Abstract
The myeloid cell–specific expression and interferon-γ (IFN-γ) induction of Fc γ receptor I (FcγRI) requires cooperation between PU.1 and signal transducer and activator of transcription 1 (Stat1) by means of mechanisms that are unknown. We found that PU.1 and Stat1 mediated distinct functions in the activation of FcγRI promoter. The basal activity of the natural FcγRI promoter was strictly dependent on PU.1, and IFN-γ induction required both PU.1 and Stat1. Recruitment of TATA-binding protein (TBP) to the FcγRI promoter did not replace PU.1 in promoter activation, suggesting that TBP is not sufficient for FcγRI activation and that PU.1 mediates additional contacts with basal transcription machinery. In contrast, Stat1 did not interact with basal transcription machinery, but the Stat1-mediated activation of FcγRI promoter critically required CREB-binding protein (CBP)/p300. These results define functional cooperativity between PU.1 and Stat1 in FcγRI promoter activation, in which PU.1 appears to serve as a bridging factor with the basal transcription machinery and IFN-γ–mediated induction of transcription occurs through recruitment of CBP/p300 by Stat1.
Introduction
Interferon-γ (IFN-γ)–induced gene responses are largely dependent on signal transducer and activator of transcription 1 (Stat1). Stat1 is phosphorylated on tyrosine 701 by Jak kinases in the receptor complex, and the phosphorylated Stat1 dimers translocate to the nucleus and bind to specific regions (γ-activated sequence [GAS]) on IFN-γ–responsive promoters.1 2
Regulation of gene expression is conferred by combinatorial use of various enhancer element–binding transcription factors and their cooperation in recruitment of general transcription factors (GTFs). The activation domains of transcription factors may function by interacting directly with GTFs, or they may employ coactivator proteins to facilitate transcription. Stat1 has been shown to cooperate with Sp1, AP1, and nuclear factor-κB transcription factors,3and Stat1-mediated transcription has been shown to involve histone acetyltransferase CREB-binding protein (CBP)/p300 as well as p300/CBP-interacting protein coactivators.3-5However, most of the studies related to Stat-mediated transcriptional activation have used artificial promoters in the context of heterologous promoters and have therefore not identified the mechanisms by which Stats are connected to basal transcription machinery.
An important cell-type–specific target for IFN-γ is the high-affinity receptor for IgG (Fc γ receptor I [FcγRI]),6 which mediates phagocytosis, respiratory burst, and antibody-mediated cytotoxic reactions. FcγRI is constitutively expressed on myeloid cells, but its expression is rapidly induced by IFN-γ. FcγRI promoter contains 2 ciselements within the 190 nucleotides upstream of the translation initiation site that confer the IFN-γ inducibility and myeloid cell–specific expression of the gene. The IFN-γ response region (GRR) binds Stat1, and myeloid expression requires a PU.1 (Spi-1)-binding element.7-9
PU.1, which belongs to the Ets family of transcription factors, regulates several genes participating in the development and function of myeloid and B cells.10 11 The function of PU.1 in cell differentiation has been shown to involve cooperation with transcription factors such as acute myelogenous leukemia 1, CCAAT/enhancer-binding protein β (C/EBPβ), C/EBPδ, and Myb, but the role of PU.1 in cytokine-induced transcriptional activation and its cooperation with Stat factors is unknown. The requirement for both PU.1 and Stat1 in transcriptional activation of FcγRI provided an experimental system for investigating the specific roles of Stat1 and PU.1 in cytokine-induced transcriptional activation.
Study design
Plasmids
Stat1, PU.1, and IFN regulatory factor–GAS–luc reporter were characterized previously.12,13 The other reporters were constructed by inserting fragments from the FcγRI promoter, made by polymerase chain reaction (PCR) from human genomic DNA, into promoterless pLuc vector14 or vector containing TATA box from IFN-β promoter.15 The 189/66-ΔGAS-TATAluc was constructed by mutating CAG (from −142 to −140) to ACT to disrupt the Stat1-binding site. The TCC sequence from the PU.1-binding site (from −94 to −92) was mutated to GAA to yield 89/66-ΔPU.1-TATAluc. The 189/66-ΔGASΔPU.1-luc harbors both mutations. Wild-type E1A, Δ conserved region 1 (ΔCR1), and ΔCR2,16 were subcloned into pCI-neo vector and tagged with C-terminal HA epitope. E1AR2G mutant17 and E1A ΔN (amino acids 45-243) were generated by PCR.
Cell culture and transfections
Mouse macrophage RAW 264.7 cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% fetal-calf serum (FCS). Human fibrosarcoma U3A cells18 were cultured in DMEM with 10% Cosmic calf serum. U3A cells were transfected by using the calcium phosphate coprecipitation method. RAW264.7 cells were transfected by electroporation with a GenePulser apparatus (250 V and 960 μF; Bio-Rad; Hercules, CA).
Reporter gene assays
RAW 264.7 cells (6 × 106) were transfected with 6 μg reporter and 4 μg pCMV–β-galactosidase plasmid. After electroporation, cells were divided on 4 3.5-cm plates per transfection and incubated for 24 hours. Cells were starved overnight in DMEM with 1% FCS, left untreated or treated for 6 hours with 20 ng/mL murine IFN-γ, and lysed in reporter lysis buffer (Promega, Madison, WI). Luciferase activity was measured with a luminometer (1254 Luminova; ThermoLabsystems, Beverly, MA) and normalized against the β-galactosidase activity of the lysates. Reporter gene assays with U3A cells were done as described previously.12
Results and discussion
A heterologous TATA-containing promoter cannot substitute for the function of PU.1 in FcγRI activation
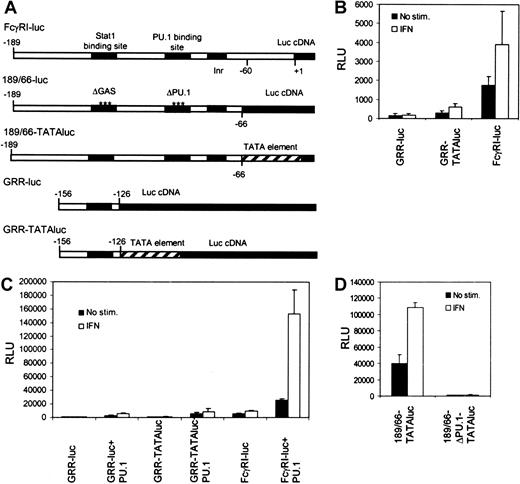
We investigated the mechanisms that underlie transcriptional regulation in IFN-γ signaling in the context of natural FcγRI promoter. A typical feature for most myeloid-specific genes, including FcγRI, is the lack of TATA element. Instead, these promoters contain an initiator sequence, and PU.1 or Stat1 might function by recruiting TBP and allowing assembly of the preinitiation complex. PU.1 was found to interact with TBP, as was shown previously,19 whereas no interaction between Stat1 and TBP was detected (S.A., unpublished data, March 2001). To analyze the functional importance of TBP recruitment in activation of the FcγRI promoter, we inserted the Stat1-binding element GRR of the FcγRI promoter (−156 to −126) into a luciferase construct with or without a TATA-box element from the IFN-β promoter (GRR-TATAluc and GRR-luc, respectively). In RAW264.7 cells, the GRR-luc construct was inactive (Figure1B). The presence of TATA box in the context of GRR element resulted in low basal and IFN-γ–induced activation of the reporter. However, activation of GRR-TATAluc was significantly lower than that of FcγRI-luc. Similar results were obtained in Stat1- and PU.1-transfected U3A cells lacking endogenous Stat1 and PU.1 (Figure1C). These results indicate that the presence of TATA box in the promoter is not sufficient for efficient Stat1-mediated transcription.
Stat1-binding site alone is not sufficient to drive IFN-γ–inducible transcription of FcγRI gene.
(A) Schematic diagram of the reporter constructs. (B) RAW264.7 cells were transfected with different reporter constructs, and luciferase activity was measured after 6 hours of treatment with IFN-γ. (C) The same reporter constructs as in panel B were analyzed in U3A cells. U3A cells were transfected with 1 μg reporter, 0.5 μg β-galactosidase plasmid, 250 ng Stat1, and 50 ng PU.1 and either not stimulated or stimulated with IFN-γ for 6 hours before measurement of luciferase activity. (D) RAW264.7 cells were transfected with 189/66-TATAluc or 189/66-ΔPU.1-TATAluc and analyzed as in shown in panel B. Values shown (± SD) are from 3 different experiments.
Stat1-binding site alone is not sufficient to drive IFN-γ–inducible transcription of FcγRI gene.
(A) Schematic diagram of the reporter constructs. (B) RAW264.7 cells were transfected with different reporter constructs, and luciferase activity was measured after 6 hours of treatment with IFN-γ. (C) The same reporter constructs as in panel B were analyzed in U3A cells. U3A cells were transfected with 1 μg reporter, 0.5 μg β-galactosidase plasmid, 250 ng Stat1, and 50 ng PU.1 and either not stimulated or stimulated with IFN-γ for 6 hours before measurement of luciferase activity. (D) RAW264.7 cells were transfected with 189/66-TATAluc or 189/66-ΔPU.1-TATAluc and analyzed as in shown in panel B. Values shown (± SD) are from 3 different experiments.
To test the functional relation between PU.1-TBP recruitment and TATA box directly, we made a FcγRI promoter construct containing GRR and PU.1 elements in the presence of TATA box (189/66-TATAluc). The 189/66-TATAluc was active in RAW264.7 cells, but mutation of the PU.1 site completely abrogated its activity (Figure 1D). These results indicate that TATA box cannot substitute the function of PU.1 and that recruitment of TBP is not sufficient for FcγRI promoter activation. It is possible that TBP is not required for FcγRI activation. An example of such regulation is the activation of major histocompatibility complex class I by class II transcriptional activator, which occurs independently of TBP-associated factor II 250 and TATA box.20 Alternatively, PU.1 may be mediating other critical interactions in FcγRI promoter activation. This hypothesis was supported by the finding that RNA polymerase II interacted with PU.1 but not with Stat1 (J.Y., unpublished data, August 2001).
E1A inhibits IFN-γ–induced transcription of FcγRI
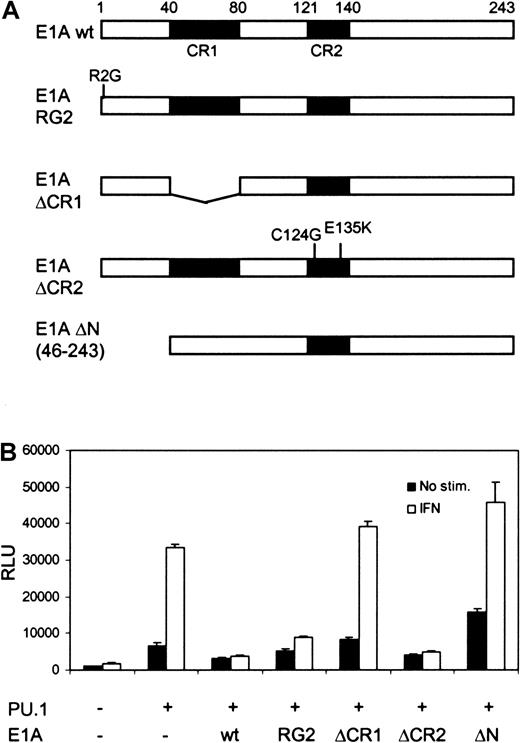
Both Stat1 and PU.1 have been shown to interact with CBP/p300,4,21 and recruitment of CBP/p300 to the promoter could be an underlying mechanism for their cooperation. To analyze the role of CBP/p300 in FcγRI activation, U3A cells were cotransfected with different E1A constructs, along with PU.1, Stat1, and FcγRI-luc reporter. Although E1A binds to and inhibits CBP/p300,16it only partly decreased basal PU.1-mediated transcription of FcγRI. In contrast, E1A completely abrogated IFN-γ–dependent transcription (Figure 2B). The specificity of E1A function was further demonstrated by using the mutant E1AR2G, which inhibits and interacts with Stat1 but not with CBP/p300.17 E1AR2G inhibited the IFN-γ–induced activity without affecting basal transcription. Additional E1A mutants lacking the ability to interact with CBP also failed to inhibit FcγRI promoter activity. E1A ΔCR1, which cannot bind CBP or retinoblastoma (Rb) protein, and E1A ΔN, which does not bind CBP/p300, did not inhibit transcription. E1A ΔCR2 (C124G, E135K), which restrained binding only to Rb, inhibited transcription in a manner similar to that of wild-type E1A. These results suggest that the function of Stat1 in transcriptional activation depends strictly on its ability to recruit CBP/p300.
The effect of E1A on basal and IFN-γ–induced transcription of FcγRI.
(A) Schematic diagram of the E1A constructs. (B) Equimolar amounts (determined by Western blotting) of E1A constructs were transfected into U3A cells together with FcγRI-luc, Stat1, and PU.1. Experimental conditions and DNA amounts were the same as in Figure 1C. Mean (± SD) normalized values from 3 experiments are shown.
The effect of E1A on basal and IFN-γ–induced transcription of FcγRI.
(A) Schematic diagram of the E1A constructs. (B) Equimolar amounts (determined by Western blotting) of E1A constructs were transfected into U3A cells together with FcγRI-luc, Stat1, and PU.1. Experimental conditions and DNA amounts were the same as in Figure 1C. Mean (± SD) normalized values from 3 experiments are shown.
Cytokine-induced transcription is assumed to require precise arrangement of promoter-binding transcription factors, hierarchical recruitment of coactivator proteins, and connection of these factors and proteins to the basal transcription machinery. The results reported here strongly suggest that PU.1 and Stat1 mediate distinct functions in transcriptional activation of the FcγRI promoter. PU.1 is responsible for basal activation and bridges with GTFs, whereas the function of Stat1 depends on CBP/p300 recruitment. It is likely that analogous functional coordination between different transcription factors to assemble GTFs and coactivator proteins to the promoter also occurs in other cytokine-responsive genes.
We thank Paula Kosonen for excellent technical assistance and Drs J. E. Darnell, T. Kouzarides, and I. Kerr for providing reagents.
Prepublished online as Blood First Edition Paper, April 30, 2002; DOI 10.1182/blood-2001-12-0236.
Supported by the Medical Research Council (Academy of Finland), the Medical Research Fund of Tampere University Hospital, the Finnish Foundation for Cancer Research, the Sigrid Juselius Foundation, and the Tuberculosis Foundation of Tampere.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Olli Silvennoinen, Institute of Medical Technology, University of Tampere, FIN-33014 Tampere, Finland; e-mail:olli.silvennoinen@uta.fi.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal