Dendritic cells (DCs) are key antigen-presenting cells for stimulating immune responses and they are now being investigated in clinical settings. Although defined as lineage-negative (Lin−) HLA-DR+ cells, significant heterogeneity in these preparations is apparent, particularly in regard to the inclusion or exclusion of CD14+, CD16+, and CD2+ cells. This study used flow cytometry and a panel of monoclonal antibodies (mAbs), including reagents from the 7th Leukocyte Differentiation Antigen Workshop, to define the cellular composition of 2 standardized peripheral blood mononuclear cell (PBMCs)–derived Lin− HLA-DR+preparations. Lin− cells were prepared from PBMCs by depletion with CD3, CD14, CD19, CD11b, and either CD16 or CD56 mAbs. Analysis of the CD16-replete preparations divided the Lin− HLA-DR+ population into 5 nonoverlapping subsets (mean ± 1 SD): CD123 (mean = 18.3% ± 9.7%), CD1b/c (18.6% ± 7.6%), CD16 (49.6% ± 8.5%), BDCA-3 (2.7% ± 1.4%), and CD34 (5.0% ± 2.4%). The 5 subsets had distinct phenotypes when compared with each other, monocytes, and monocyte-derived DCs (MoDCs). The CD85 family, C-type lectins, costimulatory molecules, and differentiation/activation molecules were also expressed differentially on the 5 Lin−HLA-DR+ subsets, monocytes, and MoDCs. The poor viability of CD123+ DCs in vitro was confirmed, but the CD16+ CD11c+ DC subset also survived poorly. Finally, the individual subsets used as stimulators in allogeneic mixed leukocyte reactions were ranked by their allostimulatory capacity as CD1b/c > CD16 > BDCA-3 > CD123 > CD34. These data provide an opportunity to standardize the DC populations used for future molecular, functional and possibly even therapeutic studies.
Introduction
Dendritic cells (DCs) are specialist antigen-presenting cells that originate from the bone marrow and play critical roles in the initiation and direction of immune responses.1,2 They are being investigated in cancer biology, transplantation, and autoimmunity. The definition of a DC, to date, has been mainly a functional one.2 The ability of DCs to take up, process, and present antigens to stimulate T (and B) lymphocytes is accompanied by certain, less consistent, phenotypic and morphologic features. DCs lack certain lineage (Lin)–specific markers and express high levels of major histocompatibility complex (MHC) class II molecules; thus, the phenotypic definition of DC as Lin−HLA-DR+ cells has become widespread. Their relatively low frequency in leukocyte preparations3,4 and, in particular, the paucity of DC-specific reagents, has hampered their investigation. This has meant that purification of DCs has and continues to depend heavily on their Lin− status. However, recent evidence suggests that one or more DC or DC-like populations may express CD14, CD2, or CD16, which have traditionally been associated with Lin+ cells.5-7 Interlaboratory variation in the particular combination of Lin monoclonal antibody (mAb) and purification methodologies (rosetting, magnetic bead depletion, flow cytometric sorting) used, is likely to result in DC preparations with different compositions.
Human blood DC preparations contain several phenotypically and functionally distinct subpopulations.8,9 Their constitution, function, and lineage of origin still require clarification. The original “myeloid” CD11c+CD123lo DC subset is now contrasted with the CD11c−CD123hi “lymphoid” DC population.10 Other markers including CD33, CD16, CD2, CD1, and CD85 (immunoglobulinlike transcript; ILT)11 have been used to further fractionate these populations. Thus, CD33 density has been suggested to define a mature and immature myeloid subset6 and recent reports of a CD16+ DC population5,12 may extend these to include the previously described CD16+CD14lo monocyte population.13,14 CD2 has been described on a subset of CD14+ leukocytes, which exhibit DC characteristics including the capacity for priming naive T lymphocytes.7 CD1a was reported to demarcate a Lin− population, which acquired Langerhans cell (LC) features in vitro15; however, this population has been reinvestigated and redefined as a CD1a−CD1b/c+population.16 The CMRF-44 mAb appeared to define 3 blood DC populations after a brief period of in vitro culture.17 BDCA-3 mAb identifies another subpopulation.18
Knowledge of the relative contributions of the DC subsets to a defined Lin−HLA-DR+ preparation, would be useful. So, too, would a direct comparison of the phenotypic and functional properties of subsets. This might assist attempts to define whether these apparent subsets represent different stages of differentiation/activation of the same lineage or the progression of DC differentiation pathways. In this study, we examined the heterogeneity of human peripheral blood mononuclear cell (PBMC)–derived DC preparations using a wide panel of mAbs, many of which were made available by colleagues participating in the DC section of the 7th Leukocyte Differentiation Antigen Workshop (LDAW).19Emphasis was placed on identifying subsets with restricted expression of molecules with known functions, which might therefore delineate functionally distinct subsets.
Materials and methods
Monoclonal and polyclonal antibodies used in the study are listed in Table 1.
mAb and polyclonal reagents used in this study
| CD/name . | Name/clone . | Conjugate . | Source . |
|---|---|---|---|
| Lineage-depletion mAb | |||
| CD3 | OKT3 | Nil | ATCC1-a |
| CD11b | OKM1 | Nil | ATCC |
| CD14 | CMRF31 | Nil | In house |
| CD16 | HuNK-2 | Nil | I. McKenzie1-b |
| CD19 | FMC63 | Nil | H. Zola1-c |
| CD56 | NKH-1 | Nil | Beckman Coulter1-d |
| HLA-DR | L243 | Nil | ATCC |
| Phenotyping mAb | |||
| BDCA-3 | — | PE, FITC | Miltenyi |
| CD1b/c* | M306 | Biotin | Immunex1-e |
| CD1c* | BDCA-1 | FITC, PE | Miltenyi |
| CD2 | T11 | PE, FITC | Coulter |
| CD4 | Leu-3a | PE | BD |
| CD7 | M-T107 | PE | BD |
| CD11c | Leu-M5 | PE | BD |
| CD11c | 3.9 | FITC | Southern Biotech1-f |
| CD14 | Leu-M3 | PE | BD |
| CD16 | Leu-11a/c | PE, FITC | BD |
| CD20 | B9E9 | PE | Immunotech1-g |
| CD33 | P67.6 | PE | BD |
| CD34 | Anti-HPCA-2 | PE, FITC | BD |
| CD40 | MAB 89 | PE | Immunotech |
| CD52 | YTH34.5 | FITC | Serotec1-h |
| CD56 | NKH-1 | PE | Immunotech |
| CD64 | 10.1 | PE | Serotec |
| CD64 | 10.1 | FITC | Pharmingen1-i |
| CD80/B7-1 | BBI | PE | Pharmingen |
| CD83 | HB15a | Nil, FITC, PE | Immunotech |
| CD85a/ILT-5 | 7H5 | Nil | A van Agthoven1-j |
| CD85d/ILT-4 | 42DI | Nil | A van Agthoven |
| CD85j/ILT-2 | HP-F1 | Nil | A van Agthoven |
| CD85j/ILT-2 | VMP55 | FITC | Dako1-k |
| CD85k/ILT-3 | ZM3.8 | Nil | A van Agthoven |
| CD86/B7-2 | 2331 | PE | Pharmingen |
| CD123 | 7G3 | PE | Pharmingen |
| CLA | HECA-452 | FITC | Pharmingen |
| CMRF-44 | — | Biotin | In house |
| CMRF-56 | — | Biotin | In house |
| DEC-205/CD205 | MMRI-7 | Biotin | In house |
| DEC-205/CD205 | M335 | Biotin | Immunex1-j |
| DC-SIGN/CD209 | AZN-D1 | Nil | Y. van Kooyk1-j |
| HLA-DR | Immu-35T | PC5 | Immunotech |
| HLA-DR | B-F1 | FITC | Serotec |
| Langerin/CD207 | DCGM-4 | Nil | Schering-Plough1-j |
| MMR/CD206 | MMR 190.BB3 | Nil | F. Sallusto1-j |
| Detecting reagents | |||
| Anti-mouse Ig | Polyclonal | FITC, PE | Silenus1-l |
| Anti-mouse Ig | Polyclonal | Biotin | Sigma1-m |
| Streptavidin | — | PE, PE-Cy5 | Dako |
| Avidin | — | FITC | BD |
| CD/name . | Name/clone . | Conjugate . | Source . |
|---|---|---|---|
| Lineage-depletion mAb | |||
| CD3 | OKT3 | Nil | ATCC1-a |
| CD11b | OKM1 | Nil | ATCC |
| CD14 | CMRF31 | Nil | In house |
| CD16 | HuNK-2 | Nil | I. McKenzie1-b |
| CD19 | FMC63 | Nil | H. Zola1-c |
| CD56 | NKH-1 | Nil | Beckman Coulter1-d |
| HLA-DR | L243 | Nil | ATCC |
| Phenotyping mAb | |||
| BDCA-3 | — | PE, FITC | Miltenyi |
| CD1b/c* | M306 | Biotin | Immunex1-e |
| CD1c* | BDCA-1 | FITC, PE | Miltenyi |
| CD2 | T11 | PE, FITC | Coulter |
| CD4 | Leu-3a | PE | BD |
| CD7 | M-T107 | PE | BD |
| CD11c | Leu-M5 | PE | BD |
| CD11c | 3.9 | FITC | Southern Biotech1-f |
| CD14 | Leu-M3 | PE | BD |
| CD16 | Leu-11a/c | PE, FITC | BD |
| CD20 | B9E9 | PE | Immunotech1-g |
| CD33 | P67.6 | PE | BD |
| CD34 | Anti-HPCA-2 | PE, FITC | BD |
| CD40 | MAB 89 | PE | Immunotech |
| CD52 | YTH34.5 | FITC | Serotec1-h |
| CD56 | NKH-1 | PE | Immunotech |
| CD64 | 10.1 | PE | Serotec |
| CD64 | 10.1 | FITC | Pharmingen1-i |
| CD80/B7-1 | BBI | PE | Pharmingen |
| CD83 | HB15a | Nil, FITC, PE | Immunotech |
| CD85a/ILT-5 | 7H5 | Nil | A van Agthoven1-j |
| CD85d/ILT-4 | 42DI | Nil | A van Agthoven |
| CD85j/ILT-2 | HP-F1 | Nil | A van Agthoven |
| CD85j/ILT-2 | VMP55 | FITC | Dako1-k |
| CD85k/ILT-3 | ZM3.8 | Nil | A van Agthoven |
| CD86/B7-2 | 2331 | PE | Pharmingen |
| CD123 | 7G3 | PE | Pharmingen |
| CLA | HECA-452 | FITC | Pharmingen |
| CMRF-44 | — | Biotin | In house |
| CMRF-56 | — | Biotin | In house |
| DEC-205/CD205 | MMRI-7 | Biotin | In house |
| DEC-205/CD205 | M335 | Biotin | Immunex1-j |
| DC-SIGN/CD209 | AZN-D1 | Nil | Y. van Kooyk1-j |
| HLA-DR | Immu-35T | PC5 | Immunotech |
| HLA-DR | B-F1 | FITC | Serotec |
| Langerin/CD207 | DCGM-4 | Nil | Schering-Plough1-j |
| MMR/CD206 | MMR 190.BB3 | Nil | F. Sallusto1-j |
| Detecting reagents | |||
| Anti-mouse Ig | Polyclonal | FITC, PE | Silenus1-l |
| Anti-mouse Ig | Polyclonal | Biotin | Sigma1-m |
| Streptavidin | — | PE, PE-Cy5 | Dako |
| Avidin | — | FITC | BD |
ATCC indicates American Type Culture Collection; BD, Becton Dickinson.
CD1b/c (M306) and CD1c (BDCA-1) used interchangeably as required. Denoted as CD1b/c in text and in figures.
Manassas, VA
Austin Research Institute, Melbourne, Australia
Child Health Research Institute, Adelaide, Australia
Gladesville, Australia
Seattle, WA
Birmingham, AL
Beckman Coulter, Gladesville, Australia
Raleigh, NC
BD Biosciences, San Diego, CA
7th LDAW c/o Nuffield Department of Clinical Biochemistry and Cellular Science
Botany, NSW, Australia
Chemicon, Boronia, Victoria, Australia
Sydney, Australia
Cell purification
Buffy coats from healthy donors were obtained from the Australian Red Cross Service (Brisbane, Australia). PBMCs were isolated by standard density gradient centrifugation over Ficoll-Paque Plus (Pharmacia, Uppsala, Sweden). PBMCs were stained with mAb cocktails designed either to include CD16+ cells (CD3 [OKT3], CD14 [CMRF-31], CD19 [FMC-63], CD11b [OKM1], CD56 [NKH-1]) or to exclude them (CD3, CD14, CD19, CD11b, CD16 [HuNK-2]). CD11b was included to ensure monocyte-negative selection was adequate and because several investigations have shown all blood DCs lack CD11b.6 20 Following washing, the cells were incubated with Biomag goat anti–mouse-immunoglobulin–coated magnetic beads (Polysciences, Warrington, PA). Labeled cells were depleted by first preclearing with a MCP-1 magnet (Dynal, Oslo, Norway) followed by passing through a magnetic-activated cell sorting (MACS) CS column on a Variomacs magnet (Miltenyi Biotech, Gladbach, Germany). For some experiments, Lin−HLA-DR+ cells were sort purified (FACS Vantage, Becton Dickinson, San Jose, CA) using 2-color labeling with phycoerythrin (PE)–conjugated CD3, CD56, and CD20 to gate out contaminating Lin+ cells in combination with HLA-DR-PE-cyanin 5.1 (Cy5). For cell activation and functional studies Lin− preparations were labeled with fluorescein isothiocyanate (FITC)–conjugated lineage mAb (CD56, CD3, CD14, and CD20) and various combinations of PE-conjugated or PE-Cy5–conjugated subset mAbs (CD123, CD16, CD1b/c, BDCA-3, and CD34), and the desired Lin− subsets sorted.
For T-cell purification, PBMCs were fractionated by incubation with neuraminidase-treated sheep red blood cells followed by separation of the rosetting (ER+) and nonrosetting (ER−) populations on Ficoll density gradients. After lysis of the ER+ fraction with 0.15M NH4Cl, pure populations of responder T cells were prepared by magnetic immunodepletion with CD14 (CMRF-31), CD19 (FMC-63), CD16 (HuNK-2), CD11b (OKM1), and HLA-DR (L243) mAbs. The resulting cells were 96% to 100% CD3+.
For generation of monocyte-derived DCs (MoDCs), the nonrosetting population was cultured at 0.3 × 106 CD14+monocytes/mL in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF; 200 U/mL, Sandoz-Pharma, Basel, Switzerland) and interleukin 4 (IL-4; 50 U/mL, Sigma, St Louis, MO) for 5 days.21
Cell staining
Cells were incubated with mAb according to the manufacturer's instructions, or at 5 to 20 μg/mL in 2% fetal calf serum (FCS) in phosphate-buffered saline (PBS), or in hybridoma culture supernatant for 20 to 60 minutes at 4°C. Cells were washed in 2% FCS in PBS. Unconjugated mAbs were detected with the appropriate conjugated secondary detection reagents diluted in 2% FCS in PBS. For phenotypic analysis, following staining the cells were washed and fixed with 1% paraformaldehyde prior to analysis on a FACS Calibur cytometer (Becton Dickinson). For cell survival studies, unfixed cells were analyzed, and propidium iodide (PI; 3 μg/mL) was used to identify dead cells. Cytoplasmic staining, using “Fix & Perm” (Caltag, Burlingame, CA), for langerin was done on PBMCs by 4-color flow cytometry using biotinylated antilangerin mAb (CD207, DCGM-4) detected with streptavidin-peridinin chlorophyll protein (PerCp). DCs were identified as surface-labeled Lin mAb-FITC−/HLA-DR-APC+ events, and DC subsets were identified with the appropriate PE conjugates.
DC culture
Sort-purified Lin−CD11c+(± CD16+ cells) or Lin−CD11c−cells were incubated overnight at 106 cells/mL in RPMI 1640 with 10% FCS, 10 ng/mL IL-3 (Gibco BRL, Grand Island, NY) and 200 U/mL GM-CSF, or were cocultured in the absence of cytokines with allogeneic CD3+ T cells at a 1:1 ratio.
Allogeneic MLRs
Allogeneic mixed leukocyte reactions (MLRs) were established using various numbers of each Lin− subset cultured in triplicate in round-bottom 96-well tissue culture plates (Costar, Acton, MA) with 105 freshly isolated allogeneic T cells, at 37°C in 5% CO2 for 5 days. T-cell proliferation was measured by the uptake of [3H]-thymidine (1 μCi/well [0.037 MBq/well]; 6.7 Ci/mM [248 MBq/mM]; Amersham, Buckinghamshire, United Kingdom), which was added 18 hours prior to harvesting. Cells were harvested onto glass fiber filter paper with an automated harvester (TomTec Mach III, Hamden, CT) and [3H]-thymidine incorporation was measured by liquid scintillation spectroscopy (Wallac, Turku, Finland). Responses are reported as mean cpm ± SEM for triplicate wells.
Results
Analysis of the composition of Lin−HLA-DR+PBMC preparations
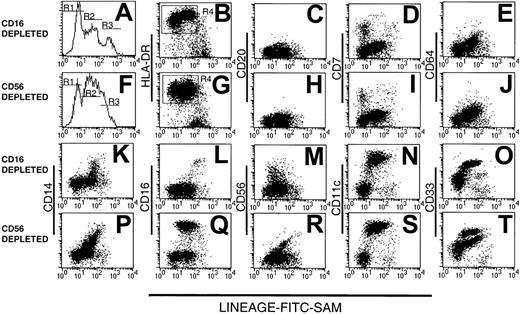
We prepared 2 types of Lin− PBMC preparations using either CD16 or CD56 mAb to deplete natural killer (NK) cells, the latter with a view to studying the CD16+ DC population.5 12 The resulting CD16- and CD56-depleted Lin− preparations were examined for purity, assessed by reactivity with FITC-conjugated sheep anti–mouse immunoglobulin (FITC-SAM) to detect contaminating Lin+ cells. The majority (75% ± 25.8% SD, n = 14) of cells within both types of Lin− preparations were Lin− (R1 in Figure1) or weakly fluorescent (within the second decade of fluorescence intensity, R2 in Figure 1). The remaining strongly labeled cells were considered to be contaminating Lin+ cells (R3, Figure 1A,F).
Human Lin− PBMCs.
Analysis of human Lin− PBMC preparations obtained using a mAb mix containing either CD16 (A-E and K-O) or CD56 mAb (F-J and P-T). Lin− cells were stained with FITC-SAM (to detect residual Lin+ cells) and HLA-DR in conjunction with one of a panel of lineage mAbs. Flow cytometry profiles of (A) CD16-depleted or (F) CD56-depleted residual lineage-labeling intensity shows 3 peaks: R1, R2, and R3. The Lin−HLA-DR+ cells (B,G) were analyzed further for CD20 (C,H), CD7 (D,I), CD64 (E,J), CD14 (K,P), CD16 (L,Q), CD56 (M,R), CD11c (N,S), and CD33 (O,T).
Human Lin− PBMCs.
Analysis of human Lin− PBMC preparations obtained using a mAb mix containing either CD16 (A-E and K-O) or CD56 mAb (F-J and P-T). Lin− cells were stained with FITC-SAM (to detect residual Lin+ cells) and HLA-DR in conjunction with one of a panel of lineage mAbs. Flow cytometry profiles of (A) CD16-depleted or (F) CD56-depleted residual lineage-labeling intensity shows 3 peaks: R1, R2, and R3. The Lin−HLA-DR+ cells (B,G) were analyzed further for CD20 (C,H), CD7 (D,I), CD64 (E,J), CD14 (K,P), CD16 (L,Q), CD56 (M,R), CD11c (N,S), and CD33 (O,T).
The composition of the Lin− preparations was further assessed by 3-color flow cytometry using FITC-SAM, PE-Cy5–HLA-DR and a panel of PE-conjugated lineage markers. Both CD16- and CD56-depleted Lin− preparations contained an exclusively HLA-DR+ population within R1 and R2, whereas the cells within R3 were largely HLA-DR− (Figure 1B,G). In CD16-depleted preparations, the cells within R2 expressed HLA-DR at a higher density than those in R1. This difference was barely evident in CD56-depleted preparations. Gating on the Lin−HLA-DR+ cells (R4, Figure 1B,G) demonstrated that cells from both types of preparation lacked CD20+ B cells (Figure1C,H), but a small population of CD7+ cells was consistently observed in R1 (Figure 1D,I). In both preparations, cells within R2 expressed low levels of CD64, with a modest correlation between the intensity of CD64 and low-density Lin+ marker staining (Figure 1E,J). No CD64+ cells were found within R1. Variable CD14 expression within both preparations contributed to the low-intensity Lin+ labeling in R2 and correlated with Lin+ intensity (Figure 1K,P). The CD56-depleted preparations contained increased numbers of CD14+ cells. As expected, CD16+ cells were present in the CD56-depleted Lin− cell populations (Figure1 L,Q). A disperse HLA-DR+CD56+ population was detected in the Lin−CD16-depleted preparations with residual positive cells also found in the CD56-depleted preparations. These were largely restricted to R2 (Figure 1M,R).
As expected, Lin−HLA-DR+ cells were divided into substantial CD11c− and CD11c+populations, with the 2 populations localized to R1 and R2, respectively (Figure 1N,S). Staining with CD33 divided CD16-depleted Lin− preparations into 2 subsets (Figure 1O). In contrast, CD56-depleted Lin− preparations contained 3 cell populations, based on CD33 expression (Figure 1T). Thus, whereas all myeloid Lin− cells uniformly expressed CD11c, the Lin−HLA-DR+CD16+ cells could be distinguished by their low-density CD33 expression from their HLA-DR+CD11c+CD16− counterparts. Lin−CD33lo cells have been described as immature blood DCs.22 Subsequent Lin− subset analyses were performed on CD16-containing, CD56-depleted Lin− preparations.
CD123, CD1b/c, CD16, BDCA-3, and CD34 subdivide the Lin−HLA-DR+ preparations into 5 distinct nonoverlapping subsets
CD16,5,12 CD1c,15,18,16 and BDCA-318 have recently been identified as markers that subdivide the CD11c+ DC population. CD123−, CD123lo, and CD123hi subpopulations are present in Lin− preparations.9 Furthermore, CD34+ cells have been documented as components of Lin−HLA-DR+ preparations.20 We, therefore, examined the relative frequency and the level of expression of HLA-DR and CD11c by each of the 5 Lin− subsets defined using these reagents (CD16, CD1b/c, BDCA-3, CD123, and CD34).
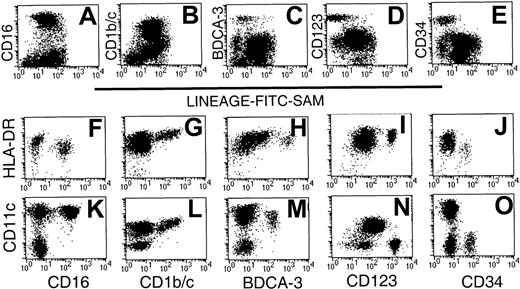
Initial 2-color analysis confirmed that each of the 5 subsets was present in the Lin− preparations. Whereas the CD16+ and CD1b/c+ subsets were largely restricted to R2, the CD123+ and CD34+ subsets were localized within R1 (Figure 1). In contrast, the BDCA-3 subset was dispersed between both regions (Figure 2A-E). The relative expression of HLA-DR and CD11c by the 5 populations was next examined on sort-purified Lin−HLA-DR+ cells by 3-color staining. The CD16+ cells were identified as CD11chi cells (Figure 2K), which exhibit a lesser overall density of HLA-DR (Figure2F). Labeling with CD1b/c identified an HLA-DRhisubpopulation (Figure 2G), which also expressed the highest levels of CD11c (Figure 2L; note that different fluorochrome conjugates of CD11c were used in Figure 2K-O). Notably, the highest levels of CD1b/c expression directly correlated with the highest levels of both HLA-DR and CD11c. BDCA-3+ DCs expressed CD11c and moderate to high levels of HLA-DR (Figure 2H,M). The CD123hi subset had moderate- to high-density HLA-DR expression but was CD11c−(Figure 2I,N). The CD34+ subset was CD11c− and expressed only low levels of HLA-DR (Figure 2J,O). Thus, the expression of CD16, BDCA-3, and CD1b/c was restricted to CD11c+ cells and CD123 and CD34 to CD11c− cells. These 5 Lin− populations expressed differential levels of HLA-DR.
Phenotypic analysis of Lin− PBMC preparations identifies 5 phenotypic subsets.
CD56-depleted Lin− preparations were stained with FITC-SAM and either CD16, CD1b/c, BDCA-3, CD123, or CD34. Live cells were gated based on forward- and side-scatter characteristics and analyzed for (A) CD16, (B) CD1b/c, (C) BDCA-3, (D) CD123, and (E) CD34 staining. Sort-purified Lin−HLA-DR+ cells were used to examine the relative intensity of HLA-DR and CD11c on each of the defined subsets: CD16 (F,K), CD1b/c (G,L), BDCA-3 (H,M), CD123 (I,N), and CD34 (J,O). Differences in fluorescence intensity for BDCA-3 (H,M) or CD11c (K-O) reflect the use of different fluorescent conjugates.
Phenotypic analysis of Lin− PBMC preparations identifies 5 phenotypic subsets.
CD56-depleted Lin− preparations were stained with FITC-SAM and either CD16, CD1b/c, BDCA-3, CD123, or CD34. Live cells were gated based on forward- and side-scatter characteristics and analyzed for (A) CD16, (B) CD1b/c, (C) BDCA-3, (D) CD123, and (E) CD34 staining. Sort-purified Lin−HLA-DR+ cells were used to examine the relative intensity of HLA-DR and CD11c on each of the defined subsets: CD16 (F,K), CD1b/c (G,L), BDCA-3 (H,M), CD123 (I,N), and CD34 (J,O). Differences in fluorescence intensity for BDCA-3 (H,M) or CD11c (K-O) reflect the use of different fluorescent conjugates.
Further 3-color analysis of a series of CD56-depleted Lin−preparations stained with HLA-DR, CD11c, and one of CD1b/c, BDCA-3, CD16, CD34, or CD123 showed that these subpopulations accounted for 94.1% of the HLA-DR+ cells. Furthermore, 93.8% of the CD11c+ population, which comprised 75.6% of the HLA-DR+ cells, was represented by 3 subsets: CD16+ (65.5%), CD1b/c+ (24.6%), and BDCA-3 (3.6%; Table 2). In these preparations, the CD123hi and CD34+ populations comprised the remaining CD11c− cells, that is, 18.3% and 5.0% of HLA-DR+ cells, respectively. Thus, based on the discreet expression of CD123, CD1b/c, CD16, BDCA-3, and CD34, these Lin−HLA-DR+ preparations could be fractionated, almost in their entirety, into 5 distinct cell populations. The data in Table 2 suggest that, on average, 5.9% of Lin−HLA-DR+ cells do not belong to one of the subsets described. The mAbs to positively define these cells have not yet been identified and they were not characterized further in this study.
DC subset frequency
| Donor no. . | % of HLA-DR+Lin− cells . | Sum of 5 subsets . | |||||
|---|---|---|---|---|---|---|---|
| CD16 . | CD1b/c . | BDCA-3 . | CD123 . | CD34 . | CD11c . | ||
| 1 | 51.4 | 13.3 | 1 | 12.4 | 6.7 | 78.7 | 84.8 |
| 2 | 51.4 | 19.9 | 1.9 | 20 | 6.9 | 72.1 | 100.1 |
| 3 | 52.7 | 8.6 | 5.9 | 38.1 | 5.8 | 72.1 | 111.1 |
| 4 | 43.6 | 23.7 | 2.5 | 16.2 | 8.8 | 75.7 | 94.8 |
| 5 | 55.9 | 18.7 | 3.3 | 10.9 | 1.4 | 83.3 | 90.2 |
| 6 | 58.4 | 13.2 | 2 | 18.5 | 2.8 | 72.5 | 94.9 |
| 7 | 46.1 | 35 | 2 | 6.8 | 4.3 | 83.8 | 94.2 |
| 8 | 56 | 15.3 | 2.3 | 13.3 | 5.3 | ND | 92.2 |
| 9 | 30.6 | 19.9 | 3.2 | 28.3 | 2.8 | 66.4 | 84.8 |
| Mean | 49.6 | 18.6 | 2.7 | 18.3 | 5.0 | 75.6 | 94.1 |
| SD | 8.5 | 7.6 | 1.4 | 9.7 | 2.4 | 6.0 | 8.0 |
| Range | 31-58 | 9-24 | 1-6 | 7-38 | 1-9 | 66-94 | 85-111 |
| Donor no. . | % of HLA-DR+Lin− cells . | Sum of 5 subsets . | |||||
|---|---|---|---|---|---|---|---|
| CD16 . | CD1b/c . | BDCA-3 . | CD123 . | CD34 . | CD11c . | ||
| 1 | 51.4 | 13.3 | 1 | 12.4 | 6.7 | 78.7 | 84.8 |
| 2 | 51.4 | 19.9 | 1.9 | 20 | 6.9 | 72.1 | 100.1 |
| 3 | 52.7 | 8.6 | 5.9 | 38.1 | 5.8 | 72.1 | 111.1 |
| 4 | 43.6 | 23.7 | 2.5 | 16.2 | 8.8 | 75.7 | 94.8 |
| 5 | 55.9 | 18.7 | 3.3 | 10.9 | 1.4 | 83.3 | 90.2 |
| 6 | 58.4 | 13.2 | 2 | 18.5 | 2.8 | 72.5 | 94.9 |
| 7 | 46.1 | 35 | 2 | 6.8 | 4.3 | 83.8 | 94.2 |
| 8 | 56 | 15.3 | 2.3 | 13.3 | 5.3 | ND | 92.2 |
| 9 | 30.6 | 19.9 | 3.2 | 28.3 | 2.8 | 66.4 | 84.8 |
| Mean | 49.6 | 18.6 | 2.7 | 18.3 | 5.0 | 75.6 | 94.1 |
| SD | 8.5 | 7.6 | 1.4 | 9.7 | 2.4 | 6.0 | 8.0 |
| Range | 31-58 | 9-24 | 1-6 | 7-38 | 1-9 | 66-94 | 85-111 |
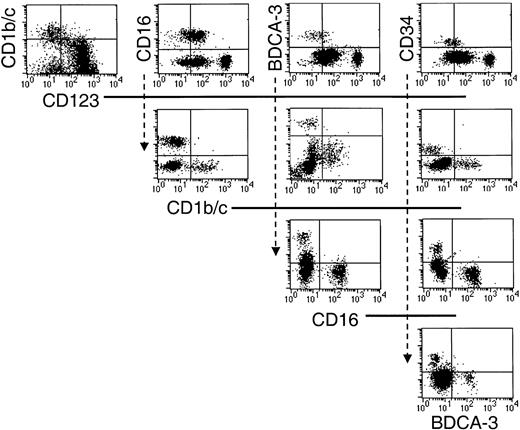
Having positively defined 5 populations, additional triple-labeling studies were undertaken to clarify that they were nonoverlapping subsets. Thus, sort-purified Lin−HLA-DR+ cells were stained with various combinations of the defining antibodies, and the restricted expression of each of these markers by the 5 Lin−HLA-DR+ populations was confirmed (Figure3).
The 5 phenotypically defined subsets in CD56-depleted Lin− PBMC preparations represent nonoverlapping populations.
Sort-purified HLA-DR+ Lin− cells were stained with various combinations of the defining mAb as indicated in the dot-plot representations.
The 5 phenotypically defined subsets in CD56-depleted Lin− PBMC preparations represent nonoverlapping populations.
Sort-purified HLA-DR+ Lin− cells were stained with various combinations of the defining mAb as indicated in the dot-plot representations.
The 5 Lin−HLA-DR+ PBMC subsets express unique phenotypes
We next examined the 5 Lin−HLA-DR+populations for their expression of markers reported to be expressed by DC subsets. Lin− preparations were labeled with FITC- or PE-SAM followed by HLA-DR–PE-Cy5, and the Lin−HLA-DR+ cells (ie,HLA-DR+ cells within R1 and R2, Figure 1) were sorted. They were then labeled with one of the 5 PE- or FITC-conjugated subset markers and one of a panel of PE- or FITC-conjugated mAbs or isotype-matched controls (Table 3). An analysis of CD14hi monocytes and immature MoDCs was also performed for comparative purposes. Of note, cutaneous lymphocyte antigen (CLA), a homing receptor for skin that has previously been reported to be widely expressed by DCs,15 23 was not detected on the CD16+ population and only minimally expressed on the MoDCs. In contrast, all other populations expressed CLA either homogeneously (CD1b/c, BDCA-3, CD123) or heterogeneously (CD34 and CD14). Additionally, CD2 expression was limited to the CD1b/c+ population, a small and variable proportion of the CD123hi and CD34+ subsets, and a small subset of CD14hi monocytes. CD64 expression was restricted to the CD1b/c subset and the CD14hi monocytes. A small proportion of each Lin−HLA-DR+CD11c+subpopulation expressed CD56, in contrast to CD7 expression, which was confined to a subset of the CD123hi population.
Phenotype of HLA-DR+ Lin−populations
| . | CD16 . | CD1b/c . | BDCA-3 . | CD123hi . | CD34 . | CD14hi . | MoDCs . |
|---|---|---|---|---|---|---|---|
| HLA-DR | +/++3-150 | ++ | ++ | +/++3-150 | + | ++3-150 | ++3-151 |
| CD11c | +++ | +++ | ++ | − | − | +++ | +++ |
| CD33 | ++ | +++ | +++ | + | + | +++ | ++ |
| CD4 | + | + | + | ++ | − | + | ++ |
| CD2 | − | + | − | +3-151 | +3-151 | +3-151 | + |
| CLA | − | ++ | ++ | + | +3-151 | +3-151 | +/− |
| CD7 | − | − | − | +3-151 | − | +3-151 | − |
| CD56 | +3-151 | +3-151 | +3-151 | +3-151 | NT | +3-151 | − |
| CD64 | − | + | − | − | − | + | − |
| CD62L | + | + | ++ | ++3-150 | + | ++3-150 | − |
| CD52 | +++ | ++ | ++ | ++ | ++ | ++ | − |
| ILT molecules | |||||||
| ILT2/CD85j | ++ | + | − | + | − | + | ++ |
| ILT3/CD85k | + | ++ | +/− | ++ | − | + | + |
| ILT4/CD85d | ++ | +/− | − | − | − | + | +/− |
| ILT5/CD85a | + | + | − | − | − | + | + |
| C-type lectins | |||||||
| DEC205/CD205 | + | ++ | +++ | ++ | ++ | ++ | + |
| MMR/CD206 | − | − | − | − | − | − | + |
| Langerin/CD207 surface | − | − | − | − | − | − | − |
| cytoplasmic | +/− | +/− | +/− | +/− | − | +/− | + (TGF-β) |
| DC-SIGN/CD209 | − | − | − | − | − | − | + |
| Costimulatory molecules | |||||||
| CD40 | + | +3-150 | ++ | +/− | + | + | ++ |
| CD80 | − | − | − | − | − | − | + |
| CD86 | ++ | + | + | + | − | + | + |
| . | CD16 . | CD1b/c . | BDCA-3 . | CD123hi . | CD34 . | CD14hi . | MoDCs . |
|---|---|---|---|---|---|---|---|
| HLA-DR | +/++3-150 | ++ | ++ | +/++3-150 | + | ++3-150 | ++3-151 |
| CD11c | +++ | +++ | ++ | − | − | +++ | +++ |
| CD33 | ++ | +++ | +++ | + | + | +++ | ++ |
| CD4 | + | + | + | ++ | − | + | ++ |
| CD2 | − | + | − | +3-151 | +3-151 | +3-151 | + |
| CLA | − | ++ | ++ | + | +3-151 | +3-151 | +/− |
| CD7 | − | − | − | +3-151 | − | +3-151 | − |
| CD56 | +3-151 | +3-151 | +3-151 | +3-151 | NT | +3-151 | − |
| CD64 | − | + | − | − | − | + | − |
| CD62L | + | + | ++ | ++3-150 | + | ++3-150 | − |
| CD52 | +++ | ++ | ++ | ++ | ++ | ++ | − |
| ILT molecules | |||||||
| ILT2/CD85j | ++ | + | − | + | − | + | ++ |
| ILT3/CD85k | + | ++ | +/− | ++ | − | + | + |
| ILT4/CD85d | ++ | +/− | − | − | − | + | +/− |
| ILT5/CD85a | + | + | − | − | − | + | + |
| C-type lectins | |||||||
| DEC205/CD205 | + | ++ | +++ | ++ | ++ | ++ | + |
| MMR/CD206 | − | − | − | − | − | − | + |
| Langerin/CD207 surface | − | − | − | − | − | − | − |
| cytoplasmic | +/− | +/− | +/− | +/− | − | +/− | + (TGF-β) |
| DC-SIGN/CD209 | − | − | − | − | − | − | + |
| Costimulatory molecules | |||||||
| CD40 | + | +3-150 | ++ | +/− | + | + | ++ |
| CD80 | − | − | − | − | − | − | + |
| CD86 | ++ | + | + | + | − | + | + |
NT indicates not tested; TGF-β, positive staining after culture with TGF-β.
Heterogeneous staining.
Subset positive.
The 5 Lin−HLA-DR+ subsets have diverse CD85 (ILT) molecular profiles
Because DCs, either ex vivo or in vitro derived, express several family members of the CD85 family,11 we examined the expression of some of the inhibitory members of this family (CD85j, k, d, a = ILT-2, -3, -4, and -5, respectively) by the Lin−HLA-DR+ subsets (Figure4; Table 3).
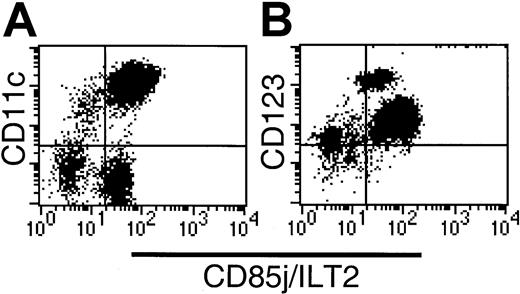
CD85j (ILT2) expression on CD56-depleted Lin−HLA-DR+ PBMCs.
(A) CD11c labeling demonstrates heterogeneous CD85j expression within both the CD11c+ and CD11c− compartments. (B) The CD123hi subset is CD85j+, indicating that the CD34+ subset lacks CD85j expression.
CD85j (ILT2) expression on CD56-depleted Lin−HLA-DR+ PBMCs.
(A) CD11c labeling demonstrates heterogeneous CD85j expression within both the CD11c+ and CD11c− compartments. (B) The CD123hi subset is CD85j+, indicating that the CD34+ subset lacks CD85j expression.
CD85j (ILT-2/LIR1/MIR7) is expressed widely by leukocytes including myeloid DCs. CD85j expression by CD16-depleted Lin−populations was initially analyzed by 3-color flow cytometry using HLA-DR, CD85-FITC (VMP55), and CD11c-PE or CD123-PE. Whereas all HLA-DR+CD123+ cells and the majority of HLA-DR+CD11c+ cells expressed CD85j, a small population of CD11c+ cells lacked the expression of CD85j (Figure 4A). Further examination of the Lin−HLA-DR+ populations revealed high levels of CD85j on the CD16+ population and moderate levels of expression on the CD1b/c+ and CD123+populations. Negligible expression of CD85j was detected on the BDCA-3+CD11clo population. As suggested by our initial screening (Figure 4B), the Lin−HLA-DR+CD34+population was CD85j−.
The expression of CD85k (ILT-3, mAb ZM3.8), CD85d (ILT-4, mAb 42D1), and CD85a (ILT-5, mAb 7H5) by the 5 populations is summarized in Table3. Again, the CD34+ population lacked expression of these 3 ILT molecules. The CD16 and CD1b/c subsets expressed varying levels of each. The BDCA-3 population expressed minimal levels of ILT-3, but none of the other molecules. The CD123hi population expressed ILT-3 at levels equivalent to the CD1b/c subset, but lacked expression of ILT-4 and ILT-5. Monocytes, as expected, expressed all the CD85 molecules tested at similar densities, whereas MoDCs expressed profiles intermediate to the CD16+ and CD1b/c+Lin−HLA-DR+ populations.
C-type lectin expression by the 5 Lin−HLA-DR+ subsets
DCs express several C-type lectin molecules, some of which may act as antigen uptake receptors. The expression of MMR (CD206), DEC-205 (CD205), DC-SIGN (CD209), and langerin (CD207) Lin−HLA-DR+ by the 5 Lin−HLA-DR+ subsets was examined in light of their potential to identify DC subpopulations. In contrast to MMR, DC-SIGN, and langerin, which were not detected on any of the Lin−HLA-DR+ subsets, DEC-205 was present on all the subsets (Table 3). Differential levels of DEC-205 expression were noted, with the BDCA-3+ subset expressing the highest levels, followed by the CD123hi, CD1b/c+, and CD34+ subsets, which expressed equivalent levels. The CD16+ subset expressed the lowest levels. Fresh monocytes expressed DEC-205 at a level similar to the CD1b/c+, CD123hi, and CD34+ subsets. Notably, MoDCs expressed lower levels of DEC-205 than the monocytes from which they were derived. Whereas MMR and DC-SIGN were absent from all Lin− populations (confirmed by reverse-transcription polymerase chain reaction, data not shown) and CD14himonocytes, both of these lectins were expressed by MoDCs. Langerin was not found on the surface of any of the freshly purified DC populations examined and cytoplasmic levels were, at most, marginally positive (Table 3). Langerin was induced on a subset of CD11c+Lin− cells after 18 hours of in vitro culture (in the absence of cytokines),19 confirming the specificity of antibody staining, but the ability to discriminate the DC subsets was lost after in vitro culture and this aspect was not pursued further.
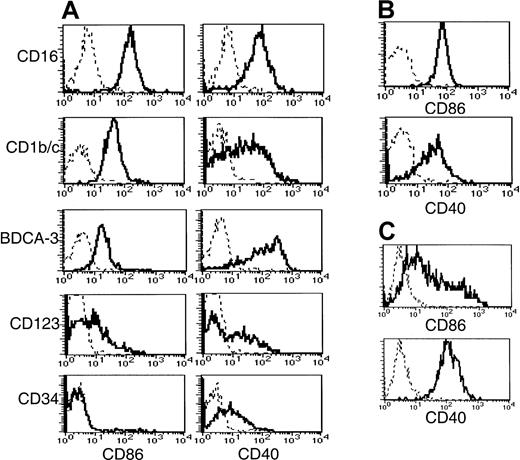
Disparate CD40 and CD86 expression on freshly purified Lin−HLA-DR+ cells
To obtain insight into the activation status and costimulatory capacity of the 5 subsets, we examined their expression of CD40, CD80, and CD86 (Figure 5). The BDCA-3+ population expressed significantly higher levels of CD40 than the other 2 CD11c+ populations. A broad range of CD40 expression by the CD1b/c+ population was noted compared to the expression by the CD16+ and BDCA-3+ populations, possibly reflecting a continuum of activation. CD86 expression did not correlate with CD40. Thus, the highest level of CD86 expression was detected on the CD16+subset, whereas the CD1b/c+ and BDCA-3+populations expressed lower levels. By comparison, monocytes expressed CD86 and CD40 (Figure 5B) at levels equivalent to the CD1b/c+ and CD16+ subsets, respectively. None of the Lin−HLA-DR+ subsets or monocytes expressed detectable levels of cell surface CD80, in contrast to MoDCs, which uniformly expressed low levels (Table 3).
Costimulatory molecule expression by Lin−HLA-DR+ subsets, monocytes, and MoDCs.
CD56-depleted Lin− PBMC preparations were stained with HLA-DR, one of the subset-defining mAbs, and one of PE-conjugated CD86, CD40, or negative control mAbs. Histograms representing CD86 and CD40 expression (solid lines) by (A) CD56-depleted HLA-DR+Lin− subsets is compared to that of (B) monocytes and (C) MoDCs. Negative control antibody staining is indicated by a dashed line.
Costimulatory molecule expression by Lin−HLA-DR+ subsets, monocytes, and MoDCs.
CD56-depleted Lin− PBMC preparations were stained with HLA-DR, one of the subset-defining mAbs, and one of PE-conjugated CD86, CD40, or negative control mAbs. Histograms representing CD86 and CD40 expression (solid lines) by (A) CD56-depleted HLA-DR+Lin− subsets is compared to that of (B) monocytes and (C) MoDCs. Negative control antibody staining is indicated by a dashed line.
Decreased viability of the CD123+ and CD16+subsets following in vitro culture
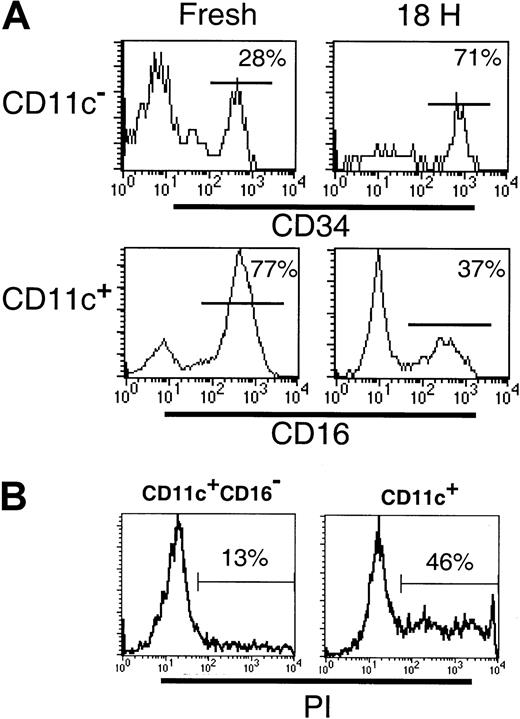
Differences between the CD11c+ and CD123+subsets in their survival in vitro have been described.24Therefore, we examined the survival of the DC subsets following 18 hours of culture. Cultured CD11c+ DCs have been observed to up-regulate cell surface CD123, such that gating CD11c+ and CD11c− populations becomes difficult. Furthermore, following 18 hours of culture, BDCA-3 was expressed by an increased proportion of CD11c+ cells and was induced on a subset of CD123+ cells.18 Thus, to facilitate subset tracking in these studies, Lin−CD11c+ and CD11c− subsets were sorted prior to culturing. Following 18 hours in the presence of GM-CSF and IL-3, or allogeneic T cells, the survival of the CD11c+CD16−(CD1b/c+ and BDCA-3+ populations), CD11c+CD16+, CD11c−CD34− (CD123+ population) and CD11c− CD34+ populations was examined. Even in the presence of GM-CSF and IL-3, cytokines reported to increase the survival of the CD11c+ and CD123+ cells, respectively,24 significant cell death occurred in both the CD11c+ and CD11c− cultures. Comparison of the relative proportion of CD16+ and CD34+cells in these cultures indicated that the CD11c+CD16+ and CD123+ populations had the poorest survival (Figure 6A). Thus, whereas the freshly sorted CD11c+ preparations contained 68% (± 6.6% SD; n = 4) CD16+ cells, following culture, this decreased to 31% (± 6.7%; n = 4) of viable (based on forward- and side-scatter characteristics) CD11c+ cells. To demonstrate that the loss of CD16+ cells was due to cell death rather than the loss of cell surface CD16 expression, CD11c+ (CD16+inclusive) and CD11c+CD16− populations were sort purified and cultured as described above. As can be seen in Figure6B, the CD11c+ (CD16+ inclusive) cultures contained an increased proportion of PI+ dead cells (46%) as compared to the CD11c+CD16− cultures (13%). Sort-purified CD11c+CD16+ cells were also examined and exhibited equally poor survival in culture (data not shown). Similarly, the relative frequency of CD34+ cells in CD11c− populations following 18 hours culture increased (from 28% to 71%) reflecting a proportionately greater decrease in the number of CD123+ cells. Addition of allogeneic T cells to the DC cultures had no effect on the relative proportion of any of the subsets examined (data not shown). Thus, as previously documented, DCs survive poorly in vitro, with the CD123+ and the CD11c+CD16+ subsets having the poorest survival rate of the 4 subsets examined.
Differential survival of Lin−HLA-DR+ PBMC subpopulations.
(A) The frequency of CD34+ or CD16+ cells was examined in freshly sorted or cultured (18 hours with GM-CSF and IL-3) Lin− CD11c− or CD11c+ cells, respectively. The preferential survival of CD34+ cells over CD123hi (top panel) and reduced frequency of the CD11c+CD16+ subset (bottom panel) following culture were noted. (B) The frequency of dead cells was examined in 18-hour cultures of sort-purified CD11c+CD16−and CD11c+ (CD16+ inclusive) populations. An increase in the percentage of PI+ (dead) cells was associated with the CD11c+ (CD16+inclusive) cultures.
Differential survival of Lin−HLA-DR+ PBMC subpopulations.
(A) The frequency of CD34+ or CD16+ cells was examined in freshly sorted or cultured (18 hours with GM-CSF and IL-3) Lin− CD11c− or CD11c+ cells, respectively. The preferential survival of CD34+ cells over CD123hi (top panel) and reduced frequency of the CD11c+CD16+ subset (bottom panel) following culture were noted. (B) The frequency of dead cells was examined in 18-hour cultures of sort-purified CD11c+CD16−and CD11c+ (CD16+ inclusive) populations. An increase in the percentage of PI+ (dead) cells was associated with the CD11c+ (CD16+inclusive) cultures.
In vitro induction of activation-associated molecules on Lin−HLA-DR+ subsets
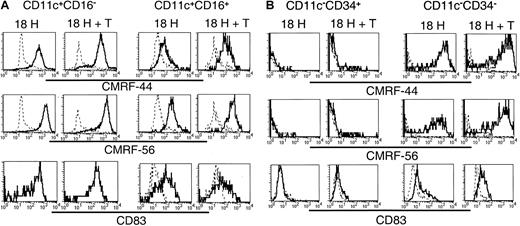
Having established that the 5 subsets exhibited distinct molecular profiles, and that, at least a proportion of each of these subsets survived in vitro, we compared their relative expression of the activation-associated antigens CMRF-44, CMRF-56, and CD83 following in vitro culture. Lin−CD11c+ (including CD16+) and CD11c− subsets were sorted and cultured as described above, and the expression of CMRF-44, CMRF-56, and CD83 was examined on the CD11c+CD16− and CD11c+CD16+ subsets (Figure7A) and the CD11c−CD34+ (ie, CD34+) and CD11c−CD34− (ie,CD123+) populations (Figure 7B). The cultured CD11c+CD16− cells expressed uniformly high levels of each of CMRF-44, CMRF-56, and CD83. The level of expression of these antigens was not increased further on this population following coculture with allogeneic T cells. CMRF-44, CMRF-56, and CD83 were induced on the CD11c+CD16+ population; however, the levels of expression of CMRF-44 and CMRF-56 were distinctly lower than that of the CD11c+CD16−subset, and only a proportion of the CD16+ cells expressed CD83. Minimal additional increases in CMRF-44 and CMRF-56 staining of both the CD16− and CD16+ subsets resulted after coculture with T cells. Cultured CD123+(CD11c−CD34−) cells expressed CMRF-44 and CMRF-56 at levels comparable to the CD11c+CD16− population, whereas CD83 expression was much less. However, coculture with allogeneic T cells induced higher levels of all 3 molecules on the CD123+population. Finally, only a very small proportion of CD34+cells was induced to express CMRF-44 or CMRF-56, which was slightly increased by T-cell coculture. CD83 was not detected on the CD34+ cells in either culture condition.
Induction of the DC-associated differentiation/activation antigens CMRF-44, CMRF-56, and CD83 on cultured Lin− cells.
Sort-purified CD11c+ and CD11c− populations were cultured for 18 hours in the presence of GM-CSF and IL-3 with or without allogeneic T lymphocytes. Following culture, cells were harvested and stained with biotinylated CMRF-44 or CMRF-56, or purified CD83 mAb followed by biotinylated anti–mouse IgG. Biotinylated antibody was detected with PE- or PE-Cy5–conjugated streptavidin, in conjunction with PE-Cy5– or PE-conjugated CD16 or CD34 for the (A) CD11c+ and (B) CD11c− populations, respectively. CD11c+CD16−, CD11c+CD16+, CD11c−CD34+, and CD11c−CD34− populations were gated and examined for their expression of activation antigens (solid lines). Dashed lines represent negative control antibody staining.
Induction of the DC-associated differentiation/activation antigens CMRF-44, CMRF-56, and CD83 on cultured Lin− cells.
Sort-purified CD11c+ and CD11c− populations were cultured for 18 hours in the presence of GM-CSF and IL-3 with or without allogeneic T lymphocytes. Following culture, cells were harvested and stained with biotinylated CMRF-44 or CMRF-56, or purified CD83 mAb followed by biotinylated anti–mouse IgG. Biotinylated antibody was detected with PE- or PE-Cy5–conjugated streptavidin, in conjunction with PE-Cy5– or PE-conjugated CD16 or CD34 for the (A) CD11c+ and (B) CD11c− populations, respectively. CD11c+CD16−, CD11c+CD16+, CD11c−CD34+, and CD11c−CD34− populations were gated and examined for their expression of activation antigens (solid lines). Dashed lines represent negative control antibody staining.
Allostimulatory capacity of Lin−HLA-DR+subsets
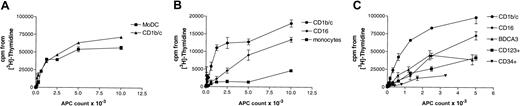
A defining property of DCs is their potent antigen-presenting function. We, therefore, examined the capacity of the 5 Lin− subsets to induce T-cell proliferation in allogeneic MLRs. Monocytes and MoDCs were included as reference populations. For comparative purposes, the CD1b/c+ population was used as the internal standard for each MLR. Each of the cell populations tested exhibited some degree of allostimulatory capacity although this was minimal for monocytes (Figure8). Comparison of the blood Lin−HLA-DR+ populations demonstrated clear differences in their allostimulatory capacity; they were ranked CD1b/c > CD16 > BDCA-3 > CD123 > CD34. The stimulator dose/T-cell proliferative response curve for the BDCA-3 subset differed significantly from the others, with an apparent early plateau. The CD123+ populations were less effective stimulators of the MLR and the CD34+ population was, as previously reported, a weak but definite stimulator of an allogeneic MLR.
Allostimulatory capacity of sort purified Lin− cell subpopulations.
Varying numbers of MoDCs, monocytes, and CD1b/c+, CD16+, BDCA-3+, CD123+, and CD34+ Lin− cells were cultured for 5 days in a 96-well plate with 105 allogeneic normal peripheral blood T cells per round-bottom well. T-cell proliferation was assessed by addition of tritiated thymidine. Results are expressed as means ± SEM of triplicate wells. Three separate representative experiments (A-C) are shown. Data are representative of a minimum of 3 experiments for each subset compared to the CD1b/c+population.
Allostimulatory capacity of sort purified Lin− cell subpopulations.
Varying numbers of MoDCs, monocytes, and CD1b/c+, CD16+, BDCA-3+, CD123+, and CD34+ Lin− cells were cultured for 5 days in a 96-well plate with 105 allogeneic normal peripheral blood T cells per round-bottom well. T-cell proliferation was assessed by addition of tritiated thymidine. Results are expressed as means ± SEM of triplicate wells. Three separate representative experiments (A-C) are shown. Data are representative of a minimum of 3 experiments for each subset compared to the CD1b/c+population.
Discussion
It is clear from recent differences in data reported in the literature10,12,15 and from data presented at the 7th LDAW16 19 that the cellular constitution of human blood DC preparations varies considerably. The main factors likely to contribute are the mAb mixtures used to select Lin− cells, the immunoselection methodology, and the gating criteria used in subsequent flow cytometric analysis. We undertook this analysis of the composition of human Lin− or putative DC preparations in an effort to define the cellular heterogeneity and thus provide a basis to compare preparations, methodology, and results from different laboratories. We prepared Lin− cells by depleting with CD3, CD14, CD19, CD11b, and CD16 or CD56 mAbs but used CD56 in preference, because this permitted the examination of a Lin−HLA-DR+CD16+ population, which has been reported to exhibit DC characteristics. We used a panel of markers previously reported to be expressed by DCs or cells within Lin− preparations to analyze the Lin−HLA-DR+ population. This analysis identified 5 distinct nonoverlapping subsets that we identified as CD123hi, CD1b/c+, CD16+, BDCA-3+, and CD34+. A more extensive phenotypic analysis of these subsets including costimulatory molecules, ILT molecules, C-type lectins, and others demonstrated a unique molecular phenotype for each of the 5 subsets. These differed from the molecular phenotypes obtained for circulating CD14hi monocytes or MoDCs. Furthermore, the culture requirements, activation potential, and allostimulatory capacity differed among these 5 Lin− populations. Having phenotypically defined these 5 discrete subsets in blood, the substantial task of assigning specialized functional roles or stages of development to these subsets remains to be addressed. We have demonstrated the basic allostimulatory capacity of each subset, but a myriad of additional functional assays are required to compare antigen uptake, cross-presentation, cytokine secretion, presentation to and activation of B and T lymphocytes (CD4 versus CD8, memory versus naive, TH1 versus TH2 polarization), and so forth.
The CD11c+ or myeloid blood DC population has been noted to be heterogeneous in our own16,25 and other studies.8 18 We show here that it includes the CD16+, CD1b/c+, and BDCA-3+subpopulations. The CD16+ population comprised a large but variable (40%-80%) proportion of the CD11c+ subset. It expressed low levels of CD14 and CD33, was the only subset to lack CLA expression, and had heterogeneous but generally lower levels of cell surface HLA-DR. Curiously, the CD16+ population had the highest levels of CD86 and relatively high levels of CD40. An important feature of this cell population was its apparently poor viability in tissue culture with only 50% surviving 12 to 18 hours in 10% FCS/RPMI-1640, even in the presence of GM-CSF and IL-3. The CD1b/c+ population represents 20% to 50% of the CD11c+ population and was noted to express the highest levels of CD11c. It expresses low to negligible levels of CD14 but is clearly CD33+ and universally expresses high levels of cell surface HLA-DR. The correlation between increasing CD1b/c expression and increasing CD11c and HLA-DR expression in conjunction with heterogeneous CD40 expression is indicative of a population of cells undergoing differentiation. The BDCA-3 subpopulation is much smaller and accounts for 2% to 3% of the CD11c+ cells. Interestingly, its CD11c expression was consistently at the lower end of the range of the CD11c+ population. It lacked CD14 but was again clearly CD33+ with moderate to high levels of cell surface HLA-DR. Notably, these BDCA-3+ cells had lower levels of CD86 but expressed the highest levels of CD40 and DEC-205.
As previously reported,9,10 25 the lymphoid or CD123hi DC population was readily distinguished from the myeloid CD11c+ DC population. However, we note that the CD11c+ population is unequivocally weakly CD123+ and that the expression of CD123 increases on in vitro culture, so that gating using this marker needs to be done carefully. Some heterogeneity in the CD123hi population was noted with differential expression of HLA-DR, CD40, CD2, and CD7.
These studies emphasize the fact that Lin−HLA-DR+ preparations may include other cell populations, which may or may not be classified as DCs or DC precursors. The CD34+ population presumably reflects, in part, the low-level circulating population of hematopoietic progenitors.26 Some or all of these have the capacity to differentiate into DCs,27,28 and it has been known for some time that these cells have allostimulatory potential.20 Our data in Table 2 suggest that one or more additional subsets of Lin−HLA-DR+ cells exist in blood. These will be CD123−/lo and CD34−and either CD11c+ or CD11c−. We did not characterize them further in this study because of the small numbers and lack of defining antibodies. We also note, in passing, that all 5 Lin−HLA-DR+ populations including the CD34+ population express CD52, a fact that may be relevant to interpreting the immunosuppressive effect of CD52 therapy in allogeneic transplantation.29 The low level of staining on CD34+ hematopoietic stem cells, or the self-renewing subset, may account for the apparent lack of influence on engraftment.
In terms of the function of these subsets, we investigated their expression of several relevant molecules and the classic definition of DC function—their ability to stimulate in an allogeneic MLR. Both the CD123hi DCs and the BDCA-3+ subpopulation of CD11c+ DCs express high levels of CD62L. It has been suggested that CD62L may mediate the migration of the CD123+ population directly from the blood via high endothelial venules (HEVs) into lymphoid tissues,30,31 thus explaining the relative paucity of this population in the normal peripheral tissues. It is possible that the BDCA-3 population behaves similarly, but the tissue distribution of this subset is only now undergoing analysis. A number of C-type lectins were investigated in relation to their potential as antigen uptake receptors. It appears that all 5 Lin−HLA-DR+populations either lack or require a signaling event to induce surface expression of CD206, CD207, or CD209. CD206 is not induced on blood DCs but is on MoDCs.32 The same is true of CD209,33 which has recently been described with DC-SIGNR and CD23 on tissue macrophages.34 The induction of langerin (CD207) on in vitro–derived DCs requires the presence of transforming growth factor β (TGF-β).35CD11c+/CDla+ blood DCs have been claimed to be direct precursors of the langerin-expressing LCs.15 The CD1 mAb used in that study was the BB5 clone but subsequent work during the 7th LDAW showed it to recognize CD1b/c rather than CD1a.16 Insignificant levels of cytoplasmic langerin were detected in the putative LC precursor, CD1b/c+ subset, and in the other subsets (Table 3). The broad expression of CD205 (DEC-205) as a potential antigen-loading receptor on all the cells is interesting. Our own unpublished and other data36suggests DEC-205 does load antigen into antigen-presenting cells, but it may have other functions on other cells, as suggested by its presence on CD34+ cells. There is also increasing awareness that DC molecular phenotype and function are highly regulated and these may differ according to the stimuli.37 The different CD85 molecule profiles on the 5 subsets is therefore highly relevant, given their likely contribution to both up- and down-regulation of intracellular signaling pathways in both DC and other leukocyte populations.11 30
The MLR studies produced unequivocal results. The CD1b/c population invariably expressed high levels of the CD83, CMRF-44, and CMRF-56 antigens, which have been associated with DC differentiation or activation. Consistent with this, the CD1b/c subset was invariably the most potent allostimulatory cell population with the CD16+population the next most effective stimulators. Curiously enough, these results were independent of the level of expression of the classic costimulator molecules CD80/CD86 and this may suggest that other costimulatory molecules such as OX40L, RANKL, or 41BBL may be involved. We confirmed that CD123hi DCs stimulate an allogeneic MLR9 as does the CD34+ cell population20 although the latter cells were the least effective stimulators in the absence of any prior in vitro differentiation. Clearly, further analysis of a number of different functional properties (including susceptibility to CD85-mediated inhibition) of these 5 potential DC subpopulations is required. The practicalities of this are daunting, but the major molecular and functional differences outlined in this work argue that each population may have to be addressed individually. It is even possible that certain subpopulations will be preferred for certain therapeutic applications. In our hands, the blood CD11c+ DCs38 and the CMRF-56+ DCs (Lopez et al, submitted manuscript) stimulate strong in vitro blood primary T-lymphocyte responses.
Finally, although it is tempting to try and assign these CD11c+ subsets to a theoretical DC differentiation/activation pathway, we think it is premature to do so. The CD11c+CD1b/c+ subset appears the most activated phenotypically and functionally but whether it is derived from a CD14+ precursor as previously suggested15 18 requires confirmation. Although the relationships of these subpopulations to accepted hematologic pathways needs to be established, no direct correlation with MoDCs was observed. The results nonetheless bring some clarity to the field and the definition of these Lin−HLA-DR+ subsets provides an opportunity to standardize the populations for further molecular and possibly even therapeutic functional studies.
Prepublished online as Blood First Edition Paper, August 15, 2002; DOI 10.1182/blood-2001-11-0097.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Derek Hart, Dendritic Cell Laboratory, Mater Medical Research Institute, Mater Misericordiae Hospitals, South Brisbane, 4101, Australia; e-mail:dhart@mmri.mater.org.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal