Fanconi anemia (FA) is an autosomal recessive chromosomal instability syndrome characterized by congenital abnormalities, progressive bone marrow failure, and cancer predisposition. Although patients with FA are candidates for bone marrow transplantation or gene therapy, their phenotypic heterogeneity can delay or obscure diagnosis. The current diagnostic test for FA consists of cytogenetic quantitation of chromosomal breakage in response to diepoxybutane (DEB) or mitomycin C (MMC). Recent studies have elucidated a biochemical pathway for Fanconi anemia that culminates in the monoubiquitination of the FANCD2 protein. In the current study, we develop a new rapid diagnostic and subtyping FA assay amenable for screening broad populations at risk of FA. Primary lymphocytes were assayed for FANCD2 monoubiquitination by immunoblot. The absence of the monoubiquitinated FANCD2 isoform correlated with the diagnosis of FA by DEB testing in 11 known patients with FA, 37 patients referred for possible FA, and 29 healthy control subjects. Monoubiquitination of FANCD2 was normal in other bone marrow failure syndromes and chromosomal breakage syndromes. A combination of retroviral gene transfer and FANCD2 immunoblotting provides a rapid subtyping assay for patients newly diagnosed with FA. These new FA screening assays would allow efficient testing of broad populations at risk.
Introduction
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility disorder characterized by diverse clinical features such as skeletal or skin abnormalities, progressive bone marrow failure, and increased risk of malignancies.1-5 FA has been reported in diverse ethnic groups, with an estimated heterozygous carrier frequency of 1 in 300,6 although this estimate may run higher in certain ethnic groups.7 Early and accurate diagnosis of Fanconi anemia is important, because it profoundly affects patient monitoring and treatment decisions and permits early genetic counseling of family members. Given the striking sensitivity of patients with FA to DNA-damaging agents,8-11 timely diagnosis is critical prior to the use of chemotherapy or radiation therapy of these patients in the bone marrow transplant setting. Diagnosis based on clinical manifestations alone can be difficult because 30% to 40% of patients lack developmental malformations or a positive family history.12,13 Patients previously not known to carry the diagnosis of FA may present with myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML) as the initial manifestation of their disease. Patients with acute leukemia have been diagnosed with FA after developing toxicity from their bone marrow transplant conditioning regimen.14
The currently used diagnostic test for FA relies on the increased chromosomal breakage and radial formation of FA cells in response to diepoxybutane (DEB) compared with cells from healthy control subjects,15,16 or from patients with other chromosomal instability disorders17 or genetic syndromes. The DEB test has excellent sensitivity, specificity, and reproducibility,16 and it is effective in prenatal diagnosis of FA.18 19 The DEB test is labor intensive and time consuming, however, and does not easily lend itself to broad screening studies. Furthermore, in some cases of Fanconi mosaicism, false-negative results have been reported with DEB screening of patient lymphocytes.
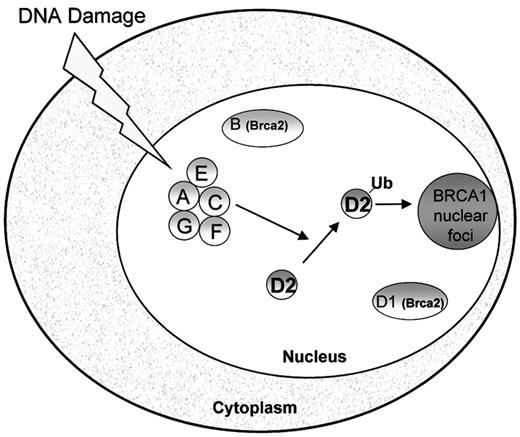
Studies of FA proteins have yielded insights into the molecular pathogenesis of FA. Fanconi anemia is a genetically heterogeneous disorder comprised of at least 8 complementation groups.20,21 The genes corresponding to each of these groups (A,22 B,23 C,24D1,23 D2,21 E,25F,26 and G27) have been cloned. The similar clinical presentation of patients of different subtypes suggests that these genes function in a common pathway. Indeed, interactions between FANCA, FANCC, FANCE, FANCF, and FANCG have been described.28-32 Activation of the FA protein complex by DNA damage or cell cycle progression results in monoubiquitination of the downstream FANCD2 protein.33 Monoubiquitination is a newly recognized class of posttranslational protein modification.34 Following monoubiquitination, FANCD2 localizes to nuclear foci where it colocalizes with other DNA-repair proteins such as BRCA1 (Figure1).33 A point mutation of FANCD2 at lysine 561, the site of monoubiquitination, abrogates its ability to correct the mitomycin C sensitivity of FA-D2 cells. Mutations in any of the upstream FA proteins are accompanied by loss of both DNA damage-inducible FANCD2 monoubiquitination and FANCD2 nuclear foci.
Model of the Fanconi anemia pathway.
The Fanconi proteins (FANCA, FANCC, FANCE, FANCF, FANCG) are required for the monoubiquitination of FANCD2. Following DNA damage, FANCD2 is monoubiquitinated and targeted to nuclear foci containing BRCA1, where it may play a role in DNA repair by homologous recombination.33 FANCD1/BRCA2 is not required for FANCD2 monoubiquitination.
Model of the Fanconi anemia pathway.
The Fanconi proteins (FANCA, FANCC, FANCE, FANCF, FANCG) are required for the monoubiquitination of FANCD2. Following DNA damage, FANCD2 is monoubiquitinated and targeted to nuclear foci containing BRCA1, where it may play a role in DNA repair by homologous recombination.33 FANCD1/BRCA2 is not required for FANCD2 monoubiquitination.
In the current study, we exploit these new advances in the molecular understanding of FA to develop an improved diagnostic screen. We demonstrate that the monoubiquitination of the FANCD2 protein, as judged by a simple Western blot, correlates with DEB-induced chromosome breakage.
Patients, materials, and methods
Lymphocyte isolation and PHA stimulation
Peripheral blood mononuclear cells from patients with known Fanconi anemia or patients referred for diagnostic testing to the Dana-Farber Comprehensive Center for Fanconi Anemia or from healthy volunteers were isolated by centrifugation over a Histopaque1077 (Sigma, St Louis, MO) cushion for 30 minutes at 1200 rpm in a Sorvall RT6000 centrifuge. Samples were stripped of identifying information and assigned a code number to maintain patient confidentiality. Lymphocytes were washed twice in RPMI 1640 (Cellgro, Herndon, VA), then stimulated with phytohemagglutinin (PHA) for 3 days in RPMI 1640 containing 15% heat-inactivated fetal bovine serum (FBS; Sigma) and 2 mM glutamine, penicillin, and streptomycin. Cell cultures were maintained in a humidified incubator containing 5% carbon dioxide at 37°C. Cell lines for chromosomal instability syndromes and for dyskeratosis congenita were obtained from the National Institute of General Medical Sciences (NIGMS, Camden, NJ) Human Genetic Cell Repository, Coriell Institute for Medical Research. Cell lines for other bone marrow failure syndromes were derived from patients seen in the Children's Hospital bone marrow failure clinic. The study was approved by the institutional review board of the Dana-Farber Cancer Institute, and informed consent was obtained from participating patients according to the Declaration of Helsinki.
Cell culture
Epstein-Barr virus (EBV)–transformed lymphoblasts were maintained in RPMI culture media as described for the primary lymphocyte cultures above.
Mitomycin C and DEB sensitivity assay
Mitomycin C (MMC) and DEB sensitivity assays were performed in the cytogenetics laboratory certified by the Clinical Laboratories Improvement Act (CLIA) in the Comprehensive Center for Fanconi Anemia at the Dana-Farber Cancer Institute. Peripheral blood was cultured in the presence of phytohemagglutinin with or without DEB or MMC. Samples were coded, and metaphase cells were blindly scored and analyzed for chromosomal breakage, including the formation of radials. Results were compared with healthy controls and positive controls run in parallel.
Immunoblotting and immunofluorescence microscopy
Cell lysates of primary and EBV-transformed lymphocytes were prepared and analyzed by immunoblotting as previously described.33 For immunofluorescent analysis, suspension cells were immobilized onto coverslips coated with poly-l-lysine (Sigma) prepared according to the manufacturer's protocol. Cells were washed in phosphate-buffered saline (PBS), spotted onto the coverslips, and allowed to settle for 10 to 15 minutes. Coverslips were washed twice in PBS then fixed and probed with the anti-FANCD2 E35 polyclonal antibody as previously described.33 For the secondary antibody, Alexis Fluor 488 (Molecular Probes, Eugene, OR) fluorescent secondary antibody was used at a dilution of 1:200. Coverslips were mounted in ProLong Antifade (Molecular Probes).
Retroviral transduction
Results
Detection of FANCD2 monoubiquitination in primary lymphocytes
We reasoned that analysis of FANCD2 protein modification by immunoblot might provide a convenient diagnostic screen for FA. To test this hypothesis, we initially examined healthy human peripheral blood lymphocytes (PBLs) for expression of the FANCD2 isoforms by immunoblotting. No FANCD2 protein was detected in primary lymphocyte lysates (Figure 2A, lane 1). Because FANCD2 expression is increased in actively cycling cells, the effect of PHA stimulation on FANCD2 expression in primary human lymphocytes was assayed (Figure 2A, lanes 2-5). After 48 to 72 hours of PHA stimulation, FANCD2 expression and monoubiquitination was evident. Increased expression of FANCA, FANCC, and FANCG was also observed following PHA stimulation.
Primary lymphocytes from patients with Fanconi anemia fail to monoubiquitinate FANCD2.
(A) Lymphocytes isolated from healthy control volunteers were stimulated with PHA for up to 72 hours prior to immunoblotting with antibodies against the indicated FANC proteins. Expression levels of all 4 Fanconi proteins rose following 72 hours of PHA stimulation. (B) Protein extracts of lymphocytes isolated from healthy control volunteers or from patients with Fanconi anemia were immunoblotted for FANCD2 following PHA stimulation. Although the lymphocytes from healthy control volunteers showed both the unmodified (FANCD2-S) and the monoubiquitinated (FANCD2-L) FANCD2 forms, the lymphocytes from the patients with Fanconi anemia showed only the lower unmodified FANCD2-S form. (C) Protein extracts from lymphocytes from obligate FA heterozygous carriers were immunoblotted for FANCD2 following PHA stimulation.
Primary lymphocytes from patients with Fanconi anemia fail to monoubiquitinate FANCD2.
(A) Lymphocytes isolated from healthy control volunteers were stimulated with PHA for up to 72 hours prior to immunoblotting with antibodies against the indicated FANC proteins. Expression levels of all 4 Fanconi proteins rose following 72 hours of PHA stimulation. (B) Protein extracts of lymphocytes isolated from healthy control volunteers or from patients with Fanconi anemia were immunoblotted for FANCD2 following PHA stimulation. Although the lymphocytes from healthy control volunteers showed both the unmodified (FANCD2-S) and the monoubiquitinated (FANCD2-L) FANCD2 forms, the lymphocytes from the patients with Fanconi anemia showed only the lower unmodified FANCD2-S form. (C) Protein extracts from lymphocytes from obligate FA heterozygous carriers were immunoblotted for FANCD2 following PHA stimulation.
Absence of FANCD2 monoubiquitination correlates with DEB sensitivity
We next compared the FANCD2 isoforms in PBLs from patients known to have FA versus healthy control subjects (Figure 2B-C). Following PHA stimulation, FANCD2 protein expression was induced in both FA and control PBLs. The FA PBLs failed to express the activated isoform of FANCD2 (FANCD2-L), consistent with an upstream defect in the FA pathway. PBLs from obligate FA heterozygotes (parents of patients with FA) exhibited normal levels of the FANCD2-L ubiquitinated isoform by immunoblotting (Figure 2C). No consistent difference in levels of the FANCD2-L isoform was noted between healthy controls and carriers of FA (data not shown). The proportion of FANCD2 that is present as the upper ubiquitinated band tends to be higher in transformed cell lines than in primary cells, most likely as a result of the higher proliferative rate of the former.33 Nonetheless, the upper FANCD2 band was easily detected in non-FA blood samples even following shipment from abroad. As an added control, blood samples simultaneously drawn from parental controls were shipped together with the patient samples, and the upper FANCD2-L form was detected in all such controls. The upper FANCD2-L form was readily detected even in samples processed up to 3 days following the blood draw. The advantage of using primary blood samples is the avoidance of the possibility of selective proliferation of any reverted wild-type cells as may be seen in mosaic patients. Furthermore, immortalization of cell lines takes up to 1 month, whereas Western blots of primary blood samples can be completed in a few days. Cell lines from patients with Fanconi anemia are often difficult to establish, so an assay amenable for primary blood samples would be advantageous.
We next compared the results of the FANCD2 immunoblot assay with the DEB/MMC chromosome breakage test. Blood samples were analyzed by both conventional DEB chromosomal breakage test and by FANCD2 immunoblotting. PBLs from 11 patients previously known to carry the diagnosis of FA failed to express the activated FANCD2 (FANCD2-L) isoform (Table 1). We also compared the results of DEB testing with the FANCD2 immunoblot in 37 patients prospectively referred for FA testing. Six referred patients were missing the FANCD2-L monoubiquitinated form and were subsequently diagnosed with FA by DEB testing. The Fanconi anemia blood samples tested included patients of subtypes A, C, E, and G. Western blot analyses for the affected Fanconi protein were performed for some of these samples and revealed full-length protein in some patients, truncated protein in others, and absent protein expression in others; nonetheless, the absence of the ubiquitinated FANCD2 band was seen in all cases. Thirty-one patients referred for FA diagnostic testing expressed the monoubiquitinated FANCD2 (FANCD2-L) form and, as predicted, did not exhibit increased chromosomal breakage in response to DEB (data not shown). DEB-resistant blood samples from 29 healthy adult control subjects all expressed the FANCD2-L form (data not shown).
Absence of FANCD2 correlates with DEB-induced chromosomal breakage
| Patient no. . | Sample no. . | DEB . | FANCD2-L . |
|---|---|---|---|
| Patients with FA | |||
| 1 | FA1064 | S | Absent |
| 2 | FA1073 | S | Absent |
| 3 | FA1082 | S | Absent |
| 4 | FA1086 | S | Absent |
| 5 | FA1087 | S | Absent |
| 6 | FA1088* | I | Absent |
| 7 | FA1089 | S | Absent |
| 8 | FA1090 | S | Absent |
| 9 | FA1123 | S | Absent |
| 10 | DF1147 | S | Absent |
| 11 | DF1152 | S | Absent |
| Patients referred for FA test | |||
| 1 | FA1094 | S | Absent |
| 2 | FA1085 | S | Absent |
| 3 | FA1114* | I | Absent |
| 4 | FA1108* | I | Absent |
| 5 | FA1124* | I | Absent |
| 6 | FA1137 | S | Absent |
| Patient no. . | Sample no. . | DEB . | FANCD2-L . |
|---|---|---|---|
| Patients with FA | |||
| 1 | FA1064 | S | Absent |
| 2 | FA1073 | S | Absent |
| 3 | FA1082 | S | Absent |
| 4 | FA1086 | S | Absent |
| 5 | FA1087 | S | Absent |
| 6 | FA1088* | I | Absent |
| 7 | FA1089 | S | Absent |
| 8 | FA1090 | S | Absent |
| 9 | FA1123 | S | Absent |
| 10 | DF1147 | S | Absent |
| 11 | DF1152 | S | Absent |
| Patients referred for FA test | |||
| 1 | FA1094 | S | Absent |
| 2 | FA1085 | S | Absent |
| 3 | FA1114* | I | Absent |
| 4 | FA1108* | I | Absent |
| 5 | FA1124* | I | Absent |
| 6 | FA1137 | S | Absent |
S indicates sensitive; and I, intermediate.
Mosaic.
Two of the patients newly diagnosed with FA had been previously tested for increased DEB-induced chromosomal breakage with negative results. These 2 patients had no FANCD2-L by immunoblotting and were subsequently diagnosed by DEB testing as mosaic for FA, with a mixed population of FA and wild-type lymphocytes. A total of 4 mosaic patients were assayed by Western blots and lacked the upper FANCD2-L isoform in our series (data not shown).
FANCD2 monoubiquitination is normal in other inherited bone marrow failure and chromosomal instability syndromes
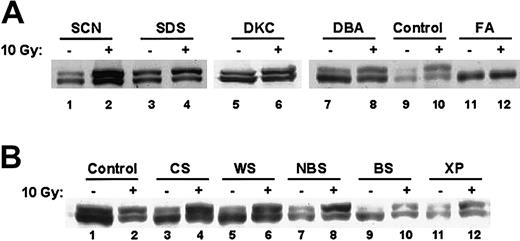
Because other bone marrow failure syndromes are also associated with physical anomalies and increased risk of malignancies, lymphocytes from patients with severe congenital neutropenia, Shwachman-Diamond syndrome, dyskeratosis congenita, and Diamond-Blackfan anemia were analyzed for FANCD2 monoubiquitination (Figure3A). Patients with these syndromes did not exhibit DEB-induced chromosomal breaks (data not shown). Western blots of these other bone marrow failure syndromes exhibited normal FANCD2 monoubiquitination that was enhanced by ionizing radiation (Figure 3A). In addition, lymphocytes from patients with other chromosomal instability syndromes (Cockayne syndrome, Werner syndrome, Nijmegen breakage syndrome, Bloom syndrome, xeroderma pigmentosa) showed normal FANCD2 monoubiquitination (Figure 3B). Thus, loss of FANCD2 monoubiquitination is specific to FA. Taken together, these data demonstrate that the FANCD2 Western blot distinguishes FA from other chromosome breakage syndromes.
FANCD2 monoubiquitination is normal in other inherited bone marrow failure and chromosomal instability syndromes.
(A) EBV-immortalized lymphocytes from patients with the indicated bone marrow failure syndromes were harvested before or 6 hours after treatment with 10 Gy radiation. Cell lysates were analyzed by immunoblotting with an anti-FANCD2 FI17 monoclonal antibody. FA indicates Fanconi anemia; SCN, severe congenital neutropenia; SDS, Shwachman-Diamond syndrome; DKC, dyskeratosis congenita; and DBA, Diamond-Blackfan anemia. (B) EBV-immortalized lymphocytes from patients with the indicated chromosomal breakage syndromes were harvested before or 6 hours after treatment with 10 Gy radiation. Cell lysates were analyzed by immunoblotting with an anti-FANCD2 FI17 monoclonal antibody. CS indicates Cockayne syndrome; WS, Werner syndrome; NBS, Nijmegen breakage syndrome; BS, Bloom syndrome; and XP, xeroderma pigmentosa.
FANCD2 monoubiquitination is normal in other inherited bone marrow failure and chromosomal instability syndromes.
(A) EBV-immortalized lymphocytes from patients with the indicated bone marrow failure syndromes were harvested before or 6 hours after treatment with 10 Gy radiation. Cell lysates were analyzed by immunoblotting with an anti-FANCD2 FI17 monoclonal antibody. FA indicates Fanconi anemia; SCN, severe congenital neutropenia; SDS, Shwachman-Diamond syndrome; DKC, dyskeratosis congenita; and DBA, Diamond-Blackfan anemia. (B) EBV-immortalized lymphocytes from patients with the indicated chromosomal breakage syndromes were harvested before or 6 hours after treatment with 10 Gy radiation. Cell lysates were analyzed by immunoblotting with an anti-FANCD2 FI17 monoclonal antibody. CS indicates Cockayne syndrome; WS, Werner syndrome; NBS, Nijmegen breakage syndrome; BS, Bloom syndrome; and XP, xeroderma pigmentosa.
FANCD2 forms nuclear foci in response to DNA damage or during the S phase of the cell cycle.33 Nuclear foci assembly correlates with the monoubiquitination of FANCD2. Primary PBLs from patients with FA failed to form FANCD2 nuclear foci, whereas FANCD2 nuclear foci formation remained intact in lymphocytes from patients with other bone marrow failure syndromes or chromosomal instability syndromes (data not shown).
Subtyping of patients with FA following retroviral-mediated gene transfer
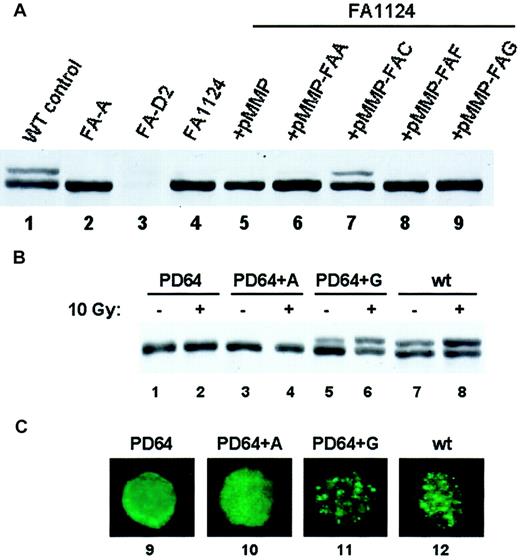
Retroviral vectors carrying a wild-type Fanconi cDNA can correct the MMC sensitivity of FA cells of the corresponding subtype.35 36 We, therefore, reasoned that assaying for restoration of FANCD2 monoubiquitination following retroviral infection of PBLs from patients newly diagnosed with FA might allow rapid FA subtyping. To test this hypothesis, we transduced EBV-immortalized lymphoblasts from a patient newly diagnosed with FA (FA1124) with retroviruses carrying different wild-type FA cDNAs and tested for restoration of the FA pathway by assaying for FANCD2 monoubiquitination (Figure 4A). The pMMP-FANCC retroviral supernatant complemented the lymphocytes from patient FA1124, restoring FANCD2 monoubiquitination (Figure 4A, lane 7). Introduction of the pMMP vector or pMMP retroviruses carrying other FA subtype cDNAs failed to restore FANCD2 monoubiquitination in this patient. FANCD2 monoubiquitination and foci formation were also restored following retroviral correction of a known FA-G patient's lymphocytes with the FANC-G retrovirus (Figure 4B, lanes 5,6; 4C, lane 11).
Restoration of FANCD2 monoubiquitination following retroviral transduction provides a rapid Fanconi anemia subtyping assay.
(A) EBV-immortalized lymphocytes from a patient newly diagnosed with FA (FA1124) were transduced with retroviruses carrying vector (lane 5) or the indicated FANC cDNAs (lanes 6-9). Cell lysates were analyzed by FANCD2 immunoblotting. The FANCC retrovirus restored FANCD2 monoubiquitination, consistent with this patient's subtype of FA-C. (B) EBV-immortalized lymphocytes from a patient with known FA-G were infected with pMMP retroviruses containing the FANCA (lanes 3-4) or the FANCG (lanes 5-6) cDNA as indicated. The restoration of FANCD2 monoubiquitination was assessed by immunoblotting. (C) Lymphocytes were also analyzed for restoration of FANCD2 nuclear foci by immunofluorescence (lanes 9-12). Original magnification × 600.
Restoration of FANCD2 monoubiquitination following retroviral transduction provides a rapid Fanconi anemia subtyping assay.
(A) EBV-immortalized lymphocytes from a patient newly diagnosed with FA (FA1124) were transduced with retroviruses carrying vector (lane 5) or the indicated FANC cDNAs (lanes 6-9). Cell lysates were analyzed by FANCD2 immunoblotting. The FANCC retrovirus restored FANCD2 monoubiquitination, consistent with this patient's subtype of FA-C. (B) EBV-immortalized lymphocytes from a patient with known FA-G were infected with pMMP retroviruses containing the FANCA (lanes 3-4) or the FANCG (lanes 5-6) cDNA as indicated. The restoration of FANCD2 monoubiquitination was assessed by immunoblotting. (C) Lymphocytes were also analyzed for restoration of FANCD2 nuclear foci by immunofluorescence (lanes 9-12). Original magnification × 600.
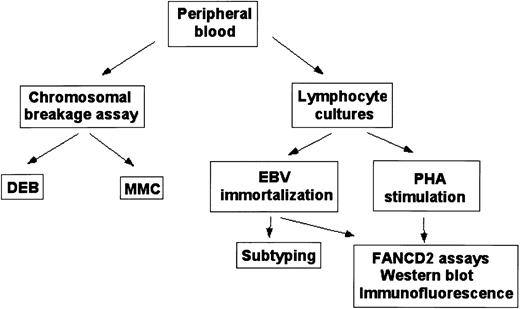
Taken together, these data demonstrate that the combination of retroviral transduction and FANCD2 activation assays facilitate the rapid subtyping of patients newly diagnosed with FA. On the basis of the FANCD2 activation assays described above, Figure5 summarizes the different assays for the work-up and subtyping of patients with Fanconi anemia.
Fanconi anemia diagnostic protocol.
Schematic outline for diagnosis and subtyping of peripheral blood samples.
Fanconi anemia diagnostic protocol.
Schematic outline for diagnosis and subtyping of peripheral blood samples.
Discussion
In the current study, we developed a rapid diagnostic and subtyping screen for FA based on recent advances in our understanding of the FA signaling pathway. The monoubiquitination of the FANCD2 protein and its subsequent translocation to nuclear foci require the function of multiple upstream FA genes. Accordingly, screening for FANCD2 monoubiquitination, by FANCD2 immunoblotting or immunofluorescence, provides a simple rapid assay for the integrity of the FA pathway.
As many as 30% to 40% of patients with FA do not have associated congenital malformations or a positive family history,12,13 and some present with malignancy as the first symptom.14 A test that does not rely on laborious cytogenetic analysis would facilitate FA screening. All patients presenting with relevant congenital anomalies, early-onset malignancies and myelodysplasias, or other nonspecific stigmata of Fanconi anemia may be readily tested. Because patients with FA are sensitive to the toxicities of alkylating agents and radiation, the diagnosis of FA prior to the initiation of chemotherapy or bone marrow transplantation would allow appropriate tailoring of their therapy.14Patients presenting with congenital anomalies could also be screened prior to the onset of pancytopenia or malignancy. FA testing may also be considered in cancer patients who have had unexpected toxicity resulting from radiation or chemotherapy.37 38 Although it is currently premature to consider the FANCD2 Western blot assay as a substitute for chromosomal breakage studies, the latter is labor intensive, requires highly trained staff, and does not lend itself to screening broad patient populations at risk. Screening such populations by Western blot would facilitate identification of previously undetected patients for whom additional DEB testing should be pursued.
Alternative tests for FA have been previously explored. FA cells exhibit cell cycle abnormalities with G2 phase prolongation and arrest by flow cytometry.39,40 Studies comparing results of DEB chromosomal breakage with cell cycle analyses of patients referred for FA testing showed close correlation between the results of these 2 tests.41 Constitutive elevation of serum alpha-fetoprotein (AFP) has been reported in patients with FA,42 although variations in AFP levels determined by different methodologies have limited the diagnostic utility of this test so far.43Furthermore, elevated AFP levels may be associated with other pathologic conditions such as malignancies,44 liver disease,45 neural tube defects,46 and Down syndrome.47 High serum AFP levels are also seen in association with ataxia-telangiectasia48 and have been used as a diagnostic screen for this disorder.49 More recently, the Comet assay50 has been used to detect patients with FA and carriers of FA.51
For patients with an initial negative DEB test but a strong clinical suspicion for FA, immunoblots may be a helpful adjunct test to identify patients with mosaicism for whom further testing would be helpful. Patients with mosaic FA have 2 populations of peripheral T cells: one with the increased chromosomal breakage typical of FA and one with wild-type cytogenetics. The wild-type clone results from somatic reversion of a mutated FA allele.52-54 In our series, the FANCD2 immunoblot indicated the diagnosis of FA in PBLs from 4 patients with mosaic FA; however, it would be reasonable to expect that PBLs from patients with a high percentage of cells reverted to wild type might manifest the FANCD2-L form. The current standard for patients with a high suspicion for Fanconi anemia but a negative DEB test is to repeat the DEB test on skin fibroblasts, which do not undergo reversion to wild type, and the same would hold true for such patients with a negative Western blot. We are currently investigating the status of FANCD2 in bone marrow fibroblasts from patients with mosaic FA. Mosaicism has not been observed in FA skin fibroblasts. Because patients with FA undergo regular bone marrow evaluations as part of their clinical care, the use of bone marrow mesenchymal cells would spare patients from undergoing an additional skin biopsy procedure.
Most patients with FA have a mutation in the FA genes corresponding to subtypes A (70%), C (10%), or G (10%).5,20,55,56Mutations in these genes disrupt the assembly of the FA protein complex and prevent the downstream activation of the FANCD2 protein. Patients with a rare form of FA may have false-negative FANCD2 immunoblots. For instance, if an FA gene encodes a protein that functions downstream of FANCD2, then the corresponding FA cells may have a normal FANCD2 immunoblot with both FANCD2 isoforms expressed. Indeed, this is the case with cells derived from patients with FA subtype D131 32; however, this subtype is extremely rare. As discussed above, some patients with a high degree of mosaicism may also be missed by DEB testing and show both FANCD2 isoforms. In such cases with a high index of suspicion, testing on patient fibroblasts is necessary.
The FANCD2 immunoblot provides a powerful screening test for acquired mutations affecting the FA pathway. Acquired mutations in FA genes may account, at least in part, for bone marrow failure and malignancies in the general (non-FA) population, through mechanisms that might include loss of heterozygosity in the setting of a germ line heterozygous mutation or from acquisition of 2 somatic mutations. In addition to the previously reported interaction between BRCA1 and FANCD2, the recent identification of the gene for FANCD1 and FANCB as the BRCA2 gene establishes a biochemical link between Fanconi anemia and tumor suppression.23 Furthermore, the ataxia telangiectasia (ATM) cell cycle checkpoint pathway has been shown to be responsible for phosphorylation of FANCD2.57FA mutations may also account for increased sensitivity to chemotherapy or radiation in some patients treated for malignancy.37 39Because 7 different FA genes each exhibit heterogeneous mutations, gene sequencing approaches to identify FA pathway disruption are laborious, and the functional significance of any novel mutations would require confirmation. Screening for the integrity of the FA biochemical pathway by FANCD2 immunoblot would provide compelling evidence for functional disruption of the FA pathway.
We thank Hanna Gazda and Colin Sieff for cells from Diamond-Blackfan anemia patients. We thank Eva Guinan for stimulating discussions.
Prepublished online as Blood First Edition Paper, August 29, 2002; DOI 10.1182/blood-2002-05-1399.
Supported by National Institutes of Health grants R01HL52725 and P01HL54785 (A.D.D.), P50 HL54785 (A.D.D., D.G.N., A.S.), and by grants from the Charles H. Hood Foundation and Doris Duke Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Akiko Shimamura, Dana-Farber Cancer Institute, Department of Pediatric Oncology, Harvard Medical School, 44 Binney St, Boston, MA 02115; e-mail:akiko_shimamura@dfci.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal