Abstract
Lymphomatoid papulosis (LyP) represents an intriguing cutaneous T-cell lymphoproliferative disorder with a histologic appearance resembling malignant lymphoma. This finding strongly contrasts with the benign clinical course of the disease. However, in 10% to 20% of cases, LyP can precede, coexist with, or follow malignant lymphoma. In these cases, the same T-cell population has been shown to be present in the LyP as well as in the associated lymphoma. In most LyP cases, there is—despite the sometimes extremely long course of the disease—no evolution of a secondary lymphoma. The investigation of these uncomplicated LyP cases for the presence of clonal T-cell receptor rearrangements has produced heterogeneous results. This might be explained by biologic or technical reasons arising from analyzing whole tissue DNA extracts. To definitively clarify whether the large atypical CD30+ cells in LyP without associated lymphoma all belong to the same clone or represent individually rearranged T cells, we analyzed the T-cell receptor–γ rearrangements of single CD30+ as well as of single CD30− cells isolated from 14 LyP lesions of 11 patients. By using this approach we could demonstrate that the CD30+ cells represent members of a single T-cell clone in all LyP cases. Moreover, in 3 patients the same CD30+ cell clone was found in anatomically and temporally separate lesions. In contrast, with only a few exceptions, the CD30− cells were polyclonal in all instances and unrelated to the CD30+ cell clone. Our results demonstrate that LyP unequivocally represents a monoclonal T-cell disorder of CD30+ cells in all instances.
Introduction
Lymphomatoid papulosis (LyP) occupies a highly interesting position within the spectrum of cutaneous lymphoproliferative disorders. The histologic picture of LyP is extremely variable and closely mimics that of malignant lymphoma.1 Three major histologic types have been recognized; these are designated as A, B, and C.2 3 Types A and C are characterized by the presence of large atypical blasts including mononucleated and binucleated or multinucleated cells resembling Reed-Sternberg cells characteristic of Hodgkin disease. The atypical blasts express one or more T-cell antigens as well as the lymphoid activation antigen CD30. While in type A these cells are embedded in a dense inflammatory background, in type C they form large sheets closely simulating CD30+ cutaneous anaplastic large cell lymphoma (ALCL). LyP type B is composed of small to medium-sized CD30− T cells showing epidermotropism, thus closely resembling classical mycosis fungoides.
The clinical features of LyP are more uniform and usually consist of repetitive episodes of self-healing papulonodular skin lesions often spread over a period of several years. From 10% to 20% of LyP cases are associated with malignant lymphoma, especially mycosis fungoides, CD30+ cutaneous ALCL, or Hodgkin disease, which can precede, coexist with, or follow LyP and can also appear in the lymph nodes.4-8 In many of these cases the same clonal T-cell receptor (TCR) rearrangements have been found in the LyP as well as in the associated lymphoma, identifying LyP as a precursor lesion.6 9-12
Most LyP cases are, however, not associated with T-cell lymphomas of various types.4,7 In these cases, clonal T-cell populations were detectable only in a proportion of analyzed lesions,13-16 which might, in addition, be different when taken at various times.13 Therefore, it was concluded that LyP represents in most instances a reactive skin disease that can, under certain circumstances, present as a localized clonal lymphoid disorder.15
Previous investigations were performed either by Southern blot analysis or by polymerase chain reaction (PCR) using DNA extracts from whole-tissue samples. However, this approach is often unable to detect small populations of clonally rearranged T cells residing in an abundant background of polyclonal cells, as frequently observed in LyP. Furthermore, the analysis of whole-tissue DNA does not allow for the assignment of the clonal rearrangements to a definite cell population.
To overcome these limitations, we have used an approved single-cell technique with which we recently successfully clarified the cellular origin and clonality of Hodgkin-Reed-Sternberg cells.17With this technique we isolated CD30+ as well as CD30− cells from 14 LyP lesions of 11 patients with LyP and analyzed their TCR-γ rearrangements. In addition, the results of single-cell analyses were compared with GeneScan analysis of TCR-γ PCR products from whole-tissue DNA extracts. With one exception, only patients without preceding or coexisting malignant lymphoma were included in our study. With this approach we were able to definitely answer the following questions: Do CD30+ cells in a single LyP lesion represent members of a single T-cell clone? Do CD30+ cells of anatomically and temporally separate lesions in the same patient belong to the same clone or to different clones? Is clonality exclusively restricted to the population of CD30+T cells? If not, do CD30− T cells also represent members of the CD30+ clone or of an own separate clone?
Materials and methods
Clinical data of the 11 cases of LyP are summarized in Table1. All 11 patients, 6 men and 5 women, presented a typical history of recurrent self-healing papulonodular eruptions.
Clinical features of 11 patients with LyP A
| Patient no. . | Age*/sex . | Duration* . | Site of biopsy . | Associated lymphoma . | Remarks . |
|---|---|---|---|---|---|
| 1† | 62/F | 1 y | A: Left groin | — | Trunk/groin: multifocal papular/nodular skin lesions that slowly subside; exclusion of systemic disease by computed tomography of thorax and abdomen; bone marrow biopsy |
| B: Left axilla | |||||
| 2† | 42/F | 10 y | C: Left flank | — | Involvement of extremities: multifocal papular/papulonecrotic skin lesions with partial spontaneous remission, good response to psoralen-UV/A therapy |
| D: Left thigh | |||||
| 3† | 76/F | 10 mo | E: ? | — | Trunk/extremities: disseminated papular lesions |
| F: Back | |||||
| 4 | 49/F | 4 mo | Right elbow | — | Elbows/thighs: papular/papulonecrotic lesions |
| 5 | 31/M | 1 mo | Abdomen | — | Abdomen: a single nodular lesion |
| 6 | 57/M | ? | Right knee | — | Trunk/extremities: disseminated papular lesions/plaques; coexisting small-plaque parapsoriasis |
| 7 | 82/M | 30 y | ? | Mycosis fungoides | Involvement of extremities: multifocal papular/papulonecrotic skin lesions; diagnosis of mycosis fungoides 28 y after the onset of LyP |
| 8 | 7/M | 3 mo | Abdomen | — | Trunk/extremities: disseminated self-healing papular lesions |
| 9 | 61/M | 2 mo | Right upper arm | — | Trunk/accentuation of extremities: multifocal papular/papulonecrotic lesions |
| 10 | 77/F | 6 mo | Right shoulder | — | Accentuation of the trunk: disseminated papular lesions |
| 11 | 24/M | 3 y | Left upper arm | — | Trunk/extremities: multifocal papular/papulonecrotic/nodular skin lesions that spontaneously subside |
| Patient no. . | Age*/sex . | Duration* . | Site of biopsy . | Associated lymphoma . | Remarks . |
|---|---|---|---|---|---|
| 1† | 62/F | 1 y | A: Left groin | — | Trunk/groin: multifocal papular/nodular skin lesions that slowly subside; exclusion of systemic disease by computed tomography of thorax and abdomen; bone marrow biopsy |
| B: Left axilla | |||||
| 2† | 42/F | 10 y | C: Left flank | — | Involvement of extremities: multifocal papular/papulonecrotic skin lesions with partial spontaneous remission, good response to psoralen-UV/A therapy |
| D: Left thigh | |||||
| 3† | 76/F | 10 mo | E: ? | — | Trunk/extremities: disseminated papular lesions |
| F: Back | |||||
| 4 | 49/F | 4 mo | Right elbow | — | Elbows/thighs: papular/papulonecrotic lesions |
| 5 | 31/M | 1 mo | Abdomen | — | Abdomen: a single nodular lesion |
| 6 | 57/M | ? | Right knee | — | Trunk/extremities: disseminated papular lesions/plaques; coexisting small-plaque parapsoriasis |
| 7 | 82/M | 30 y | ? | Mycosis fungoides | Involvement of extremities: multifocal papular/papulonecrotic skin lesions; diagnosis of mycosis fungoides 28 y after the onset of LyP |
| 8 | 7/M | 3 mo | Abdomen | — | Trunk/extremities: disseminated self-healing papular lesions |
| 9 | 61/M | 2 mo | Right upper arm | — | Trunk/accentuation of extremities: multifocal papular/papulonecrotic lesions |
| 10 | 77/F | 6 mo | Right shoulder | — | Accentuation of the trunk: disseminated papular lesions |
| 11 | 24/M | 3 y | Left upper arm | — | Trunk/extremities: multifocal papular/papulonecrotic/nodular skin lesions that spontaneously subside |
? indicates unknown.
At the moment of biopsy (in relation to the first biopsy).
Investigation of 2 anatomically and temporally separate biopsies.
Tissue samples and immunostaining
Fourteen tissue samples of 11 patients with clinical and biopsy-proven LyP were studied. The histologic diagnosis of LyP was confirmed by at least 3 independent experts (H.S., P.K., I.A.). Histologic classification was done for all biopsies and reviewed according to established criteria.3
Immunohistochemistry was performed, as previously described, by using the immunoalkaline phosphatase-antiphosphatase technique18and streptavidin-biotin technique. Antibodies against the following antigens were applied: CD30, CD2, CD3, CD4, CD5, CD8, CD20, TCR-β chain, perforin, granzyme B, and ALK1. To differentiate CD30− T cells, double staining using antibodies against CD30 and CD2 was carried out.
Isolation of single cells
Single CD30+ LyP cells as well as single CD30−/CD2+ cells were isolated from immunostained frozen tissue sections using a hydraulic micromanipulation device as previously described.17 In cases of insufficient CD30/CD2 double staining, 2 to 6 single CD30− cells were pooled and isolated from tissue sections stained with antibodies against CD30. Buffer aliquots covering the sections were aspirated as negative controls for PCR analysis. All cases were analyzed at least twice in completely independent cell isolation and PCR assays.
Single-cell PCR
Isolated cells were digested with proteinase K (1 hour, 50°C, 0.1 mg/mL) and, after heat denaturation of the enzyme, subjected to PCR. For the detection of TCR-γ rearrangements, 4 different sets of primers were established. For amplification of rearrangements involving Vγ1 to Vγ8 gene segments, 2 seminested PCRs were carried out in separate reactions employing a consensus Vγ1 to Vγ8 primer (VG-1) in conjugation with the Jγ-specific primers JGT1/2 and JGT3, respectively. For reamplification the VG-1 primer was replaced by a nested Vγ1 to Vγ8 consensus primer (VG-2) whereas the same Jγ-specific primers were used. The buffer (TaqGold [2 U] and TaqGold buffer [Perkin-Elmer], 1.37 mM MgCl2, 0.2 mM each deoxyribonucleoside triphosphate, 200 ng each primer) and cycling conditions (95°C for 30 seconds, 64°C [first 5 cycles] and 61°C [remaining 35 cycles] for 30 seconds, 72°C for 30 seconds) were as previously described.19
For the amplification and reamplification of the rearrangements involving Vγ9, Vγ10, or Vγ11 gene segments, we created new nested Vγ primers that were used in combination with the above-mentioned Jγ-specific primers. The annealing conditions for the detection of Vγ9 rearrangements were 5 cycles at 68°C (30 seconds) and 35 cycles at 64°C (30 seconds) for the first round of PCR and 40 cycles at 65°C for reamplification. The remaining buffer and cycling conditions were as described for the Vγ1 to Vγ8 amplification with the exception of 100 ng of each primer and 2.0 mM MgCl2. Vγ10 and Vγ11 rearrangements could be amplified using the same annealing and cycling conditions: 35 cycles at 65°C for the first round of amplification and 35 cycles at 60°C (30 seconds, Vγ9) and 57°C (30 seconds, Vγ10), respectively, for reamplification.
PCR products were separated on polyacrylamide gels (PAGE 6%) stained with ethidium bromide.
PCR of whole-tissue DNA extracts
The data obtained from single-cell PCR were confirmed by PCR analysis of the corresponding whole tissue DNA. For this, DNA was extracted from frozen skin specimens by following the manufacturer's recommendations (QIAamp DNA Mini Kit; QIAGEN, Hilden, Germany). Whole-tissue DNA extracts were analyzed for rearrangements of Vγ1 to Vγ8, Vγ9, Vγ10, and Vγ11 gene segments as previously described.20
Fluorescent fragment analysis
For GeneScan analysis, PCR amplification was carried out under the conditions described in detail above. The amplification was performed with 5-carboxyfluorescein–labeled VG-2 primer or with 5-carboxyfluorescein–labeled JGT1/2 primer and JGT3 primer, respectively, for amplification of Vγ9, Vγ10, or Vγ11 gene segments. A total of 2.0 μL of diluted PCR product (dilution dependent on intensity of the amplificate) was mixed with 2.0 μL foramide and 0.5 μL 6-carboxyrhodamine dye–labeled DNA size standard (GeneScan-500-[ROX], Applied Biosystems, Weiterstadt, Germany) and 0.5 μL loading buffer (applied with the size standard). After denaturation (2 minutes at 90°C) and cooling on wet ice (3 minutes), 2.5 μL was size-separated on a high-resolution polyacrylamide gel and analyzed using an automated 373A DNA sequencer. The size of the PCR products was determined using computer software GeneScan 672 (Applied Biosystems).
DNA sequence analysis
Unlabeled PCR products were isolated and sequenced directly by fluorescence chain termination technique using fluorescence-labeled dideoxynucleotide triphosphates (BigDye; Applied Biosystems). The sequencing reactions were analyzed on an automated DNA sequencer (377A; Applied Biosystems). To detect possible contamination, sequences were compared with each other and with our own TCR-γ sequences collected in our institute over the last 7 years. Published germline sequences were used to determine the type of TCR-γ rearrangement. International Immunogenetics database [IMGT]; (http://imgt.cines.fr.:8104; initiator and coordinator, Marie-Paule Lefranc, Montpellier, France).
Results
Clinical characteristics
Clinical data are summarized in Table 1. All 11 patients, 6 male and 5 female, presented with a typical history of recurrent self-healing papulonodular eruptions. In 5 of the 11 patients, biopsies taken within 6 months after first manifestation of LyP were studied. There was a history of lymphoma in 1 patient only (case no. 7). This patient developed mycosis fungoides 28 years after the onset of LyP (tissue sample of the mycosis fungoides lesion was not available). One patient (no. 6) suffered from coexisting small-plaque parapsoriasis.
Histologic and immunophenotypical analysis
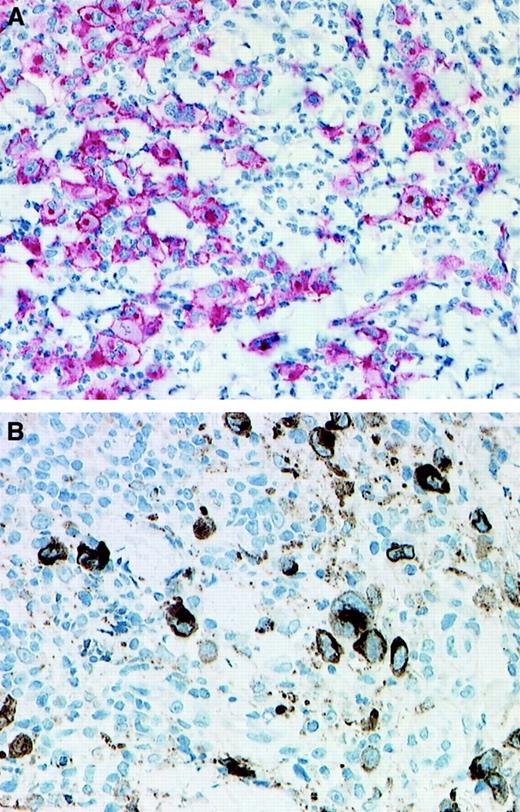
The 14 biopsies from the 11 patients with LyP included in this study showed the features of LyP type A (Table2). They were characterized by the presence of large atypical CD30+ cells with abundant cytoplasm, hypochromatic nuclei and prominent nucleoli, and a dense inflammatory background (Figure 1A). All CD30+ cells were positive for at least one T-cell antigen; they were negative for the anaplastic lymphoma kinase (ALK) protein and for B-cell antigens. In 8 of the 14 biopsies, the cytotoxic molecules perforin and/or granzyme B were detectable in more than 50% of the large CD30+ atypical cells (Figure 1B).
Immunophenotypical features of 14 biopsies from 11 patients with LyP
| Patient no. . | CD30 . | CD2 . | CD3 . | CD4 . | CD5 . | CD8 . | CD20 . | βF1 . | Perforin . | Granzyme B . | ALK1 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A | + | + | + | + | nd | − | − | nd | nd | nd | − |
| 1B | + | + | −/+ | + | + | − | − | −/+ | − | E | − |
| 2C | + | + | + | + | + | − | − | + | + | nd | − |
| 2D | + | + | + | + | + | − | − | ni | +/− | ni | − |
| 3E | + | +/− | − | + | − | − | − | − | + | − | − |
| 3F | + | − | − | + | − | − | − | E | + | − | − |
| 4 | + | + | E | E | − | − | − | − | E | + | − |
| 5 | + | + | + | E | + | + | − | −/+ | − | −/+ | − |
| 6 | + | +/− | + | + | ni | − | − | ni | −/+ | + | − |
| 7 | + | ni | + | + | ni | − | − | ni | ni | ni | − |
| 8 | + | + | − | + | + | − | − | − | + | + | − |
| 9 | + | + | −/+ | +/− | − | − | − | − | − | E | − |
| 10 | + | + | + | + | + | − | − | ni | ni | ni | − |
| 11 | + | −/+ | +/− | +/− | + | − | − | ni | + | + | − |
| Patient no. . | CD30 . | CD2 . | CD3 . | CD4 . | CD5 . | CD8 . | CD20 . | βF1 . | Perforin . | Granzyme B . | ALK1 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A | + | + | + | + | nd | − | − | nd | nd | nd | − |
| 1B | + | + | −/+ | + | + | − | − | −/+ | − | E | − |
| 2C | + | + | + | + | + | − | − | + | + | nd | − |
| 2D | + | + | + | + | + | − | − | ni | +/− | ni | − |
| 3E | + | +/− | − | + | − | − | − | − | + | − | − |
| 3F | + | − | − | + | − | − | − | E | + | − | − |
| 4 | + | + | E | E | − | − | − | − | E | + | − |
| 5 | + | + | + | E | + | + | − | −/+ | − | −/+ | − |
| 6 | + | +/− | + | + | ni | − | − | ni | −/+ | + | − |
| 7 | + | ni | + | + | ni | − | − | ni | ni | ni | − |
| 8 | + | + | − | + | + | − | − | − | + | + | − |
| 9 | + | + | −/+ | +/− | − | − | − | − | − | E | − |
| 10 | + | + | + | + | + | − | − | ni | ni | ni | − |
| 11 | + | −/+ | +/− | +/− | + | − | − | ni | + | + | − |
+ indicates positive; nd, not done; −, negative; −/+, less than 50% of CD30+ cells; E, less than 10% of CD30+cells; ni, not interpretable; +/−, more than 50% of CD30+ cells.
CD30 and granzyme B expression.
Immunohistologic detection of CD30 expression (A; immunoalkaline phosphatase–antiphosphatase staining) and granzyme B expression (B; streptavidin-biotin staining) in 2 cases of LyP. (A) Patient no. 8; (B) patient no 4. (Original magnification × 130.)
CD30 and granzyme B expression.
Immunohistologic detection of CD30 expression (A; immunoalkaline phosphatase–antiphosphatase staining) and granzyme B expression (B; streptavidin-biotin staining) in 2 cases of LyP. (A) Patient no. 8; (B) patient no 4. (Original magnification × 130.)
Amplification of the TCR-γ rearrangements in single cells and whole-tissue DNA extracts
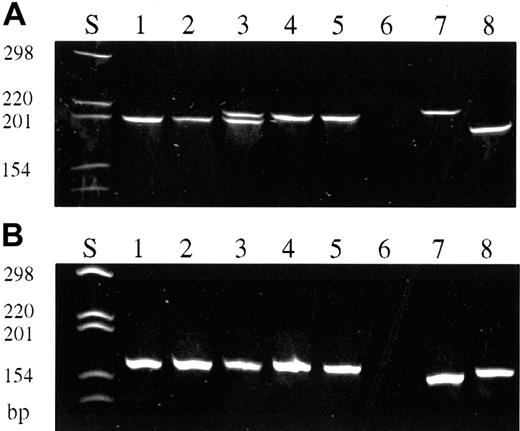
A total of 387 single CD30+ cells were isolated from the 14 frozen tissue specimens of 11 patients (Table3). The cells of all but 2 cases gave rise to TCR-γ–specific products, ranging from 3 to 17 amplificates per biopsy (total 123 PCR products). In case nos. 1 to 7, TCR-γ PCR products revealed rearrangements using Vγ gene segments 1 to 8 (Figure 2A), whereas in case nos. 8 and 9 TCR-γ amplificates demonstrated a rearrangement of Vγ9 and Vγ10 gene segments, respectively (Figure 2B). In the remaining 2 cases, no PCR products could be obtained from single cells. Both cases, however, led to the detection of unequivocal dominant PCR products when whole tissue DNA extracts in conjugation with GeneScan analysis were used for TCR-γ PCR.
Detection of TCR-γ rearrangements in single CD30+ as well as in single CD30− cells
| Patient no. . | CD30+cells . | CD30− cells . | |||
|---|---|---|---|---|---|
| No. of isolated cells . | Sequences identical/unrelated3-150 . | Vγ family . | No. of isolated cells . | Sequences identical3-151/unrelated3-150 . | |
| 1A3-152 | 34 | 6;53-153/3 | V4/V4 | 18 | 1/7 |
| 1B3-152 | 36 | 11;63-153/0 | V4/V4 | 16 | 0/3 |
| 2C3-152 | 24 | 10;73-153/0 | V4/V8 | 123-155 | 0/5 |
| 2D3-152 | 18 | 5;73-153/0 | V4/V8 | 183-155 | 1/16 |
| 3E3-152 | 34 | 11/0 | V4 | 163-155 | 0/3 |
| 3F3-152 | 18 | 5/0 | V4 | 243-155 | 0/2 |
| 4 | 36 | 15/0 | V2 | 14 | 0/6 |
| 5 | 36 | 8/2 | V5 | 163-155 | 0/9 |
| 6 | 36 | 5;43-153/0 | V3/V8 | 183-155 | 1/10 |
| 7 | 24 | 3/0 | V8 | 183-155 | 1/7 |
| 8 | 34 | 7/0 | V10 | 163-155 | 0/4 |
| 9 | 13 | 3/0 | V9 | 163-155 | 0/3 |
| 10 | 26 | 0/0 | 3-154 | 73-155 | 0/0 |
| 11 | 18 | 0/0 | 3-154 | 0 | — |
| Total | 387 | 118/5 | 209 | 4/75 | |
| Patient no. . | CD30+cells . | CD30− cells . | |||
|---|---|---|---|---|---|
| No. of isolated cells . | Sequences identical/unrelated3-150 . | Vγ family . | No. of isolated cells . | Sequences identical3-151/unrelated3-150 . | |
| 1A3-152 | 34 | 6;53-153/3 | V4/V4 | 18 | 1/7 |
| 1B3-152 | 36 | 11;63-153/0 | V4/V4 | 16 | 0/3 |
| 2C3-152 | 24 | 10;73-153/0 | V4/V8 | 123-155 | 0/5 |
| 2D3-152 | 18 | 5;73-153/0 | V4/V8 | 183-155 | 1/16 |
| 3E3-152 | 34 | 11/0 | V4 | 163-155 | 0/3 |
| 3F3-152 | 18 | 5/0 | V4 | 243-155 | 0/2 |
| 4 | 36 | 15/0 | V2 | 14 | 0/6 |
| 5 | 36 | 8/2 | V5 | 163-155 | 0/9 |
| 6 | 36 | 5;43-153/0 | V3/V8 | 183-155 | 1/10 |
| 7 | 24 | 3/0 | V8 | 183-155 | 1/7 |
| 8 | 34 | 7/0 | V10 | 163-155 | 0/4 |
| 9 | 13 | 3/0 | V9 | 163-155 | 0/3 |
| 10 | 26 | 0/0 | 3-154 | 73-155 | 0/0 |
| 11 | 18 | 0/0 | 3-154 | 0 | — |
| Total | 387 | 118/5 | 209 | 4/75 | |
Unique sequence different from the clonal CD30+ cells.
Identical to clonal CD30+ cells.
Interval between the biopsies: A-B: 8 months; C-D: 52 months; E-F: 3 months.
Biallelic rearrangement.
Results from 2 to 6 pooled CD30− cells.
No amplificate by single-cell analysis, but dominant TCR-γ gene rearrangement demonstrated by GeneScan analysis.
TCR-γ PCR of isolated single CD30+ cells and single CD30− cells (polyacrylamide gel stained with ethidium bromide, PAGE 6%).
(A) TCR-γ PCR products obtained with primers covering Vγ gene segments 1 to 8. CD30+ cells (lanes 1-5). Initial biopsy of patient no. 2 (lanes 1, 3, 4); biopsy taken 52 months later (lanes 2, 5). Note the biallelic rearrangement in this patient. Simultaneous detection of both rearrangements is only in lane 3, whereas lanes 1 and 2 and lanes 4 and 5 represent either 1 of the 2 rearranged alleles, respectively. Negative control (lane 6). Reactive CD30−cells of patient no. 2 (lanes 7, 8). (B) TCR-γ PCR products obtained with primers covering Vγ gene segment 10. CD30+ cells of patient no. 8 (lanes 1-5). Negative control (lane 6). Reactive CD30− cells of patient no. 8 (lanes 7, 8).
TCR-γ PCR of isolated single CD30+ cells and single CD30− cells (polyacrylamide gel stained with ethidium bromide, PAGE 6%).
(A) TCR-γ PCR products obtained with primers covering Vγ gene segments 1 to 8. CD30+ cells (lanes 1-5). Initial biopsy of patient no. 2 (lanes 1, 3, 4); biopsy taken 52 months later (lanes 2, 5). Note the biallelic rearrangement in this patient. Simultaneous detection of both rearrangements is only in lane 3, whereas lanes 1 and 2 and lanes 4 and 5 represent either 1 of the 2 rearranged alleles, respectively. Negative control (lane 6). Reactive CD30−cells of patient no. 2 (lanes 7, 8). (B) TCR-γ PCR products obtained with primers covering Vγ gene segment 10. CD30+ cells of patient no. 8 (lanes 1-5). Negative control (lane 6). Reactive CD30− cells of patient no. 8 (lanes 7, 8).
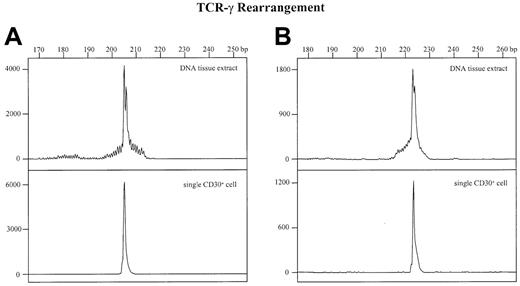
For comparison, GeneScan analyses were also performed from whole-tissue DNA extracts of all other cases as well as from the single-cell amplificates. In all of these cases a more or less prominent dominant amplificate embedded in a varying polyclonal background was found (Figure 3, upper panels) that was identical in size and sequence to the corresponding single-cell amplificate (Figure 3, lower panels). This indicates that the dominant PCR products in the 2 cases without amplificable single cells are derived from the clonal CD30+ T cells.
Detection of TCR-γ rearrangements by PCR and GeneScan analysis in whole tissue DNA extracts and single CD30+cells.
The x-axes represent molecular size (base pairs) and the y-axes fluorescence intensity. (A) Whole-tissue DNA extract (upper panel) and corresponding CD30+ single cell (lower panel) of patient no. 1. (B) Whole tissue DNA extract (upper panel) and corresponding CD30+ single cell (lower panel) of patient no. 5.
Detection of TCR-γ rearrangements by PCR and GeneScan analysis in whole tissue DNA extracts and single CD30+cells.
The x-axes represent molecular size (base pairs) and the y-axes fluorescence intensity. (A) Whole-tissue DNA extract (upper panel) and corresponding CD30+ single cell (lower panel) of patient no. 1. (B) Whole tissue DNA extract (upper panel) and corresponding CD30+ single cell (lower panel) of patient no. 5.
In 10 cases (patient nos. 1-10) lesional single CD30−cells or pooled CD30− cells were isolated from 13 frozen tissue sections (Table 3). Seventy-nine TCR-γ–specific PCR products were produced, which were different in their sizes in nearly all instances.
Sequence analysis
All PCR products were sequenced and compared with each other and with our own and published data bank sequences (IMGT). In each case identical sequences were found, indicating a clonal CD30+T-cell population (Tables 3 and 4). In most cases all CD30+ cells belonged to the same T-cell clone, whereas in 2 cases (patient nos. 1 and 5) a very small portion of the CD30+ cells were unrelated to the clonal population (5 [4%] of a total of 123 CD30+ cells). Furthermore, the TCR-γ rearrangements of the CD30+ cells isolated from biopsies of 3 patients (patient nos. 1-3), which were taken at different times, were identical in all instances, indicating the presence of the same T-cell clone in all lesions.
DNA nucleotide sequences of clone-specific TCR-γ rearrangements
| Patient no. . | V region . | N region . | J region . |
|---|---|---|---|
| 14-150 | Vγ4—GATGGGCA | GG | TAAGAAAC–Jγ2 |
| Vγ4—CACCTGGG | TTTTCGGGGCCGAAAGAGG | GTAGTGAT—JγP2 | |
| 24-150 | Vγ8—CCTGGGAT | TC | ATTATTAT—Jγ2 |
| Vγ4—GGATGGGC | T | GAAACTCT—Jγ2 | |
| 3 | Vγ4—CACCTGGG | TGCCC | TTATTATA—Jγ2 |
| 4 | Vγ2—ACCTGGGA | ACCG | ATTATAAG—Jγ2 |
| 5 | Vγ5—GGACAGGC | CGGTTG | GTGATTGG—JγP2 |
| 64-150 | Vγ3—CTGGGACA | CTTAGGG | ATTATAAG—Jγ2 |
| Vγ8—CTGGGATA | — | ACTCTTTG—Jγ2 | |
| 7 | Vγ8—ACTGTGCC | TTTGG | GTGATTGG—JγP2 |
| 8 | Vγ10–GCGTGGGA | CTACCTAGTAG | ATAAGAAA—Jγ2 |
| 9 | Vγ9—TTGTGGGA | CGG | GAAACTCT—Jγ2 |
| Patient no. . | V region . | N region . | J region . |
|---|---|---|---|
| 14-150 | Vγ4—GATGGGCA | GG | TAAGAAAC–Jγ2 |
| Vγ4—CACCTGGG | TTTTCGGGGCCGAAAGAGG | GTAGTGAT—JγP2 | |
| 24-150 | Vγ8—CCTGGGAT | TC | ATTATTAT—Jγ2 |
| Vγ4—GGATGGGC | T | GAAACTCT—Jγ2 | |
| 3 | Vγ4—CACCTGGG | TGCCC | TTATTATA—Jγ2 |
| 4 | Vγ2—ACCTGGGA | ACCG | ATTATAAG—Jγ2 |
| 5 | Vγ5—GGACAGGC | CGGTTG | GTGATTGG—JγP2 |
| 64-150 | Vγ3—CTGGGACA | CTTAGGG | ATTATAAG—Jγ2 |
| Vγ8—CTGGGATA | — | ACTCTTTG—Jγ2 | |
| 7 | Vγ8—ACTGTGCC | TTTGG | GTGATTGG—JγP2 |
| 8 | Vγ10–GCGTGGGA | CTACCTAGTAG | ATAAGAAA—Jγ2 |
| 9 | Vγ9—TTGTGGGA | CGG | GAAACTCT—Jγ2 |
Biallelic.
In contrast, CD30− cells isolated from the same cases displayed unrelated TCR-γ rearrangements in all instances with only a few exceptions (Table 3). In 4 biopsies (patient nos. 1, 2, 6, and 7) 1 single CD30− cell each showed an identical TCR-γ rearrangement as detected in the CD30+ population (4 [5%] of 79 CD30− cells).
Controls
To evaluate the reliability of the single-cell approach, aliquots from the buffer covering the tissue sections were drawn after every other single-cell isolation and were analyzed for the presence of TCR-γ gene rearrangements. None of the 335 buffer controls gave rise to a specific product. The PCR analysis of 78 reactive T cells isolated from a tonsil led to the detection of nonidentical rearrangements of the TCR-γ gene in 22 cells (28%), whereas the 15 PCR products obtained from 36 neoplastic cells (41%) of a peripheral T-cell lymphoma case showed identical rearrangements without exceptions. All PCR products were sequenced.
Discussion
The nosological position of LyP is a long-standing enigma. LyP was included in the European Organization for Research and Treatment of Cancer classification for primary cutaneous lymphomas, taking into account its association with other lymphomas in 10% to 20% of cases.3 It was shown that LyP cases with associated lymphomas shared the same clonal rearrangement in most instances.6,9-12 Several attempts to demonstrate the presence of clonal T-cell populations in LyP lesions without complicating T-cell lymphomas, however, produced conflicting results.11,13-16 This might, besides the clinical benign course, be one reason why LyP is still assigned to the heterogeneous group of cutaneous pseudolymphomas, a group of benign reactive lymphoproliferative processes that clinically and/or histologically mimic cutaneous lymphomas.21
These contradictory findings may be due to biologic and/or technical reasons. On one hand, LyP might represent a continuum beginning as a polyclonal and/or oligoclonal condition that in 10% to 20% of cases becomes a clonal malignant T-cell proliferation leading to T-cell lymphoma with high probability. On the other hand, in previous studies, Southern blot technique or PCR technique analyzing whole-tissue DNA extracts was used. This approach often fails to detect a small number of clonal lymphoid cells embedded in a polyclonal background. Moreover, it does not allow for the assignment of molecular features to a morphologically distinct cell population within the lesion. In particular, the question of whether clonality is restricted to the population of CD30+ cells or whether some CD30− T cells also belong to the clone could not be answered by this approach.
To circumvent these methodically based difficulties and to conclusively clarify the clonality of atypical cells in LyP, we analyzed the TCR-γ gene rearrangements of single CD30+ as well as of single CD30− T cells micromanipulated from frozen tissue sections of lesional skin.
Fourteen biopsies of 11 patients with characteristic clinical features of LyP were included in our study. Five of these patients had only a brief history of LyP (1 to 6 months), whereas the remaining 6 patients all had a long-standing course of the disease, ranging from 10 months to 30 years. All but 1 patient presented a history without a complicating lymphoma. This contrasts with previous studies that often include patients with prior or coexisting lymphoma and therefore generates the question as to whether the clonal lesions were true LyP or LyP-like manifestations of the associated cutaneous T-cell lymphoma.10,13 The histologic differentiation of LyP and CD30+ cutaneous ALCL is, in particular, often unreliable. Furthermore, patients with associated lymphoma were not representative of the vast majority of patients with LyP, because transformation into malignant lymphoma only rarely occurs.4
A total of 387 single CD30+ cells isolated from 14 biopsies gave rise to 123 specific PCR products. Nucleotide sequence analyses of all PCR products showed identical TCR-γ gene rearrangements within each individual case. By GeneScan analysis of whole-tissue DNA extracts, a dominant population within a polyclonal background could be detected in all 14 biopsies. GeneScan analysis of the corresponding single-cell PCR product produced an identical band showing that the dominant band demonstrated in whole-tissue DNA is derived from atypical CD30+ cells. By this finding the presence of a single dominant clonal CD30+ cell population within each single lesion was proven.
Repetitive attempts to produce TCR-γ–specific PCR products in the remaining 2 cases (patient nos. 10 and 11) consistently failed. However, a dominant TCR-γ gene rearrangement could be demonstrated by GeneScan analysis of whole-tissue DNA extracts in these 2 cases as well as in all other cases where the dominant PCR product could be unequivocally assigned to the CD30+ population. Therefore, we conclude that the failure to demonstrate TCR-γ rearrangements in single CD30+ cells of 2 cases is not due to the absence of rearranged TCR-γ genes but due to a partial degradation of the DNA, which thus became unsuitable for single-copy PCR. This conclusion is further confirmed by the absence of detectable TCR-γ gene rearrangements in CD30− T cells and supported by the highly variable amplification rate of TCR-γ gene rearrangements in single CD30+ cells of the other cases (12.5%-70%), thus reflecting the different levels of DNA degradation of the tissue samples.
Weiss et al13 described different TCR rearrangements within separate LyP lesions of the same patient using the Southern blot technique and suggested a multiclonal origin of the disease. The idea of different cell clones in separate lesions that wax and wane would also be in line with the clinical course of LyP. In our study, however, we could demonstrate that in 3 of our patients the identical dominant CD30+ cell clone was also found in temporally (up to 52 months) and anatomically separate LyP lesions. This clearly indicates that the same clonal CD30+ population is responsible for the outgrowth of papular skin lesions arising at different points in time and/or at various anatomic sites. This finding proves the observations of previous studies where the same TCR rearrangement was found within separate LyP lesions of one patient.11 16
In 2 cases (patient nos. 1 and 5) a few single CD30+ cells revealed unique TCR-γ gene rearrangements unrelated to the clonal cell population (5 of a total of 123 CD30+ cells). Interestingly, the appearance of unrelated unique CD30+cells was restricted to the first of 2 biopsies in patient no. 1, whereas only clonally related CD30+ cells were found in the second biopsy obtained 8 months later. This indicates that at least in the initial phase of the disease—as also seen 4 weeks after the onset of LyP in patient no. 5—unrelated CD30+ cells can be present in addition to the clonal population. These unrelated cells seem to disappear in the further course of the disease, leading to a CD30+ population consisting of exclusively clonally related cells.
To answer the question of whether clonality is restricted to the population of CD30+ cells, we also analyzed the TCR-γ gene rearrangements of neighboring single CD30− cells. In nearly all instances nucleotide sequence analyses of PCR products from isolated CD30− cells were polyclonal and unrelated to the CD30+ cell population. In 4 cases (patient nos. 1, 2, 6, and 7), 1 single CD30− cell each displayed the same TCR-γ gene rearrangement as detected in the CD30+population (4 of a total of 79 CD30− cells). This finding demonstrates that the clonal population is not completely restricted to CD30+ cells but may in addition comprise a small number of CD30− cells. It can be suggested that these cells are either not fully activated and consequently still not CD30+or have lost their CD30 expression for unknown reasons. Technical reasons for the presence of clonally related CD30− cells such as contaminations or erroneously picked cells are unlikely because all controls displayed the expected results, thus demonstrating the reliability of our approach.
In keeping with previous data,22-24 immunophenotypical analyses of the 14 reported biopsies indicated that the atypical cells in LyP A represent CD30+/CD4+ activated helper T cells that in a few cases show a loss of CD2, CD3, and/or CD5. Moreover, in 8 (57%) of the 14 biopsies, more than 50% of the clonal CD30+ T cells additionally express cytotoxic proteins (perforin and/or granzyme B). In agreement with previous studies,25,26 these findings provide further evidence for the derivation of atypical cells in LyP from CD4+ T cells with cytotoxic activity. Today, however, the physiologic role of this subpopulation that constitutes less than 5% of CD4+ T cells in peripheral blood of healthy individuals27 is still not clear. It has been suggested that CD4-mediated cytotoxicity might be of immunomodulatory importance by eliminating antigen-presenting cells.28 Expression of cytotoxic proteins on CD4+ T cells could be induced by chronic stimulation in vitro.29 30 This observation fits with the common hypothesis of lymphomagenesis postulating that lymphoma development might be associated with a persistent antigenic stimulus.
In conclusion, these results definitely demonstrate that LyP represents a monoclonal disorder. Clonal expansion is not restricted to an individual lesion but encloses both anatomically and temporally separate lesions. Furthermore, we could show for the first time that clonality constitutes the population of CD30+ T cells. These results indicate that the prolonged course of LyP with its typical features of waxing and waning is due to the expansion and regression of one single CD30+ cell clone that shows cytologic signs of malignancy. By fulfilling 2 classic criteria of malignancy (clonality and cytologic atypia) but showing a benign clinical course, LyP takes up a highly interesting position in tumor biology. Similar findings can be observed in monoclonal gammopathy of undetermined significance, which is found in about 1% of the population over 50 years of age.31 Despite a significant risk of progression to multiple myeloma, some patients remain asymptomatic for decades. Chromosomal and gene methylation analyses of monoclonal gammopathy of undetermined significance and multiple myeloma support the hypothesis of a multistep process for the oncogenesis of multiple myeloma.32,33 Also, in LyP other factors (for example, alterations in receptor signaling as previously shown) seem to be necessary for the progression of LyP into malignant lymphoma.34 35 The likelihood of accumulating a sufficient number of such relevant genetic lesions is increased by the prolonged life span of a cell clone as it is observed in LyP.
We thank D. Jahnke, H.-H. Müller, and H. Protz for their excellent technical assistance and L. Udvarhelyi for his editorial assistance. This paper is dedicated to Prof O. Braun-Falco on the occasion of his 80th birthday.
Prepublished online as Blood First Edition Paper, April 17, 2002; DOI 10.1182/blood-2001-12-0199.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michael Hummel, Institute of Pathology, University Medical Center Benjamin Franklin, The Free University of Berlin, Hindenburgdamm 30, 12200 Berlin, Germany; e-mail:michael.hummel@medizin.fu-berlin.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal