consist of 4 proteins termed MCP-1, MCP-2, MCP-3, and MCP-4 (CCL-2, CCL-8, CCL-7, and CCL-13, respectively) that target multiple leukocyte subsets (monocytes, basophils, eosinophils, dendritic cells, and natural killer cells) whereas, in the initial phases of inflammation, CXC chemokines attract polymorphonuclear leukocytes. Therefore, chemokines are important mediators of many pathologies, including chronic inflammatory and autoimmune diseases where the coordinated expression of MCPs and resultant leukocyte infiltration correlate with disease progression.3 4
An important question in the pathogenesis of inflammation and disease is how chemoattractant signals are squelched to restrict new cell recruitment and to promote clearance of the inflammatory infiltrate as a prelude to tissue resolution. Proteolysis of chemokines may provide a mechanism for loss of chemoattractant signaling. Indeed, production of proteolytically processed chemokines in cell culture has been reported.5-8 We have demonstrated an N-terminal–truncated form of MCP-3 in human rheumatoid synovial fluid that had been cleaved at Gly4-Ile5, and the effects of engineered truncated chemokines on cell behavior have been characterized.9 Hence, the identification of proteinases that may process chemokines in vivo is important, but as yet poorly characterized. Very recently, the matrix metalloproteinase (MMP) gelatinase A (MMP-2) was shown to process the N-terminus of MCP-3 and stromal cell–derived factor-1α (SDF-1) and β.7,8 Prior to these studies, considerable emphasis was placed on the role of the serine protease dipeptidyl peptidase IV/CD 26,10 which can process many bioactive molecules with a penultimate N-terminal proline residue, including chemokines. The serine proteinase cathepsin G was also recently reported to process SDF-1α.11 However, the activity of these serine proteases in chemokine cleavage and inactivation in vivo has not been demonstrated.
MMPs are either secreted or cell-membrane–bound proteinases with broad substrate specificity that have traditionally been proposed to degrade most components of the extracellular matrix12 although in vivo evidence for this is generally lacking. Matrix proteolysis is a hallmark of inflammation with MMPs considered to be important effectors of this process and also essential for leukocyte extravasation and migration. In particular, leukocyte-derived collagenase–2 (MMP-8) and gelatinase B (MMP-9) are the prominent early proteolytic mediators of matrix degradation and allow effector cell egress to the site of tissue damage.13,14 Following this initial leukocyte proteolytic phase, stromal cells, in response to proinflammatory cytokines secreted by the cellular infiltrate, produce MMPs that amplify the acute tissue-destructive phase. Notably, interstitial collagenase (MMP-1), stromelysin-1 (MMP-3), matrilysin (MMP-7), and collagenase-3 (MMP-13) have been suggested to play important roles in inflammatory tissue destruction.12 Hence, persistence of the inflammatory infiltrate results in continued tissue destruction that can progress to chronic inflammatory disease. Although MMPs may play a role in extracellular matrix degradation in inflammation, it is gradually becoming appreciated that these proteases have a wider substrate repertoire that includes many bioactive molecules. Indirectly, the MMP-mediated inactivation of serpins15,16and chemokines7,8 can modulate healing with direct biological effects of MMP cleavage manifested by the activation and release of tumor necrosis factor–α (TNF–α),17fibroblast growth factor receptor,18Fas-ligand,19,20 and α-defensin.21 The MMP-mediated cleavage and inactivation of the complement protein mannose binding lectin22 was also very recently reported. MMP-9 is known to activate IL-8 and to cleave and inactivate SDF-1.8 23 Hence, MMP activity on nontraditional substrates may exert a biological impact on inflammatory processes as profound as those of MMP tissue-degradation effects.
We have recently reported the use of the yeast 2-hybrid system to screen for potential MMP substrates. Exosite scanning resulted in the identification of MCP-3 as a binding protein and substrate for MMP-2.7 This was a specific interaction, as MMP-2 did not cleave MCP-1, MCP-2, or MCP-4. We have also very recently identified SDF-1α and SDF-1β as MMP substrates that are cleaved by a number of MMPs, including MMP-2.8 Because MMP substrate specificity overlaps to form a robust extracellular proteolytic system, we investigated in the present report whether other MMPs also process MCP-3 and if the MCP family is generally susceptible to proteolysis by MMPs. Here we establish that MCP-1, MCP-2, MCP-3, and MCP-4 are all processed by multiple MMPs to typically remove an N-terminal tetrapeptide, resulting in reduced agonist activity and conversion to CC chemokine receptor (CCR) antagonists that are effective both in vitro and in vivo. Hence, the chemokine superfamily represents an emerging and important class of substrates for MMPs that, when cleaved by these inflammatory proteases, have novel modulatory effects in regulating inflammatory processes.
Materials and methods
Production of MMPs and MCPs
Recombinant human MMPs were expressed in Chinese hamster ovary (CHO) cells and purified from the conditioned CHO-SFM culture medium.8 Briefly, conditioned culture medium was collected and enzymatic activity was purified by gelatin-sepharose chromatography for gelatinases. The other MMPs were purified by combinations of heparin-agarose and dye-ligand chromatography as appropriate.24 Membrane type–1 MMP (MT1-MMP [MMP-14]) was produced as a soluble, transmembrane segment–deleted form. Proteolytic activity was confirmed by cleavage of quenched fluorescent peptide substrates and inhibition by the tissue inhibitor of metalloproteinase-1 (TIMP-1).25 Human MCP-1, MCP-2, MCP-3, MCP-4, and truncated derivatives were chemically synthesized by means of solid-phase methods; the polypeptides were purified by reverse-phase high-performance liquid chromatography (HPLC) and folded by means of air oxidation as previously described.26Homogeneity and fidelity of synthesis were confirmed by electrospray ionization mass spectrometry.
Electrospray ionization mass spectrometry
Chemokine samples were introduced into the mass spectrometer through a microbore PLRP column (1 × 50 mm) on the Michrom HPLC system (Michrom BioResources, Auburn, CA) (solvent system: 20% to 100% solvent B [0.045% trifluoroacetic acid, 80% acetonitrile in water] for 10 minutes; 100% solvent B for 2 minutes). The quadrupole mass analyzer (in the single-quadrupole mode) was scanned over a mass-to-charge ratio range of 300 to 10 000 d with a step size of 0.5 d and a dwell time of 1 millisecond per step. The ion source voltage was set at 5 kV, and the orifice energy was 80 V. Protein molecular weights were determined from these data by means of the deconvolution software supplied by P E Sciex (Thornhill, ON, Canada).
Proteinase assays
P-aminophenylmercuric acetate (APMA)–activated MMP (1 ng) and 1 μg chemokine were mixed in buffer (100 mM NaCl, 5 mM CaCl2, 20 mM Tris, pH 8.0) and incubated at 37°C. Aliquots were removed at 1-hour intervals, and product accumulation was monitored by densitometric analysis of Coomassie-stained Tris-tricine sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels. The overall catalytic rate/Km(kcat/Km) specificity constant was calculated from graphical determination of the kobs.7 Electrospray ionization mass spectrometry of the reaction products was used to confirm cleavage, and the data were deconvoluted to identify the scissile bond.
Chemotaxis assays
Cell migration of THP-1 monocytic leukemia cells (ATCC, Manassas, VA) or a pre-B cell line (murine B300 19; ATCC) stably transfected with either human CCR-2 or human CCR-3 (designated B300-CCR2 and B300-CCR3, respectively) was evaluated in disposable 12-well Transwell polystyrene trays (Corning Costar, Cambridge, MA) across a polycarbonate membrane with 5-μm size pores. Full-length and N-terminal–truncated synthetic chemokines corresponding to the MMP-cleaved form of the chemokines were diluted in Hepes-buffered RPMI 1640 supplemented with 10 mg/mL bovine serum albumin. Chemokine samples were added to the lower chamber, and THP-1 cells or B19 300 cell transfectants (1 × 107 cells per milliliter), in the same media without the MCPs, were added to the upper chamber. After 3-hour incubation at 37°C in 5% CO2 humidified atmosphere, migrated cells in the lower chamber were counted. Migrated cells in 5 separate fields per well from duplicate wells were enumerated on a hemacytometer by means of light microscopy.
125I-labeling of MCP-1 and measurement of chemokine receptor binding affinities
The binding affinity (dissociation constant [Kd]) of MCP-2, MCP-2(5-76), MCP-4, and MCP-4(8-75) was determined by competition for 125I-labeled MCP-1 on CCR-2 and Scatchard analysis. The 125I-labeling of MCP-2 and MCP-4 resulted in precipitation of the chemokines, which could not be used for binding assays. MCP-1 (10 μg) was labeled with 9.25 × 105 Bq (250 μCi) 125I-Bolton Hunter reagent (NEN Life Science Products, Boston, MA) at 4°C for 30 minutes in 0.1 M borate buffer, and the reaction was terminated by incubation in 0.1 M glycine. The labeled MCP-1 was separated from free125I-Bolton Hunter reagent by Sephadex G-25 size-exclusion chromatography. Binding assays were performed by means of CCR-2–transfected pre-B cells (5 × 106) suspended in 200 μL HEPES-buffered RPMI 1640 media, 10 mg/mL bovine serum albumin, and 0.1% NaN3. The 125I–MCP-1 (4 nM) and increasing concentrations of unlabeled competitor were added and incubated for 30 minutes. Cell-bound 125I–MCP-1 was separated from the unbound 125I–MCP-1 by pelleting the cells through a 2:3 mixture of diacetylphthalate and dibutylphthalate. The specifically bound counts per minute in the cell pellet were calculated after subtracting the nonspecifically bound counts per minute (counts per minute bound in the presence of 100-fold molar excess of unlabeled competitor) and dividing by the total counts per minute bound in the absence of competitor. Assays were performed in duplicate, and the experiments repeated.
Rat paw inflammation model
Male Wistar rats (200 to 225 g) (Charles River Breeding Farms, Montreal, QC, Canada) were housed in a biocontainment facility according to the Canadian Animal Care Committee with the use of institutionally approved animal care protocols. Inflammation was induced in rat paws by intraplantar injection of 1% lambda carrageenan (Sigma, St Louis, MO) in sterile saline (total volume, 100 μL) under halothane anesthesia. Paw volumes were measured by means of a hydroplethysmometer (Ugo Basile, Milan, Italy) before and 24 hours after injection of carrageenan and the induction of inflammation.27 After 24 hours, the rats were anesthetized with halothane, and an intraplantar injection of chemokine analogs in sterile saline (50 μg in 50 μL) or saline alone was administered according to a double-blind protocol. Five rats were used per treatment, and the experiment was repeated 2 or 3 times for each peptide. At 12 hours after peptide administration, paw volumes were measured in a randomized sequence by an observer unaware of the treatments. Changes in paw volume were then calculated by subtracting the final paw volume from the volume at the 24-hour time point.
Statistical analysis
Statistical analysis was performed by nonparametric (Kruskal-Wallis) analysis of variance followed by a Dunn multiple comparison test.
Results
MCP-3 is cleaved by multiple MMPs
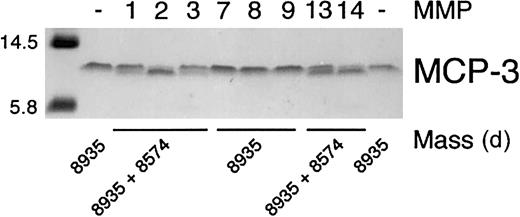
The specificity of 8 MMPs that have been associated with wound healing12 28 in the proteolysis of MCP-3 was determined. Incubation of the chemokine with recombinant MMP-1, MMP-2, MMP-3, MMP-13, and MT1-MMP resulted in a small but distinct increase in electrophoretic mobility on Tris-tricine gels, whereas MMP-7, MMP-8, and MMP-9 did not, even with prolonged incubation (Figure1). Cleavage of full-length chemokine was confirmed by electrospray ionization mass spectroscopic identification of the new lower molecular mass product (Figure 1). Deconvolution of the mass spectrometry data revealed that the MCP-3 scissile bond for each proteolytically competent MMP was Gly4-Ile5 (Table1). We designated this cleavage product MCP-3(5-76). MMP-mediated processing of MCP-3 was efficient, as shown by the high turnover rates (Table 1), with MMP-2 and MT1-MMP showing highest activity.
Specificity of MMP cleavage of MCP-3.
Tris-tricine gel analysis and masses obtained by electrospray ionization mass spectrometry of MCP-3 cleavage products produced by MMP activity. APMA-activated MMP (1 ng) was incubated with 1 μg MCP-3 in CAB assay buffer at 37°C for 8 hours. Aliquots were taken for mass spectroscopic analysis, and the reaction was stopped by the addition of SDS-PAGE sample buffer prior to electrophoresis. The mass of the molecular weight markers is indicated.
Specificity of MMP cleavage of MCP-3.
Tris-tricine gel analysis and masses obtained by electrospray ionization mass spectrometry of MCP-3 cleavage products produced by MMP activity. APMA-activated MMP (1 ng) was incubated with 1 μg MCP-3 in CAB assay buffer at 37°C for 8 hours. Aliquots were taken for mass spectroscopic analysis, and the reaction was stopped by the addition of SDS-PAGE sample buffer prior to electrophoresis. The mass of the molecular weight markers is indicated.
Analysis of MCP-3 cleavage by recombinant MMPs
| MMP . | kcat/Km(M−1s−1) . | Measured mass of MCP-3, d* . |
|---|---|---|
| MMP-1 | 2600 | 8574 |
| MMP-2 | 8000 | 8574 |
| MMP-3 | 2700 | 8574 |
| MMP-7 | 0 | 8935 |
| MMP-8 | 0 | 8935 |
| MMP-9 | 0 | 8935 |
| MMP-13 | 2600 | 8574 |
| MT1-MMP | 6000 | 8574 |
| Buffer control | 0 | 8935 |
| MMP . | kcat/Km(M−1s−1) . | Measured mass of MCP-3, d* . |
|---|---|---|
| MMP-1 | 2600 | 8574 |
| MMP-2 | 8000 | 8574 |
| MMP-3 | 2700 | 8574 |
| MMP-7 | 0 | 8935 |
| MMP-8 | 0 | 8935 |
| MMP-9 | 0 | 8935 |
| MMP-13 | 2600 | 8574 |
| MT1-MMP | 6000 | 8574 |
| Buffer control | 0 | 8935 |
Eight MMPs were assayed for their efficiency at processing MCP-3 after incubation for up to 18 hours at 37°C. For buffer control, chemokine in assay buffer was incubated in the absence of MMP for 18 hours at 37°C. Graphical analysis of time course reactions to monitor production of cleaved MCP-3 was used to estimate the kcat/Km values. Also shown is the mass of the reaction products as determined by electrospray ionization mass spectroscopy.
Mass of full-length (uncleaved) MCP-3 = 8935 d. A mass of 8574 d corresponds exactly to MCP-3(5-76) with a cleavage site at Gly4-Ile5.
Multiple MMPs cleave MCP-1, MCP-2, and MCP-4
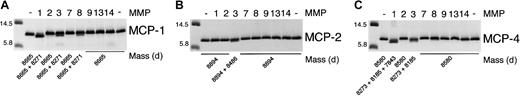
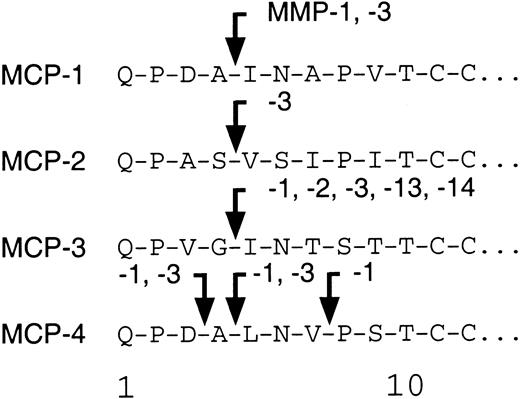
We tested the ability of the MMPs to cleave MCP-1, MCP-2, and MCP-4. As found before,7 MMP-2 was highly specific for MCP-3 with no activity against MCP-1, MCP-2, or MCP-4. Figure2 shows that MCP-1 is cleaved at Ala4-Ile5, designated MCP-1(5-76); by MMP-1; to a lesser extent (20% of total by densitometric analysis) by MMP-3; and to a very minor extent (5% of total) by MMP-8. In contrast, MCP-2 was susceptible to cleavage only by MMP-3 at Ser4-Val5, designated MCP-2(5-76). Interestingly, MCP-4 was processed at 2 sites, Asp3-Ala4 and Ala4-Leu5, by MMP-3. Both sites were also susceptible to MMP-1 activity, with proteolysis proceeding by a further cleavage at Val7-Pro8, designated MCP-4(8-75). Cleavage site susceptibility for MCP-1, MCP-2, MCP-3, and MCP-4 is summarized schematically in Figure 3.
MMP cleavage of MCP-1, MCP-2, and MCP-4.
Tris-tricine gel analysis and masses obtained by electrospray ionization mass spectrometry of MCP-1, MCP-2, and MCP-4 cleavage products produced by MMP activity. APMA-activated MMP (1 ng) was incubated with 1 μg MCP in CAB buffer at 37°C for 8 hours. Reaction was stopped by the addition of SDS-PAGE sample buffer. The mass of the molecular weight markers is indicated.
MMP cleavage of MCP-1, MCP-2, and MCP-4.
Tris-tricine gel analysis and masses obtained by electrospray ionization mass spectrometry of MCP-1, MCP-2, and MCP-4 cleavage products produced by MMP activity. APMA-activated MMP (1 ng) was incubated with 1 μg MCP in CAB buffer at 37°C for 8 hours. Reaction was stopped by the addition of SDS-PAGE sample buffer. The mass of the molecular weight markers is indicated.
MMP cleavage sites of MCP-1, MCP-2, and MCP-4.
A schematic summarizing the sites of MMP-mediated processing of the 4 human MCPs.
MMP cleavage sites of MCP-1, MCP-2, and MCP-4.
A schematic summarizing the sites of MMP-mediated processing of the 4 human MCPs.
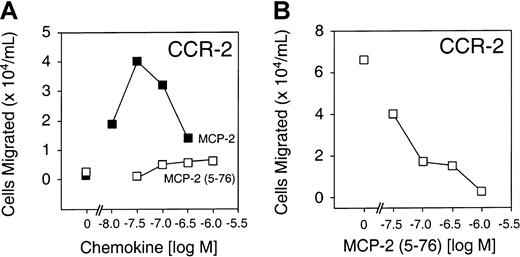
MCP-2(5-76) is a CCR-2 receptor antagonist
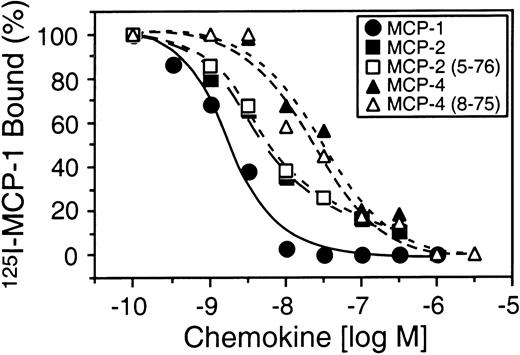
To compare the biological activity of full-length MCP-2 and MCP-2(5-76), we performed chemotaxis assays using synthetically prepared MCP-2(5-76) and a pre-B cell line stably transfected with the predominant MCP-2 chemokine receptor CCR-2. Figure4A shows that, unlike full-length MCP-2, MCP-2(5-76) was inactive as a CCR-2 agonist at all concentrations up to 10−6 M in the chemotaxis assay. However, MCP-2(5-76) retained receptor-binding affinity (Kd, 4.5 nM) very similar to that for MCP-2 (Kd, 4.4 nM) (Figure 5) and was effective as a CCR-2 antagonist in cell migration assays (Figure 4B).
Receptor activity of full-length MCP-2 and MCP-2(5-76).
(A) Cell migration assays of B300-CCR2 showing loss of receptor agonist activity of MCP-2(5-76) compared with full-length chemokine. Cells that migrated in the absence of chemokine are indicated at 0 M. (B) Cell migration assay of B300-CCR2 in the presence of full-length MCP-2 (30 nM) and phosphate-buffered saline vehicle alone (0 M) or increasing concentrations of MCP-2(5-76), as indicated. Mean values of duplicate assays are shown.
Receptor activity of full-length MCP-2 and MCP-2(5-76).
(A) Cell migration assays of B300-CCR2 showing loss of receptor agonist activity of MCP-2(5-76) compared with full-length chemokine. Cells that migrated in the absence of chemokine are indicated at 0 M. (B) Cell migration assay of B300-CCR2 in the presence of full-length MCP-2 (30 nM) and phosphate-buffered saline vehicle alone (0 M) or increasing concentrations of MCP-2(5-76), as indicated. Mean values of duplicate assays are shown.
Cell binding of MCP-2(5-76) and MCP-4(8-75).
CCR-2 binding of 125I–MCP-1 in the presence of increasing amounts of unlabeled MCP-1, MCP-2, MCP-4, or vehicle, and synthetic analogs corresponding to MCP-2(5-76), the MMP-cleaved form of MCP-2, and corresponding to MCP-4(8-75), the MMP-cleaved form of MCP-4. Mean values of duplicate assays are shown.
Cell binding of MCP-2(5-76) and MCP-4(8-75).
CCR-2 binding of 125I–MCP-1 in the presence of increasing amounts of unlabeled MCP-1, MCP-2, MCP-4, or vehicle, and synthetic analogs corresponding to MCP-2(5-76), the MMP-cleaved form of MCP-2, and corresponding to MCP-4(8-75), the MMP-cleaved form of MCP-4. Mean values of duplicate assays are shown.
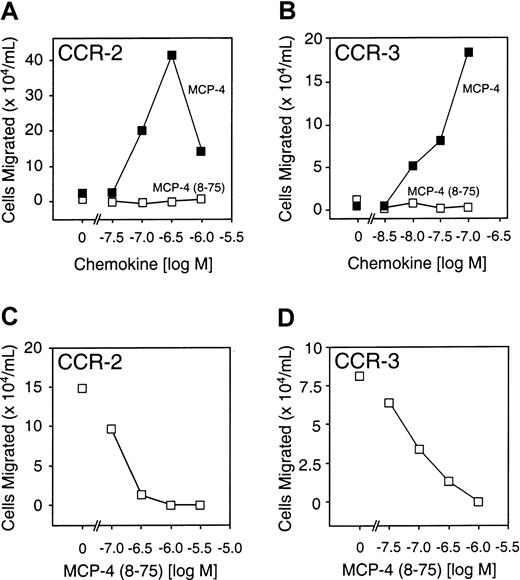
MCP-4(8-75) is a CCR-2 and CCR-3 receptor antagonist
The agonist and antagonist activity of MCP-4(8-75) was compared with that of MCP-4 with the use of 2 pre-B cell lines stably transfected with either CCR-2 or CCR-3, to which full-length MCP-4 binds with high affinity. Compared with the full-length chemokine, MCP-4(8-75) was unable to direct chemotaxis in either cell line (Figure6A,B). Like the MMP-cleaved forms of MCP-2 and MCP-3, MCP-4(8-75) was a potent receptor antagonist of both CCR-2 and CCR-3 (Figure 6C,D). MCP-4(8-75) was calculated to bind CCR-2 with a Kd of 14 nM, an affinity very similar to that of the full-length form (Kd, 16 nM) (Figure 5).
Receptor activity of full-length MCP-4 and MCP-4(8-75).
(A) (B) Cell migration assays of B300-CCR2 (panel A) or B300-CCR3 (panel B) showing activity of MCP-4(8-75), MCP-4, or vehicle alone (0 M). (C) Cell migration assay of B300-CCR2 showing, as indicated, MCP-4 (300 nM) in the presence of increasing concentrations of MCP-4(8-75) or MCP-4 alone—ie, with 0 M MCP-4(8-75). (D) Cell migration assay of B300-CCR3 in response, as indicated, to full-length MCP-4 (100 nM) and in the presence of increasing concentrations of MCP-4(8-75) or MCP-4 alone, ie, with 0 M MCP-4(8-75). Mean values of duplicate assays are shown.
Receptor activity of full-length MCP-4 and MCP-4(8-75).
(A) (B) Cell migration assays of B300-CCR2 (panel A) or B300-CCR3 (panel B) showing activity of MCP-4(8-75), MCP-4, or vehicle alone (0 M). (C) Cell migration assay of B300-CCR2 showing, as indicated, MCP-4 (300 nM) in the presence of increasing concentrations of MCP-4(8-75) or MCP-4 alone—ie, with 0 M MCP-4(8-75). (D) Cell migration assay of B300-CCR3 in response, as indicated, to full-length MCP-4 (100 nM) and in the presence of increasing concentrations of MCP-4(8-75) or MCP-4 alone, ie, with 0 M MCP-4(8-75). Mean values of duplicate assays are shown.
THP-1 cell migration in response to MCP-2(5-76) and MCP-4(8-75)
As a model of inflammatory monocytes, we used THP-1 monocytic cells, a transformed leukemic cell line that expresses CCR-2, to further assess the effects of MMP cleavage of MCP-2 and MCP-4. Whereas the full-length MCPs directed chemotaxis across transwell filters, MCP-2(5-76) and MCP-4(8-75) showed no chemotactic activity (Figure7A,B), but both were receptor antagonists as evident by the dose-dependent reduction in THP-1 cell chemotaxis (Figure 7C,D), confirming the previous results using the pre-B cells transfected with CCR-2.
The effects of MMP-cleavage of MCP-2 and MCP-4 on THP-1 monocytic cell chemotaxis.
(A) (B) Cell migration assay of THP-1 cells in response to MCP-2 and MCP-2(5-76) (panel A) or MCP-4 and MCP-4(8-75) (panel B), as indicated. Cells migrated in the absence of chemokine are shown at 0 M chemokine. Mean values of duplicate assays are presented. (C) (D) Antagonist assays are shown for MCP-2(5-76) (panel C) and MCP-4(8-75) (panel D). Chemotaxis of THP-1 cells in response to 30 nM MCP-2 or 100 nM MCP-4 in the absence and presence of MMP-cleaved chemokine analog at the indicated concentrations is shown in the corresponding panels. Mean values of duplicate assays are presented.
The effects of MMP-cleavage of MCP-2 and MCP-4 on THP-1 monocytic cell chemotaxis.
(A) (B) Cell migration assay of THP-1 cells in response to MCP-2 and MCP-2(5-76) (panel A) or MCP-4 and MCP-4(8-75) (panel B), as indicated. Cells migrated in the absence of chemokine are shown at 0 M chemokine. Mean values of duplicate assays are presented. (C) (D) Antagonist assays are shown for MCP-2(5-76) (panel C) and MCP-4(8-75) (panel D). Chemotaxis of THP-1 cells in response to 30 nM MCP-2 or 100 nM MCP-4 in the absence and presence of MMP-cleaved chemokine analog at the indicated concentrations is shown in the corresponding panels. Mean values of duplicate assays are presented.
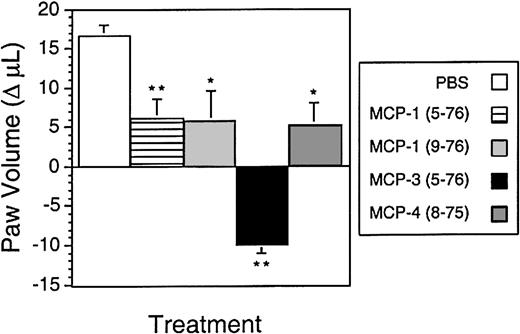
MMP-cleaved MCPs reduce inflammatory edema in vivo
To determine the biological effects of MMP cleavage of MCPs in vivo, the extent of inflammatory edema induced in a rat paw model following injection with carrageenan was measured after administration of synthetic N-terminal–truncated MCPs corresponding to the MMP-cleaved forms. Results are shown in Figure8 as the change in paw volume from the 24-hour time point, when the chemokines were injected, to the 36-hour postcarrageenan time point. Without chemokine, the paw volume continued to increase from the 24-hour value (1.80 ± 0.02 mL) as the inflammatory response progressed. MCP-1(5-76) and MCP-4(8-75) showed an approximately 66% reduction in the increase in inflammatory edema from 24 to 36 hours compared with the vehicle control (Figure 8). An engineered form of MCP-1(9-76), a potent CCR-2 antagonist,9 was found to reduce the paw volume to a similar extent. Most notable though was MCP-3(5-76), which reduced the paw volume 1.6-fold to below that at the time of injection. Hence, MMP inactivation of MCPs also generates CCR antagonists that retain cellular binding affinity and that modulate cell migration in vitro and dampen the inflammatory response to carrageenan in vivo.
Anti-inflammatory effects of MMP-cleaved MCP analogs in vivo.
Rat paws were injected with carrageenan at time 0. After 24 hours, the volume of the paws was measured, and the paws were injected with either the indicated chemokine analog (50 μg, equivalent dose 250 μg/kg) or phosphate-buffered saline alone. Shown is the change in paw volume from 24 to 36 hours. The negative value for MCP-3(5-76) indicates a reduction in volume at 36 hours compared with 24 hours. *Group differed significantly from vehicle group (P < .05). **Group differed significantly from vehicle (P < .01). Data presented as the mean ± SD; n = 5 per group.
Anti-inflammatory effects of MMP-cleaved MCP analogs in vivo.
Rat paws were injected with carrageenan at time 0. After 24 hours, the volume of the paws was measured, and the paws were injected with either the indicated chemokine analog (50 μg, equivalent dose 250 μg/kg) or phosphate-buffered saline alone. Shown is the change in paw volume from 24 to 36 hours. The negative value for MCP-3(5-76) indicates a reduction in volume at 36 hours compared with 24 hours. *Group differed significantly from vehicle group (P < .05). **Group differed significantly from vehicle (P < .01). Data presented as the mean ± SD; n = 5 per group.
Discussion
MMPs are implicated in many physiological processes involving matrix turnover,12,28 but direct evidence for this is limited.29 In contrast, MMPs are clearly identified with pathologies, including arthritis and tumor metastasis30where they have been assumed to primarily play a matrix degradative role. However, a broader MMP substrate degradome is now being revealed by recent studies (reviewed in Overall31) that show proteolytic susceptibility of signaling proteins such as MCP-3,7 SDF-1α and SDF-1β,8TNF-α,17 and Fas ligand.19,20 This indicates an equally important role for MMP activity in regulating inflammation and immune processes in which proteolytic targets include molecules involved in information networks. In view of the efficient cleavage of MCP-3, but not MCP-1, MCP-2, or MCP-4, by MMP-2,7 it was important to determine whether processing and inactivation of chemokines by MMPs are limited to a few isolated examples or are more general phenomena that may have broad biological implications for many pathological and physiological processes.
Because MMPs are differentially expressed in many tissues, in many cell types, and during inflammation and healing, we determined the specificity of 8 MMPs against the CC chemokines MCP-1, MCP-2, MCP-3, and MCP-4. In addition to MMP-2, MMP-1, MMP-3, MMP-8, MMP-13, and MT1-MMP cleave MCPs and all characteristically perform this at position 4-5, but the MMPs are not always functionally interchangeable and each MCP showed a different profile of proteolytic susceptibility. The pattern of MMP proteolysis of the MCP family is striking. MMP-3, a protease with broad extracellular matrix substrate specificity (seewww.clip.ubc.ca/mmps.shtm for a comprehensive updated list of MMP substrates), can cleave all MCP chemokines, but MMP-2, MMP-13, and MT1-MMP are active only on MCP-3. The similar specificity of MMP-2 and MT1-MMP is interesting because MT1-MMP is the major cellular activator of proMMP-2, and both proteases assemble and function together in activation complexes on the cell surface.25,33,35 Most notable was the general inability of MMP-7 and of the primarily leukocytic enzymes MMP-8 and MMP-9 to process MCPs. Although MMP-8 could cleave MCP-1, catalytic efficiency was very low. MMP-9 can activate IL-8 by N-terminal processing, but does not cleave MCP-2,36 or any other MCP as shown here. It is important to distinguish between efficient and precise proteolytic processing, as shown for these chemokines, and the more general catabolic actions of proteases during protein degradation. MMP-9 has been reported to slowly degrade the chemokines PF-4, GRO-α, and CTAP-III, with the degraded products lacking activity.23 Although in vitro biochemical assays do not replicate in vivo conditions and are often not performed in the presence of extracellular matrix components that may modulate protease activity, the slow turnover rates in vitro of PF-4, GRO-α, and CTAP-III, even at high enzyme-to-substrate ratios,23 indicate that these may be poor MMP-9 substrates in vivo, reducing the physiological relevance of any such activity of MMP-9.
In addition to the amino acid sequence of the substrate, auxiliary elements of substrate specificity are pivotally important, including substrate binding by exosites on the MMP hemopexin C domain.7,31 The absence of a hemopexin C domain in MMP-7 may explain the lack of activity against these chemokines, and the hemopexin C domain of MMP-9 is suggested to be positioned away from the catalytic domain by an extended carbohydrate-rich linker peptide,34 possibly accounting for its lack of ability to process the N-terminus of the MCPs. Therefore, the same position of the MMP cleavage site in MCPs reported here and SDF-1α and SDF1-β8 suggests that chemokine processing is a more general property of MMPs and not an isolated example limited to MCP-3 by MMP-2, first reported by McQuibban et al.7
Neoepitope antibodies have been used to show the generation of MCP-3(5-76) in human arthritis in vivo.7 Processing of SDF-1α has been observed by monocytic U937 cells in culture8 and MCP processing reported in cell culture following treatment with cytokines5,36 and concanavalin A,7 which activates the MMP-2 zymogen, conditions that model the degradative aspects of inflammation. The studies by Proost et al36 revealed that N-terminal truncations of MCP-1 and MCP-2 occurred in cell culture to generate MCP-1(5-76), MCP-1(6-76), and MCP-2(6-76), with C-terminal truncations also occurring: MCP-1(1-69) and MCP-2(1-74), but the proteinases responsible were not identified. From our data, it is likely that the in vitro production of MCP-1(5-76) observed was due to MMP activity.
Previous protein engineering studies9,26,32 have demonstrated that modification of the N-terminus of MCP chemokines alters receptor binding and activation. In this report, we have shown that MMP processing of MCP-1, MCP-2, MCP-3, and MCP-4 reduces cell migration of THP-1 monocytic leukemia cells and pre-B cells transfected with CCR-1, CCR-2, and CCR-3. Cell surface receptor binding of the cleaved chemokines still occurred with only slightly alteredKds. Thus, MMP-processing of MCP-2, MCP-3, and MCP-4 generated stable and potent CCR-1, CCR-2, and CCR-3 receptor antagonists. The antagonistic effects are due to competition for receptor occupancy, with subsequent desensitization the result of ligated receptor trafficking from the cell surface to endosomes.9 Although primary cultures of monocytes were not tested for the effects of MMP cleavage on chemotaxis, the potential for the in vivo generation of N-terminally truncated forms of MCPs by MMPs that may modify inflammatory and immune responses was demonstrated with the use of THP-1 monocytic cells.
Our previous characterization of synthetic MCP-1(5-76),9corresponding to the MMP-cleaved form generated here, revealed that MCP-1(5-76) exhibited a 10-fold reduction in receptor agonist activity. MCP-1(5-76) bound CCR-2 with a Kd of 20 nM and desensitized the receptor to subsequent chemokine treatment.9 Although MCP-1(5-76) can be detected as a weak agonist in sensitive in vitro assays, this can be outweighed in vivo by its antagonistic properties, as evident by the reduction in inflammatory edema 12 hours after its injection into inflamed paw pads. Comparable effects were found for MCP-4(8-75) and for MCP-2(5-76). The effectiveness of MCP-3(5-76), a broad-spectrum CCR antagonist, in reducing inflammation in vivo in other models7 previously suggested the potential importance of generating a broad-spectrum chemotactic antagonist for CCR-1, CCR-2, and CCR-3 in modulating inflammatory processes. Our present data support this hypothesis. Compared with the other MCP antagonists, which cover a more restricted CCR spectrum, MCP-3(5-76) was found to have the strongest anti-inflammatory effects, reducing the paw volume to below that at the time of injection: MCP-3(5-76) not only prevented new edema, but resolved some of the pre-existing inflammatory exudate present after 24 hours of inflammation. The cleavage and conversion of MCP-1, MCP-2, and MCP-4 to additional antagonists are likely to augment this response in different diseases and processes. Hence, the pathophysiological cleavage of MCPs by MMPs reduces CCR agonist activity and generates effective antagonist derivatives that may regulate inflammatory and immune processes in vivo. However, whether a connection exists between MMP activity and the modulation of CCR-1–, CCR-2–, and CCR-3–dependent cellular responses in disease remains to be elucidated and is currently under investigation in our laboratories.
The physiological likelihood of in vivo chemokine cleavage, specifically MCP-3, was revealed by the high kinetic turnover rates (Table 1). Notably, MMP-2 and MMP-14 were the most efficient at cleaving MCP-3 in vitro, but interestingly neither cleaved MCP-1, MCP-2, or MCP-4. This points to a unique role for MCP-3 cleavage by MT1-MMP and MMP-2. MT1-MMP is the physiological activator of MMP-2 and forms a cell-surface receptor for this enzyme.35 MT-MMPs are also critical for cell migration in collagen,37 an important feature of remodeling wounds. We have previously postulated that modulation of the MMP-2/MT1-MMP proteolytic axis at the cell surface changes the proteolytic profile of stromal cells from collagenolytic (predominantly manifested by MT1-MMP activity) to gelatinolytic (by MMP-2) during the conversion from a cytokine-stimulated resorptive cell to a matrix-depositing cell.31 33 Proteolysis of MCP-3 by MT1-MMP and MMP-2 recruited to the cell surface in both the collagenolytic and gelatinolytic phases of tissue remodeling is likely to also be augmented by the action of collagenase and stromelysin, which may process MCP-1, MCP-2, and MCP-4 during the tissue resorptive phase, to reduce chemokine activity directly and indirectly by creating CCR-1, CCR-2, and CCR-3 antagonist gradients. Although TIMPs present in the tissue reduce net proteolytic activity by MMPs, particularly at the interface between the inflammatory lesion and normal tissue, TIMP-2 binding to the hemopexin C domain of MMP-2 interferes with neither MCP-3 binding and cleavage nor MMP-2 activation by MT1-MMP (G.A.M., C.M.O., unpublished data, May 2000).
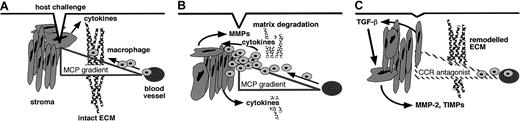
Our data support the following model (Figure9), which connects the activity of chemokines and MMPs in the stages that define the inflammatory reaction. Chemoattractant-directed leukocytes secrete MMPs, predominantly MMP-8 and MMP-9 but not MMP-2,38-40 that may degrade matrix and promote migration but do not cleave MCPs. As the inflammatory reaction progresses, cytokines such as interleukin-1 and TNF-α are also released by macrophages.38 The stromal cells respond by secreting MMPs and expressing cell surface MT1-MMP, which contributes toward the bulk removal of matrix. In addition, we propose that an important function of MMPs is to regulate the inflammatory response by processing MCPs to reduce agonist activity and to form CCR-1, CCR-2, and CCR-3 antagonist gradients. Ultimately this would deplete the cellular infiltrates of leukocytes expressing these receptors. In the tissue-resolution phase, transforming growth factor (TGF)–β1 elevates extracellular matrix deposition and stabilizes new matrix by repressing MMP and stimulating TIMP expression.41,42 TGF-β1 also stimulates MMP-2 levels,41 the most efficient MMP in the cleavage and inactivation of MCP-3. This may maintain cleaved MCP-3 levels despite the reduction in collagenase41 and stromelysin42 expression. Thus, MMPs not only are effectors of the inflammatory response but are important for its regulation.
Model of MMP regulation of chemokine action and inflammation.
(A) In response to host challenge, cytokines are released, and a chemotactic gradient is bound in the matrix to attract leukocytes from the vasculature. (B) Infiltration requires matrix proteolysis, predominantly by leukocyte-derived MMP-8 and MMP-9. As the cellular infiltrate builds, growth factors and cytokines are released by infiltrating cells and induced stromal cells. Stromal cells respond to autocrine and paracrine cytokine stimulation by increasing MMP expression. (C) In addition to clearance of matrix protein fragments, MMP activity converts MCP chemoattractants to CCR-1, CCR-2, and CCR-3 antagonistic derivatives, which both disrupt retention and reduce recruitment of the cellular components of the inflammatory infiltrate. With time, general MMP expression is suppressed, concomitant with increased MMP-2 and TIMP levels under the influence of transforming growth factor–β1 (TGF-β1), which orchestrates the final remodeling of the extracellular matrix.
Model of MMP regulation of chemokine action and inflammation.
(A) In response to host challenge, cytokines are released, and a chemotactic gradient is bound in the matrix to attract leukocytes from the vasculature. (B) Infiltration requires matrix proteolysis, predominantly by leukocyte-derived MMP-8 and MMP-9. As the cellular infiltrate builds, growth factors and cytokines are released by infiltrating cells and induced stromal cells. Stromal cells respond to autocrine and paracrine cytokine stimulation by increasing MMP expression. (C) In addition to clearance of matrix protein fragments, MMP activity converts MCP chemoattractants to CCR-1, CCR-2, and CCR-3 antagonistic derivatives, which both disrupt retention and reduce recruitment of the cellular components of the inflammatory infiltrate. With time, general MMP expression is suppressed, concomitant with increased MMP-2 and TIMP levels under the influence of transforming growth factor–β1 (TGF-β1), which orchestrates the final remodeling of the extracellular matrix.
The present and recent data from our group7 showing the efficient cleavage of MCPs 1, 2, 3, and 4 and SDF-1α and SDF-1β8 by MMPs indicate that the chemokine superfamily represents a large and biologically important new class of MMP substrate and that MCP-3 cleavage by MMP-2 is not a rare example. Notably, activity always includes cleavage at position 4-5, indicating that this site in other chemokines may also be susceptible to MMP processing. There are approximately 54 human chemokines presently identified,43 and we have initiated a comprehensive screen to determine chemokine susceptibility to MMP cleavage. It is difficult to predict from sequence analysis alone which chemokines will also be processed because we have found that both scissile bond sequence and precise domain structure contribute to the proteolytic susceptibility of the chemokine (G.A.M., C.M.O., unpublished data, May 2000). Overall, our data suggest that inflammatory-induced MMP proteolysis of chemokines may contribute to the normal dissipation of inflammatory cell infiltrates, which in turn results in reduced MMP expression in a homeostatic feedback loop—aberrations of which may contribute to chronic inflammatory pathologies.
We thank Shouming He for technical assistance in mass spectrometry analysis.
Supported by grants from the Canadian Arthritis Network of Centers of Excellence, the National Cancer Institute of Canada with funds provided in part by the Canadian Cancer Society, and the Canadian Institutes for Health Research. G.A.M. is supported by a National Cancer Institute of Canada Studentship; I.C.-L. is supported by a Canadian Institutes for Health Research Scientist Award; and C.M.O. is supported by a Canadian Research Chair in Metalloproteinase Biology.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Christopher M. Overall, 2199 Wesbrook Mall, Vancouver, BC, V6T 1Z3, Canada; e-mail: chris.overall@ubc.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal