In this study, we evaluated the prognostic significance of multiple myeloma-1/interferon regulatory factor-4 (MUM1/IRF4) expression in B-cell chronic lymphocytic leukemia (B-CLL). Our results demonstrated that the absence of MUM1/IRF4 expression showed the highest relative risk among the factors analyzed in determining the probability for death in patients with B-CLL using univariate and multivariate Cox regression analysis. Patients without MUM1/IRF4 expression had significantly worse overall survival than did those with MUM1/IRF4 expression (52% cumulative survival, 63 months vs not reached, Kaplan-Meier survival analysis; P < .03, log-rank test). Patients with MUM1/IRF4 expression were more likely to have disease at low Rai stage and interstitial/nodular marrow involvement. Furthermore, only 1 of 11 patients with MUM1/IRF4 expression and interstitial/nodular marrow involvement died during a 100-month follow-up. Our results suggest that B-CLL with expression of MUM1/IRF4, indicative of postgerminal center origin, has a more favorable clinical course and that MUM1/IRF4 is an important prognostic marker in B-CLL.
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) is the most common form of leukemia in the Western world,1 and it follows a heterogeneous clinical course.2 Available staging systems and prognostic markers, including lymphocyte doubling time, bone marrow histology, and cytogenetic abnormalities, have limited usefulness in predicting outcomes in individual patients.3-7
Recently, the histogenesis of B-CLL has been shown to be a promising prognostic indicator for patients.8-10 These studies have revealed that B-CLL cells are derived from lymphocytes at 2 different stages of differentiation, one with mutated immunoglobulin V (IgV) genes corresponding to postgerminal center (post-GC) origin, and the other with unmutated IgV genes, corresponding to pregerminal center (pre-GC) origin. Patients with unmutated IgV genes have worse prognoses than do those with mutated IgV genes.
Recent studies have demonstrated that the late-stage–GC B cells and post-GC B cells express multiple myeloma-1/interferon regulatory factor-4 (MUM1/IRF4) cells but not pre-GC B cells.11-15 By immunohistochemistry, MUM1/IRF4 is expressed in a morphologic spectrum of B-lymphocytes ranging from centrocytes to plasmablasts/plasma cells.11,14,16 Polymerase chain reaction analysis of MUM1/IRF4+ cells has shown that they contain rearranged immunoglobulin heavy-chain (IgH) genes with a varying number of VH somatic mutations.14 Functionally, MUM1/IRF4 gene expression has been shown to be essential for the function and homeostasis of mature B cells and cytotoxic T lymphocytes.17
The prognostic significance of MUM1/IRF4 expression has not been systematically studied in B-CLL, although its expression may serve as an important prognostic marker because of the linkage between MUM1/IRF4 expression and histogenesis of B lymphocytes as described above. The goal of the current study was to examine whether MUM1/IRF4 expression correlated with clinical outcome in B-CLL patients.
Study design
We retrospectively studied 30 patients with B-CLL (18 men, 12 women; age, 62.4 ± 12.7 years) diagnosed from January 1990 to December 1999. These patients were selected on the basis of the availability of marrow core biopsy samples and complete clinical data. All patients had the typical morphology and classic immunophenotypic pattern for B-CLL.
Pertinent clinical information for these patients was obtained by reviewing the records from the tumor registry, from patient charts, or both (Table 1). Rai stages were defined according to the parameters at the time of initial diagnosis. Median length of follow-up was 62.5 months (range, 1-107 months). All surviving patients were followed up for at least 25 months. The end point of follow-up was either the date of the last visit or the date of death until September 2001.
Clinical presentations and outcomes and results of laboratory evaluation of marrow specimens, including MUM1/IRF4
| Patient . | MUM1 . | Age at diagnosis . | Sex . | Rai stage . | Chemotherapy . | Marrow pattern . | Marrow involvement, % . | Vital status . | Follow-up (mo) . | Mutation status . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Neg | 55 | M | Low | Yes | D | 100 | Dead | 107 | Nonmutated |
| 2 | Neg | 44 | M | Low | Yes | D | 100 | Dead | 69 | No clonal sequence* |
| 3 | Neg | 72 | F | Low | Yes | I/N | 50 | Dead | 23 | Mutated |
| 4 | Neg | 67 | F | Low | No | I/N | 70 | Alive | 62 | Mutated |
| 5 | Neg | 67 | F | Low | Yes | D | 100 | Dead | 103 | Nonmutated |
| 6 | Neg | 66 | M | High | Yes | I/N | 70 | Dead | 89 | Nonmutated |
| 7 | Neg | 79 | M | High | Yes | I/N | 50 | Dead | 41 | No clonal sequence |
| 8 | Neg | 70 | F | High | Yes | D | 100 | Dead | 86 | Mutated |
| 9 | Neg | 77 | M | High | No | D | 80 | Dead | 2 | No clonal sequence |
| 10 | Neg | 67 | M | Low | Yes | I/N | 60 | Dead | 80 | Nonmutated |
| 11 | Neg | 63 | F | Low | No | I/N | 70 | Dead | 87 | Mutated |
| 12 | Neg | 78 | M | Low | Yes | I/N | 60 | Dead | 57 | Nonmutated |
| 13 | Neg | 69 | M | High | Yes | I/N | 50 | Alive | 40 | Mutated |
| 14 | Neg | 61 | F | High | Yes | D | 100 | Dead | 17 | Nonmutated |
| 15 | Neg | 69 | M | High | No | D | 85 | Dead | 1 | No clonal sequence |
| 16 | Neg | 79 | M | High | No | D | 90 | Dead | 7 | Mutated |
| 17 | Neg | 44 | M | High | Yes | D | 100 | Alive | 35 | Mutated |
| 18 | Neg | 63 | M | Low | Yes | D | 70 | Dead | 63 | Nonmutated |
| 19 | Pos | 66 | M | Low | No | I/N | 70 | Alive | 80 | Mutated |
| 20 | Pos | 69 | M | Low | Yes | I/N | 25 | Alive | 100 | Mutated |
| 21 | Pos | 71 | M | Low | Yes | I/N | 40 | Alive | 25 | Mutated |
| 22 | Pos | 29 | F | Low | Yes | I/N | 60 | Alive | 95 | Mutated |
| 23 | Pos | 47 | F | Low | No | I/N | 10 | Alive | 70 | Mutated |
| 24 | Pos | 50 | F | Low | Yes | I/N | 20 | Alive | 37 | Nonmutated |
| 25 | Pos | 55 | F | Low | No | I/N | 50 | Alive | 28 | Mutated |
| 26 | Pos | 60 | F | Low | No | I/N | 60 | Alive | 64 | Mutated |
| 27 | Pos | 60 | F | Low | No | I/N | 5 | Alive | 84 | No clonal sequence |
| 28 | Pos | 35 | M | Low | Yes | I/N | 10 | Dead | 67 | Not done |
| 29 | Pos | 74 | M | Low | Yes | D | 80 | Dead | 40 | No clonal sequence |
| 30 | Pos | 65 | M | High | No | I/N | 10 | Alive | 76 | No clonal sequence |
| Patient . | MUM1 . | Age at diagnosis . | Sex . | Rai stage . | Chemotherapy . | Marrow pattern . | Marrow involvement, % . | Vital status . | Follow-up (mo) . | Mutation status . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Neg | 55 | M | Low | Yes | D | 100 | Dead | 107 | Nonmutated |
| 2 | Neg | 44 | M | Low | Yes | D | 100 | Dead | 69 | No clonal sequence* |
| 3 | Neg | 72 | F | Low | Yes | I/N | 50 | Dead | 23 | Mutated |
| 4 | Neg | 67 | F | Low | No | I/N | 70 | Alive | 62 | Mutated |
| 5 | Neg | 67 | F | Low | Yes | D | 100 | Dead | 103 | Nonmutated |
| 6 | Neg | 66 | M | High | Yes | I/N | 70 | Dead | 89 | Nonmutated |
| 7 | Neg | 79 | M | High | Yes | I/N | 50 | Dead | 41 | No clonal sequence |
| 8 | Neg | 70 | F | High | Yes | D | 100 | Dead | 86 | Mutated |
| 9 | Neg | 77 | M | High | No | D | 80 | Dead | 2 | No clonal sequence |
| 10 | Neg | 67 | M | Low | Yes | I/N | 60 | Dead | 80 | Nonmutated |
| 11 | Neg | 63 | F | Low | No | I/N | 70 | Dead | 87 | Mutated |
| 12 | Neg | 78 | M | Low | Yes | I/N | 60 | Dead | 57 | Nonmutated |
| 13 | Neg | 69 | M | High | Yes | I/N | 50 | Alive | 40 | Mutated |
| 14 | Neg | 61 | F | High | Yes | D | 100 | Dead | 17 | Nonmutated |
| 15 | Neg | 69 | M | High | No | D | 85 | Dead | 1 | No clonal sequence |
| 16 | Neg | 79 | M | High | No | D | 90 | Dead | 7 | Mutated |
| 17 | Neg | 44 | M | High | Yes | D | 100 | Alive | 35 | Mutated |
| 18 | Neg | 63 | M | Low | Yes | D | 70 | Dead | 63 | Nonmutated |
| 19 | Pos | 66 | M | Low | No | I/N | 70 | Alive | 80 | Mutated |
| 20 | Pos | 69 | M | Low | Yes | I/N | 25 | Alive | 100 | Mutated |
| 21 | Pos | 71 | M | Low | Yes | I/N | 40 | Alive | 25 | Mutated |
| 22 | Pos | 29 | F | Low | Yes | I/N | 60 | Alive | 95 | Mutated |
| 23 | Pos | 47 | F | Low | No | I/N | 10 | Alive | 70 | Mutated |
| 24 | Pos | 50 | F | Low | Yes | I/N | 20 | Alive | 37 | Nonmutated |
| 25 | Pos | 55 | F | Low | No | I/N | 50 | Alive | 28 | Mutated |
| 26 | Pos | 60 | F | Low | No | I/N | 60 | Alive | 64 | Mutated |
| 27 | Pos | 60 | F | Low | No | I/N | 5 | Alive | 84 | No clonal sequence |
| 28 | Pos | 35 | M | Low | Yes | I/N | 10 | Dead | 67 | Not done |
| 29 | Pos | 74 | M | Low | Yes | D | 80 | Dead | 40 | No clonal sequence |
| 30 | Pos | 65 | M | High | No | I/N | 10 | Alive | 76 | No clonal sequence |
D indicates diffuse; dx, diagnosis; I/N, interstitial and/or nodular; Neg, negative; and Pos, positive.
No clonal band was identified for sequencing
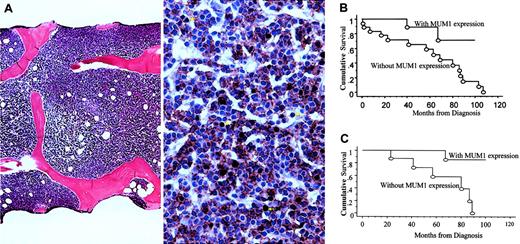
Dual immunohistochemical stains for MUM1/IRF4 (ISCAT/M-17; Santa Cruz Biotechnology, CA) and CD20 (L-26; DAKO, Carpinteria, CA) were performed on Bouin or B5-fixed, paraffin-embedded, decalcified marrow core biopsy samples. Dual staining was performed to confirm that the expression of MUM-1/IRF4 was in B cells (CD20+) but not in activated T cells or plasma cells14 (Figure1A).
Morphologic evaluation and survival analysis.
(A) Example of marrow biopsy sample with diffuse involvement pattern (left) and absence of MUM1/IRF4 expression (right), defined as less than 20% of CD20+ B cells stained positively for MUM1/IRF4. Double staining for MUM1/IRF4 (brown; nuclear staining) and CD20 (red; membrane staining) shows that most CD20+ B cells do not express MUM1/IRF4. Cells that are positive for MUM1/IRF4 but negative for CD20 represent activated T cells or plasma cells (arrows). Double staining was performed, after heat retrieval, by incubating the marrow biopsy sections with monoclonal antibody against MUM1/IRF4 (1:200 dilution) for 30 minutes, detected by DAB chromogen (DAKO), followed by the second incubation with monoclonal antibody against CD20 (1:800 dilution) for 30 minutes, detected by Vector-Nova Red (Vector, Burlingame, CA). All slides were stained using the DAKO automated immunostainer. In some patients, the monoclonal antibody (MUM1p, a kind gift from Dr B. Falini, University of Perugia, Italy) was used for double staining. Results were comparable between the 2 antibodies. (B) Patients without MUM1/IRF4 expression had significantly worse OS than those with MUM1/IRF4 expression (Kaplan-Meier survival analysis;P < .03, log-rank test). (C) Among patients with interstitial/nodular marrow involvement, patients with MUM1/IRF4 expression had significantly better OS than those without (Kaplan-Meier survival analysis; P < .02, log-rank test).
Morphologic evaluation and survival analysis.
(A) Example of marrow biopsy sample with diffuse involvement pattern (left) and absence of MUM1/IRF4 expression (right), defined as less than 20% of CD20+ B cells stained positively for MUM1/IRF4. Double staining for MUM1/IRF4 (brown; nuclear staining) and CD20 (red; membrane staining) shows that most CD20+ B cells do not express MUM1/IRF4. Cells that are positive for MUM1/IRF4 but negative for CD20 represent activated T cells or plasma cells (arrows). Double staining was performed, after heat retrieval, by incubating the marrow biopsy sections with monoclonal antibody against MUM1/IRF4 (1:200 dilution) for 30 minutes, detected by DAB chromogen (DAKO), followed by the second incubation with monoclonal antibody against CD20 (1:800 dilution) for 30 minutes, detected by Vector-Nova Red (Vector, Burlingame, CA). All slides were stained using the DAKO automated immunostainer. In some patients, the monoclonal antibody (MUM1p, a kind gift from Dr B. Falini, University of Perugia, Italy) was used for double staining. Results were comparable between the 2 antibodies. (B) Patients without MUM1/IRF4 expression had significantly worse OS than those with MUM1/IRF4 expression (Kaplan-Meier survival analysis;P < .03, log-rank test). (C) Among patients with interstitial/nodular marrow involvement, patients with MUM1/IRF4 expression had significantly better OS than those without (Kaplan-Meier survival analysis; P < .02, log-rank test).
We defined positive MUM1/IRF4 expression as consisting of more than 20% CD20+ B cells stained positively for MUM1/IRF4 according to the literature—small amounts of residual normal B cells may express MUM1/IRF4.11 18 Other morphologic findings, including percentages and patterns of involvement, in the marrow biopsy samples were evaluated. The above evaluation was performed without knowledge of the patient's clinical information.
DNA was extracted from the Giemsa-stained bone marrow aspirate smear of each patient19 (further details are outlined in Table2). An IgH gene mutation study was then performed, as outlined in Table 2. AVH gene with somatic mutations was defined as reported by Fais et al.20
Results of IgH gene mutation status study
| Patient . | Most similar germline VH gene . | Total mutation (CDR) . | Total mutation (FR) . | VH gene difference from germline, % . | Mutation status . | ||
|---|---|---|---|---|---|---|---|
| R Mut . | S Mut . | R Mut . | S Mut . | ||||
| 1 | VH1-69 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 2 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 3 | VH3-21 | 6 | 2 | 0 | 1 | 3.29 | Mutated |
| 4 | VH3-7 | 3 | 2 | 10 | 2 | 6.22 | Mutated |
| 5 | VH3-15/VH4-41 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 6 | VH3-11 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 7 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 8 | VH3-23 | 5 | 5 | 9 | 3 | 8.05 | Mutated |
| 9 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 10 | VH4-39 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 11 | VH3-48 | 4 | 2 | 1 | 6 | 4.76 | Mutated |
| 12 | VH1-69 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 13 | VH3-49 | 5 | 5 | 7 | 8 | 8.96 | Mutated |
| 14 | VH1-69 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 15 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 16 | VH3-33 | 10 | 6 | 7 | 5 | 10.25 | Mutated |
| 17 | VH3-7 | 2 | 1 | 7 | 6 | 5.86 | Mutated |
| 18 | VH1-8 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 19 | VH1-69 | 5 | 3 | 7 | 8 | 8.42 | Mutated |
| 20 | VH3-23 | 3 | 1 | 8 | 5 | 6.22 | Mutated |
| 21 | VH3-48 | 3 | 2 | 0 | 3 | 2.93 | Mutated |
| 22 | VH5-51 | 2 | 1 | 4 | 1 | 3.06 | Mutated |
| 23 | VH1-18 | 3 | 1 | 5 | 2 | 4.02 | Mutated |
| 24 | VH1-69 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 25 | VH3-23 | 10 | 1 | 8 | 3 | 8.05 | Mutated |
| 26 | VH4-59 | 4 | 2 | 6 | 6 | 6.59 | Mutated |
| 27 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 28 | NA | NA | NA | NA | NA | NA | Not done |
| 29 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 30 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| Patient . | Most similar germline VH gene . | Total mutation (CDR) . | Total mutation (FR) . | VH gene difference from germline, % . | Mutation status . | ||
|---|---|---|---|---|---|---|---|
| R Mut . | S Mut . | R Mut . | S Mut . | ||||
| 1 | VH1-69 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 2 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 3 | VH3-21 | 6 | 2 | 0 | 1 | 3.29 | Mutated |
| 4 | VH3-7 | 3 | 2 | 10 | 2 | 6.22 | Mutated |
| 5 | VH3-15/VH4-41 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 6 | VH3-11 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 7 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 8 | VH3-23 | 5 | 5 | 9 | 3 | 8.05 | Mutated |
| 9 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 10 | VH4-39 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 11 | VH3-48 | 4 | 2 | 1 | 6 | 4.76 | Mutated |
| 12 | VH1-69 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 13 | VH3-49 | 5 | 5 | 7 | 8 | 8.96 | Mutated |
| 14 | VH1-69 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 15 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 16 | VH3-33 | 10 | 6 | 7 | 5 | 10.25 | Mutated |
| 17 | VH3-7 | 2 | 1 | 7 | 6 | 5.86 | Mutated |
| 18 | VH1-8 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 19 | VH1-69 | 5 | 3 | 7 | 8 | 8.42 | Mutated |
| 20 | VH3-23 | 3 | 1 | 8 | 5 | 6.22 | Mutated |
| 21 | VH3-48 | 3 | 2 | 0 | 3 | 2.93 | Mutated |
| 22 | VH5-51 | 2 | 1 | 4 | 1 | 3.06 | Mutated |
| 23 | VH1-18 | 3 | 1 | 5 | 2 | 4.02 | Mutated |
| 24 | VH1-69 | 0 | 0 | 0 | 0 | 0 | Nonmutated |
| 25 | VH3-23 | 10 | 1 | 8 | 3 | 8.05 | Mutated |
| 26 | VH4-59 | 4 | 2 | 6 | 6 | 6.59 | Mutated |
| 27 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 28 | NA | NA | NA | NA | NA | NA | Not done |
| 29 | NA | NA | NA | NA | NA | NA | No clonal sequence |
| 30 | NA | NA | NA | NA | NA | NA | No clonal sequence |
In this study, DNA was extracted from the Giemsa-stained bone marrow aspirate smear of each patient using the protocol modified from Court et al.19 Bone marrow material was scraped from the slide, using a sterile scalpel blade, directly to an Eppendorf centrifuge tube containing the digestion buffer, as described,19 and was incubated at 55°C overnight. After overnight digestion, DNA extraction was performed using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN) following the manufacturer's protocol. The amount of DNA was quantified spectrometrically (Lambda 3A; Perkin-Elmer, Norwalk, CT). As a positive control for the quality of the extracted DNA, all DNA samples were tested by an in-house real-time quantitative polymerase chain reaction (PCR) assay targeting on β-globin. This ensured that the extracted DNA was suitable for further PCR analysis of IgH mutation status. PCR amplification of VH genes included preamplification and amplification steps. For preamplification, approximately 25 to 250 ng DNA, 2 pmol each VH leader primer, and the JH primer were used. The reaction was carried out in a 25-μL volume with Advantage 2 PCR enzyme system (Clontech, Palo Alto, CA). The amplification program consisted of 30 cycles of 45 seconds at 94°C, 45 seconds at 55°C, and 60 seconds at 72°C, followed by a final incubation step at 72°C for 5 minutes. For the second amplification, the 25-μL reaction was carried out, using 2 μL first-round reaction mixture, 2 pmol each VH family-specific forward primers, and the JH
primer. The amplification program consisted of 25 cycles of 45 seconds at 94°C, 45 seconds at 60°C, and 60 seconds at 72°C, followed by a final incubation step at 72°C for 5 minutes. Then 4 μL PCR product was analyzed by electrophoresis in 2% agarose gel and ethidium bromide staining. The PCR products were directly sequenced after purification with MultiScreen Filtration System (catalog no. MAFBN0B10; Millipore, Danvers, MA) using an automated ABI PRISM 310 Genetic Analyzer. Nucleotide sequences were aligned to EMBL/GenBank and V-BASE sequence directory. A VH gene with somatic mutations was defined as previously reported.20 VH leader primers: VH1 and VH7, CACCATGG ACTG(G/C)ACCTGGA; VH2, CAC(A/G)CTC CTGCTGCTGACCA; VH3, CACCATGGAGTTTGGGCTGA; VH3*, GGCTGAGCTGG(C/G)T TTT(C/T)CT(C/T)GT; VH4, T(C/G)CTGGTGGC(A/G)GCTCCCAGA; VH5, TCCTCCTGGCT GTTCTCCAA; VH6, ACAATGTCTGTCTCCTTCCTCAT. VHfamily-specific forward primers: VHF1/VHF7, CAGGT(C/G)CAGCTGGT(A/G)CAGTCTGG; VHF2, CAG(A/G)TCACCTTGA AGGAGTCTGG; VHF3, AGGTGCAGCTGGTGGAGTCTGG; VHF4, CAGGTGCAGCTGCA GGAGTCG; VHF5, GA(A/G)GTGCAGCTGGTGCAGTCTG; VHF6, CAGGTACAGCTGCA GCAGTCAGG. J-chain primers: JH primer, ACCTGAGGAGACGGTGACC; JH* primer, TGAGGAGACGGTGACC(A/G)(T/G)(T/G)GT.
NA indicates not applicable; CDR, complementarity-determining regions; FR, framework region; S Mut, silence mutation; and R mut, replacement mutation.
Results and discussion
In the current study, the frequency of MUM1/IRF4 expression was 40% (12 of 30 patients). Our study is the first to evaluate the expression of MUM1/IRF4 in marrow core biopsy samples of B-CLL patients. In agreement with our results, but with much smaller sample sizes, Tsuboi et al,11 Natkunam et al,18 and Falini et al14 observed that MUM1/IRF4 expression was seen in 3 (43%) of 7, 3 (43%) of 7 , and 10 (66%) of 15 small lymphocytic lymphoma (B-CLL/SLL) lymph node biopsy samples, respectively.
There are clear differences between patients with and without MUM1/IRF4 expression. MUM1/IRF4 expression was more frequent in patients with disease at low Rai stage (stages 0, 1, and 2) than at high Rai stage (stages 3 and 4) (11 of 20 vs 1 of 10; P < .02, χ2 analysis). Additionally, MUM1/IRF4 expression occurred more frequently in patients with interstitial or nodular marrow involvement than in patients with diffuse marrow involvement (11 of 19 vs 1 of 11; P < .009, χ2 analysis). Patients whose neoplastic cells expressed MUM1/IRF4 were slightly younger than those without MUM1/IRF4 expression (56.8 ± 14.2 years vs 66.1 ± 10.3 years; P = .045, Student ttest). However, no difference in sex was noted between patients with and without MUM1/IRF4 expression.
Our results showed that absence of MUM1/IRF4 expression was associated with the highest relative risk (RR) among the factors analyzed in determining the probability for death in patients with B-CLL using univariate Cox proportional hazards regression analysis (Table3). Most important, when controlling for MUM1/IRF4 expression status using multivariate Cox proportional hazards regression analysis, none of the other factors demonstrated a significant impact on overall survival (OS). Patients without MUM1/IRF4 expression had significantly worse OS than those with MUM1/IRF4 expression (52% cumulative survival, 63 months vs not reached, Kaplan-Meier survival analysis; P < .03, log-rank test; Figure 1B).
Results of univariate Cox regression analysis
| Variable . | No. patients . | P . | RR . | 95% CI . | |
|---|---|---|---|---|---|
| Lower bound . | Upper bound . | ||||
| Age (as a continuous variable) | NA | .0464 | 1.070 | 1.001 | 1.144 |
| MUM1 negative (vs positive) | 18 of 30 | .0394 | 4.808 | 1.080 | 21.28 |
| Diffuse marrow pattern (vs nondiffuse) | 11 of 30 | .0840 | 2.457 | 0.886 | 6.812 |
| Marrow involvement as a continuous variable, % | NA | .0782 | 1.018 | 0.998 | 1.039 |
| Rai stage high (vs low) | 10 of 30 | .0412 | 2.904 | 1.043 | 8.038 |
| Male (vs female) | 18 of 30 | .2830 | 1.789 | 0.619 | 5.171 |
| Chemotherapy no (vs yes) | 11 of 30 | .7922 | 0.854 | 0.264 | 2.760 |
| Variable . | No. patients . | P . | RR . | 95% CI . | |
|---|---|---|---|---|---|
| Lower bound . | Upper bound . | ||||
| Age (as a continuous variable) | NA | .0464 | 1.070 | 1.001 | 1.144 |
| MUM1 negative (vs positive) | 18 of 30 | .0394 | 4.808 | 1.080 | 21.28 |
| Diffuse marrow pattern (vs nondiffuse) | 11 of 30 | .0840 | 2.457 | 0.886 | 6.812 |
| Marrow involvement as a continuous variable, % | NA | .0782 | 1.018 | 0.998 | 1.039 |
| Rai stage high (vs low) | 10 of 30 | .0412 | 2.904 | 1.043 | 8.038 |
| Male (vs female) | 18 of 30 | .2830 | 1.789 | 0.619 | 5.171 |
| Chemotherapy no (vs yes) | 11 of 30 | .7922 | 0.854 | 0.264 | 2.760 |
Chemotherapy consisted of chlorambucil, fludarabine, or both. NA indicates not applicable.
Furthermore, our results demonstrate that MUM1/IRF4 expression with concurrent interstitial or nodular marrow involvement in B-CLL indicates a more favorable prognosis. Among patients with interstitial or nodular marrow involvement, those with MUM1/IRF4 expression had a significantly better OS than those without (58% cumulative survival, not reached vs 57 months, Kaplan-Meier survival analysis;P < .02, log-rank test; Figure 1C). Only 1 of 11 patients with MUM1/IRF4 expression and interstitial/nodular marrow involvement died during the follow-up of this study. The maximal length of follow-up in these 11 patients was 100 months.
MUM1/IRF4 expression is highly predictive of post-GC origin of B-CLL. Most patients (7 of 8, or 88%; Table 4) have mutated IgH genes. In addition, the absence of MUM1/IRF4 expression was observed in most patients (7 of 8, or 88%) with nonmutated IgH genes. These findings agree with those of previous study showing that MUM1/IRF4 expression is present in lymphocytes containing the mutated IgH gene in normal lymphoid tissue.14 However, the absence of MUM1/IRF4 expression was observed in 7 (50%) of 14 of the patients with mutated IgH genes. We hypothesize that these patients may lose or down-regulate the expression of MUM1/IRF4 through gene mutation/deletion or through unknown regulatory mechanisms during disease progression. This is partially supported by our finding that the absence of MUM1/IRF4 expression in patients with mutatedIgH genes is associated with advanced disease at a high Rai stage (4 of 7 vs 0 of 7 patients with mutated IgH gene and MUM1/IRF4 expression; P < .07, Fisher exact test) at presentation. Further follow-up of our patients to examine whether any of the MUM1/IRF4-positive patients may lose MUM1/IRF4 expression during their disease course will be important for understanding the role of MUM1/IRF4 expression in B-CLL progression.
Correlation between MUM1/IRF4 expression andIgH gene mutation status
| IgH gene . | MUM1/IRF4 positive . | MUM1/IRF4 negative . |
|---|---|---|
| Nonmutated | 1 | 7 |
| Mutated | 7 | 7 |
| IgH gene . | MUM1/IRF4 positive . | MUM1/IRF4 negative . |
|---|---|---|
| Nonmutated | 1 | 7 |
| Mutated | 7 | 7 |
In summary, our results suggest that B-CLL with MUM1/IRF4 expression, indicative of post-GC origin, has a more favorable clinical course and that patients are more likely to have disease at low Rai stage and nodular or interstitial marrow involvement. Additional studies are suggested to confirm this observation and to investigate the mechanisms of MUM1/IRF4 expression in B-CLL progression.
We thank Dr Carl G. Becker (Medical College of Wisconsin, Milwaukee) for his critical review of the manuscript and his thoughtful input. We also thank Kathryn L. Reith (Cancer Registry, Froedtert Hospital, Milwaukee, WI) for her help in organizing patient information, Glen Dawson for his help in immunohistochemical staining, and Bernard Schur for his help in preparing DNA samples.
Prepublished online as Blood First Edition Paper, August 1, 2002; DOI 10.1182/blood-2002-01-0104.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
C. Chang, Department of Pathology, Medical College of Wisconsin, 9200 W Wisconsin Ave, Milwaukee, WI 53226; e-mail: jeffchang@pol.net.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal