The molecular backgrounds of variants encountered in Afro-Caribbean black individuals and associated with the production of clinically significant antibodies against high-incidence antigens (anti-RH18, anti-RH34) and against Rhe epitopes were determined. We showed that RH:−18 phenotypes are produced by 3 distinct RHCEalleles: ceEK carrying 48G>C (exon 1), 712A>G, 787A>G, 800T>A (exon 5); ceBI carrying 48G>C (exon 1), 712A>G (exon 5), 818C>T (exon 6), 1132C>G (exon 8); and the already knownceAR allele carrying 48G>C (exon 1), 712A>G, 733C>G, 787A>G, 800T>A (exon 5), and 916A>G (exon 6). The RH:−34 phenotype is produced by the (C)ces haplotype described previously and composed of a hybrid D-CE(3-8)-D gene with 4 extra mutations next to a ces allele (733C>G; exon 5) with an extra mutation in exon 7 (1006G>T). Partial Rhe with risk of immunization against lacking epitopes can be produced by the new ces allele carrying an extra mutation in exon 3 (340C>T) and by the ceMO allele described previously. A population of sickle cell disease patients was screened to estimate the incidence of these rare alleles, with the conclusion that a procedure is required to detect the associated phenotypes in black donors to ensure transfusion safety for patients. We also described a new variant [ces(748)] and variants carrying different altered alleles in nonimmunized patients and for whom the risk of immunization is discussed.
Introduction
The RH blood group is the most polymorphic and immunogenic blood group. The RH locus is composed of 2 highly homologous genes: the RHD gene, encoding the D polypeptide; and the RHCE gene, encoding C or c together with either E or e polypeptides.1-3 Besides the 5 major antigens (D, C, E, c, e), more than 50 Rh antigens, identified by the corresponding antibodies, are described. They are schematically divided into low- and high-incidence antigens.4
Some rare Rh phenotypes are found exclusively in the black population: the RH:−46 phenotype (RN), the RH:−18 phenotype (Hr-negative), and the RH:−34 phenotype (HrB-negative). The RH:−46 phenotype is limited to the Peul ethnic group in Africa.5 Decreased expression of both C and e antigens is associated with the lack of expression of the RH46 high-incidence antigen and the appearance of the RH32 low-incidence antigen. A hybridCe-D(4)-Ce gene generates the RNphenotype.6 RH:−18 and RH:−34 phenotypes were first identified by the antibodies produced.7 Anti-RH18 and anti-RH34 react with red blood cells (RBCs) carrying the common products of the RHCE gene, but react more strongly with Rhe-positive RBCs. When the anti-RH18 is adsorbed on Rhe-negative RBCs (DccEE), the remaining reactivity is an anti-e or ce-like antibody named anti-RH19 (anti-hrS). When the anti-RH34 is adsorbed on Rhe-negative RBCs, the result is an anti–Ce-like antibody, named anti-RH31 (anti-hrB).8-10 RH:−34 RBCs were recently associated with the expression of the RH20 (VS) low-incidence antigen.11 Production of anti-RH18, anti-RH34, or anti-RH46 imposes to use equivalent rare antigen-negative RBCs or deleted-Rh RBCs (Rhnull or D- -) for transfusion, as the antibodies may be clinically significant, especially in sickle cell disease (SCD) patients, but also during pregnancy.10,12-14Despite rarity of the blood, transfusion safety may be organized for RH:−46 individuals. The phenotype is easily recognizable, therefore donor blood can be kept frozen for patients. Problems are more complex in terms of supply and demand for RH:−18 and RH:−34 phenotypes because they are not well characterized and molecular backgrounds are unknown.15
Besides RH:−18 and RH:−34 phenotypes, numerous Rhe variants are encountered in the black population.16-21 Some variants are associated with the production of anti-e in a way similar to the production of anti-D by partial D individuals, the abnormality being restricted to the partial e situation.12 Indeed, transfusion safety is also a problem for Dccee individuals producing allo–anti-e, as the Rhe-negative RBCs that would be required are RhE- positive (e and E antigens have an antithetical relationship) and cannot be transfused to those RhE-negative individuals. Therefore, only similar variant or deleted-Rh RBCs can be used.
In this work, we studied the serological reactivity and the molecular background of RBCs from Rhe-positive individuals of Afro-Caribbean origin producing anti-RH18, anti-RH34, anti-e, anti-ce, and anti-Ce. We demonstrate the clinical significance of the anti-RH18 for 2 patients who encountered a lethal outcome following incompatible transfusions and evaluated the incidence of these rare alleles in a population of SCD patients. We also describe variants with a decreased expression of Rhe antigen and for which the risk of immunization against the lacking e epitopes is unknown, making transfusion counseling difficult.
Patients, materials, and methods
Samples
Blood samples from Afro-Caribbean blacks living in France were referred to our laboratory and selected for study on different bases: production of anti-Rh antibodies and/or depressed Rhe phenotype.
Samples 1 to 12 were obtained from unrelated individuals producing either an antibody reacting with all RBCs expressing the RhCE polypeptides, or allo–anti-e, anti-ce, and anti-Ce antibodies. Cord blood was available from newborns of patients 2, 3, and 7. Individuals 7, 8, and 10 were SCD patients. Patients 7 and 8 died after incompatible transfusions. DNA from the parents of individual 8 was used for molecular analysis. Samples from siblings and parents of individual 1 also were available for study.
Samples 13 to 17 exhibited a depressed Rhe antigen that was not correlated to any known alteration. There was no antibody in the serum.
Blood samples of 146 SCD patients were randomly collected by the Center of Sickle Cell Disease in Créteil, France. Patients were from the West Indies and west and central Africa.
Serological analysis
For individuals 1 to 12, antibody specificity was characterized by testing sera against a panel of RBCs (CNRGS) discriminating antibodies against common immunogenic antigens by IgG indirect antiglobulin test (IAT) with nontreated and papain-treated RBCs on a gel matrix (DiaMed, Morat, Switzerland). Sera were also tested against Rhnull and D- - RBCs. Sera reacting with all common RBCs but not with D- - and/or Rhnull RBCs (1-10) were evaluated after adsorption on DccEE RBCs. Other rare RBCs were tested (RH:−18 and ceMO) but were not available for each serum.
Titers were evaluated for all sera against papain-treated RBCs of different phenotypes (ddccee, DCCee, DccEE) by IgG IAT. Direct antiglobulin test (DAT) and reactivity of each serum against RBCs from the antibody maker (auto-control) were performed to distinguish alloantibodies from autoantibodies. For some samples, autoadsorption studies confirmed that the antibodies were alloantibodies (data not shown).
D, C, E, c, and e status of all erythrocytes was established with routine reagents. Rhe antigen reactivity was further analyzed with separate clones (monoclonal antibodies [MoAbs]) from Serologicals (Livingston, United Kingdom): IgM (MS16, MS21, MS63, MS62, MS69) and IgG (MS70). IgG was tested using the IgG IAT on a gel matrix (DiaMed). IgMs were tested on a neutral gel matrix from the same manufacturer.
Expression of low-incidence antigens was evaluated with human sera: a serum from the CNRGS containing both anti-RH10 (V) and anti-RH20 (VS) and referred to in this study as anti-RH10/20, and a serum from the CNRGS containing only anti-RH32. Anti-RH50 (FPTT) was obtained by adsorption-elution from Mol serum.22
The high-incidence RH46 antigen was tested with a polyclonal antibody from the CNRGS. An anti-RH19 was obtained as described by Shapiro, by adsorbing the serum of an immunized RH:−18 patient (serum of individual 1 in this study) on DccEE RBCs.7 This anti-RH19 was tested on RBCs from patients 1 to 17.
Compatibility testing between the sera and RBCs of immunized patients was performed for individuals 3 and 5. The reactivity of serum 3 also was tested after adsorption-elution on RBCs from sample 5. Sera 1, 11, and 12 were tested against RBCs 9. ABO incompatibility, other antibodies, or the small number of samples did not allow compatibility testing for the other samples.
For weak D samples, phenotyping for partial D was performed with selected MoAbs known to discriminate categories of partial D. MoAbs were obtained through the Third and Fourth Workshops on MoAbs (Nantes, 1996; Paris, 2001; France).
cDNA sequence analysis
Reticulocyte RNAs were prepared from 50-mL whole blood as previously described.23 RNA was reverse transcribed using the first-strand cDNA synthesis kit (Amersham Pharmacia Biotech, Uppsala, Sweden). RHCE and RHD cDNA products were amplified by polymerase chain reaction (PCR) with the Klentaq polymerase (Clontech, Palo Alto, CA), using, respectively, primer sets P1-P3 and P4-P5, then reamplified using, respectively, sets P2-P3 and P2-P5 (primers and conditions in Table1). PCR was performed in a Thermocycler (Robocycler Genomic Gradient 96, Stratagene, La Jolla, CA). PCR products were subcloned into a PCRII vector (TA cloning kit, Invitrogen, Leek, The Netherlands). Recombinant clones and/or PCR products were sequenced with nested sequencing primers (Eurogentec, Herstal, Belgium) and on both strands by the DNA Sequencing Kit (Applied Biosystems, Foster City, CA). Sequences were analyzed on an automated fluorescence-based ABI Prism 310 (Applied Biosystems). Sequences were submitted to GenBank under the accession numbers bankit 464322, 464376, 464386, 464388, 4760047, and 467051.
Primers and conditions of PCR assays
| Primers . | Specificity . | Localization . | Sequence 5′-3′ . | PCR assays . | PCR products . | †PCR conditions . |
|---|---|---|---|---|---|---|
| P1-s | CE | 5′UT | CTCCATAGACAGGCCAGCACAG | P1-P3 | 1537 bp | 1 min 95°C, 1 min 62°C, 1 min 30 sec 72°C: 30 cycles |
| P2-s | D-CE | 5′UT | ATGCCTGGTGCTGGTGGAACCCC | P2-P318 | 1427 bp | 1 min 95°C, 1 min 62°C, 1 min 30 sec 72°C: 30 cycles |
| P3-a | CE | 3′UT | CTGTCTCTGACCTTGTTTCATTATAC | |||
| P4-s | D | 5′UT | CTCCATAGAGAGGCCAGCACAA | P4-P5 | 1590 bp | 1 min 95°C, 1 min 62°C, 1 min 30 sec 72°C: 30 cycles |
| P5-a | D | 3′UT | ATGGTGAGATTCTCCTCAAAGAGTG | P2-P5 | 1480 bp | 1 min 95°C, 1 min 62°C, 1 min 30 sec 72°C: 30 cycles |
| P6-s | D-CE | In 2 | tcagtcatcctggctctcc | |||
| P7-a | D-CE | In 3 | aggtccctcctccagcac | P6-P718 | 210 bp | 30 sec 95°C, 30 sec 62°C, 30 sec 72°C: 25 cycles |
| P8-s | D | Ex 4 | GACTACCACATGAACATGAT | |||
| P9-a | D-CE | In 5 | aatatgtgtgctagtcctgt | P8-P9 | 790 bp | 1 min 95°C, 1 min 60°C, 1 min 30 sec 72°C: 30 cycles |
| P10-s | CE | Ex 4 | ACTACCACATGAACCTGAG | P10-P918 | 1460 bp | 1 min 95°C, 1 min 60°C, 1 min 30 sec 72°C: 30 cycles |
| P11-s | CE | Ex 5 | CCCAAAGGAAGATCAGCAT | |||
| P12-a | D-CE | In 6-Ex 6 | tgtctagtttcttacCGGCA | P11-P12 | 1807 bp | 30 sec 95°C, 3 min 68°C: 32 cycles |
| P13-s | D | Ex 5 | CCCAAGGGAAGATCAGCAA | P13-P12 | 1807 bp | 30 sec 95°C, 3 min 68°C: 32 cycles |
| P14-a | CE | In 6-Ex 6 | agtttcttacCGGCAGGC | P13-P14 | 1807 bp | 30 sec 95°C, 3 min 68°C: 32 cycles |
| P15-s | D-CE | In 6 | tgttagaaatgctgttagacc | |||
| P16-a | D | In 7-Ex 7 | cacATGCCATTGCCGGCT | P15-P16 | 435 bp | 1 min 95°C, 1 min 60°C, 1 min 30 sec 72°C: 30 cycles |
| P17/P17*-a | D/CE* | Ex 5 | GCATAGTAGGTGTTGAACAC(T*) | P18-P17/17* | 1400 bp | 30 sec 95°C, 30 sec 62°C, 30 sec 72°: 30 cycles |
| P18-s | CE | In 4 | gcaacagagcaagagtcca | |||
| P19/P19*-a | D/CE* | Ex 5 | TGTCACCACACTGACTGCTAC(G*) | P18-P19/19*17 | 428 bp | 30 sec 95°C, 30 sec 65°C, 30 sec 72°C: 26 cycles |
| P20/P20*-s | 1006T/G* | Ex 7 | ACTCCATCTTCAGCTTGCTGT(G*) | |||
| P21-a | CE | In 7-Ex 7 | acccacATGCCATTGCCGTTC | P20/20*-P2117 | 94 bp | 30 sec 95°C, 30 sec 65°C, 30 sec 72°C: 26 cycles |
| P22-s | D | Ex 3 | TCGGTGCTGATCTCAGTGGA | |||
| P23/P23*-a | CE/D* | Ex 3 | ACTGATGACCATCCTCAGGG(T*) | P22-P23/23*17 | 110 bp | 30 sec 95°C, 30 sec 65°C, 30 sec 72°C: 26 cycles |
| P24/P24*-s | D/CE* | Ex 5 | TGGATGTTCTGGCCAAGTT(G*) | |||
| P25-a | CE | In 5-Ex 5 | tcacCATGCTGATCTTCCT | P24/24*-P2517 | 150 bp | 30 sec 95°C, 30 sec 60°C, 30 sec 72°C: 25 cycles |
| P26/P26*-s | 340T/C* | In 2-Ex 3 | ccttctcacccccagTATTT(C*) | |||
| P27-a | CE | Ex 3 | CTGATGACCATCCTCAGGG | P26/26*-P2727 | 152 bp | 30 sec 95°C, 30 sec 63°C, 30 sec 72°C: 30 cycles |
| P28-s | D-CE | 5′UT | ATAGTCCCTCTGCTTCCG | |||
| P29-a | D-CE | In 1-Ex 1 | ccaatgaactctcacCTTG | P28-P2927 | 340 bp | Internal control: variable conditions |
| Primers . | Specificity . | Localization . | Sequence 5′-3′ . | PCR assays . | PCR products . | †PCR conditions . |
|---|---|---|---|---|---|---|
| P1-s | CE | 5′UT | CTCCATAGACAGGCCAGCACAG | P1-P3 | 1537 bp | 1 min 95°C, 1 min 62°C, 1 min 30 sec 72°C: 30 cycles |
| P2-s | D-CE | 5′UT | ATGCCTGGTGCTGGTGGAACCCC | P2-P318 | 1427 bp | 1 min 95°C, 1 min 62°C, 1 min 30 sec 72°C: 30 cycles |
| P3-a | CE | 3′UT | CTGTCTCTGACCTTGTTTCATTATAC | |||
| P4-s | D | 5′UT | CTCCATAGAGAGGCCAGCACAA | P4-P5 | 1590 bp | 1 min 95°C, 1 min 62°C, 1 min 30 sec 72°C: 30 cycles |
| P5-a | D | 3′UT | ATGGTGAGATTCTCCTCAAAGAGTG | P2-P5 | 1480 bp | 1 min 95°C, 1 min 62°C, 1 min 30 sec 72°C: 30 cycles |
| P6-s | D-CE | In 2 | tcagtcatcctggctctcc | |||
| P7-a | D-CE | In 3 | aggtccctcctccagcac | P6-P718 | 210 bp | 30 sec 95°C, 30 sec 62°C, 30 sec 72°C: 25 cycles |
| P8-s | D | Ex 4 | GACTACCACATGAACATGAT | |||
| P9-a | D-CE | In 5 | aatatgtgtgctagtcctgt | P8-P9 | 790 bp | 1 min 95°C, 1 min 60°C, 1 min 30 sec 72°C: 30 cycles |
| P10-s | CE | Ex 4 | ACTACCACATGAACCTGAG | P10-P918 | 1460 bp | 1 min 95°C, 1 min 60°C, 1 min 30 sec 72°C: 30 cycles |
| P11-s | CE | Ex 5 | CCCAAAGGAAGATCAGCAT | |||
| P12-a | D-CE | In 6-Ex 6 | tgtctagtttcttacCGGCA | P11-P12 | 1807 bp | 30 sec 95°C, 3 min 68°C: 32 cycles |
| P13-s | D | Ex 5 | CCCAAGGGAAGATCAGCAA | P13-P12 | 1807 bp | 30 sec 95°C, 3 min 68°C: 32 cycles |
| P14-a | CE | In 6-Ex 6 | agtttcttacCGGCAGGC | P13-P14 | 1807 bp | 30 sec 95°C, 3 min 68°C: 32 cycles |
| P15-s | D-CE | In 6 | tgttagaaatgctgttagacc | |||
| P16-a | D | In 7-Ex 7 | cacATGCCATTGCCGGCT | P15-P16 | 435 bp | 1 min 95°C, 1 min 60°C, 1 min 30 sec 72°C: 30 cycles |
| P17/P17*-a | D/CE* | Ex 5 | GCATAGTAGGTGTTGAACAC(T*) | P18-P17/17* | 1400 bp | 30 sec 95°C, 30 sec 62°C, 30 sec 72°: 30 cycles |
| P18-s | CE | In 4 | gcaacagagcaagagtcca | |||
| P19/P19*-a | D/CE* | Ex 5 | TGTCACCACACTGACTGCTAC(G*) | P18-P19/19*17 | 428 bp | 30 sec 95°C, 30 sec 65°C, 30 sec 72°C: 26 cycles |
| P20/P20*-s | 1006T/G* | Ex 7 | ACTCCATCTTCAGCTTGCTGT(G*) | |||
| P21-a | CE | In 7-Ex 7 | acccacATGCCATTGCCGTTC | P20/20*-P2117 | 94 bp | 30 sec 95°C, 30 sec 65°C, 30 sec 72°C: 26 cycles |
| P22-s | D | Ex 3 | TCGGTGCTGATCTCAGTGGA | |||
| P23/P23*-a | CE/D* | Ex 3 | ACTGATGACCATCCTCAGGG(T*) | P22-P23/23*17 | 110 bp | 30 sec 95°C, 30 sec 65°C, 30 sec 72°C: 26 cycles |
| P24/P24*-s | D/CE* | Ex 5 | TGGATGTTCTGGCCAAGTT(G*) | |||
| P25-a | CE | In 5-Ex 5 | tcacCATGCTGATCTTCCT | P24/24*-P2517 | 150 bp | 30 sec 95°C, 30 sec 60°C, 30 sec 72°C: 25 cycles |
| P26/P26*-s | 340T/C* | In 2-Ex 3 | ccttctcacccccagTATTT(C*) | |||
| P27-a | CE | Ex 3 | CTGATGACCATCCTCAGGG | P26/26*-P2727 | 152 bp | 30 sec 95°C, 30 sec 63°C, 30 sec 72°C: 30 cycles |
| P28-s | D-CE | 5′UT | ATAGTCCCTCTGCTTCCG | |||
| P29-a | D-CE | In 1-Ex 1 | ccaatgaactctcacCTTG | P28-P2927 | 340 bp | Internal control: variable conditions |
Coding sequences are given in capital letters, noncoding sequences in capital italics, and intron sequences in lowercase letters. When already published, references are given for PCR assays.
s indicates sense; a, antisense; UT, untranslated region; In, intron; and Ex, exon.
Wild-type mutation.
PCR conditions: a denaturation step of 4 minutes at 95°C and an elongation step of 5 minutes at 72°C were performed for all PCR assays.
Genomic DNA analysis
Genomic DNA was isolated from peripheral blood leukocytes with a DNA isolation kit (Wizard, Genomic DNA Purification Kit, Promega, Madison, WI).
For sequence analysis, exon-specific PCR was performed on genomic DNA. Primer sequences and conditions are described in Table 1. The following primer sets were used: set P6-P7 for nonspecific exon 3; set P8-P9 forRHD exons 4 to 5; set P10-P9 for RHCE exons 4 to 5; set P11-P12 for RHCE exon 6; set P13-P12 for RHD exon 6 (set P13-P14 also was used to amplify RHCE exon 6 from individuals 1 to 8); and set P15-P16 for RHD exon 7. PCR products were subcloned or directly sequenced as described for cDNA analysis.
PCR assays
Allele-specific primer PCR (ASP-PCR) was designed for the detection of specific mutations in the population of SCD patients. The wild-type (indicated by *) ASP-PCR was performed in parallel to determine the homozygous or heterozygous status of the mutation. Sets P18-P17 and P18-P17* were used to detect G712 and A712*, respectively, in RHCE exon 5. Published oligonucleotides were used in the following ASP-PCR: sets P18-P19 and P18-P19* ASP-PCR detected G733 and C733*, respectively, in RHCE exon 5; sets P20-P21 and P20*-P21 ASP-PCR detected T1006 and G1006*, respectively, inRHCE exon 7; sets P22-P23 and P22-P23* ASP-PCR amplified a hybrid RHD-CE exon 3 and a RHD specific exon 3, respectively.17 Sets P24-P25 and P24*-25 ASP-PCR were designed to detect T667 and G667*, respectively, in RHCEexon 5. Sets P26-P27 and P26*-P27 ASP-PCR were designed to detect T340 and C340*, respectively, in RHCE exon 3. For all PCR assays, an internal control was added, amplifying a common sequence of theRHD and the RHCE genes (set P28-P29).
Results
Serological analysis
Rh terminology for RH18 (Hr), RH19 (hrS), RH34 (HrB), and RH31 (hrB) is as follows: RH18 and RH34 define high-incidence antigens produced by the RHCEgene. Anti-RH18 and anti-RH34 are the antibodies produced by RH:−18 and RH:−34 individuals, respectively. As defined by Shapiro,7 these antibodies can be distinguished after adsorption of anti-RH18 and anti-RH34 on DccEE RBCs. The remaining specificity of anti-RH18 resembles anti-e or anti-ce and is named anti-RH19. The remaining specificity of anti-RH34 resembles anti-Ce and is named anti-RH31.7 In this study, anti-RH19 is used as a reagent that is negative with all RH:−18 RBCs but also with some rare variants of Rhe antigen.18
We categorized blood samples of individuals 1 to 12 into 3 groups according to the antibody specificities (Table2) and the RBC Rh phenotypes (Table3): group A: samples 1 to 8; group B: samples 9 and 10; group C: samples 11 and 12. Origin of immunization is indicated in Table 2. A fourth group (group D: samples 13 to 17) had heterogeneous Rh reactivity (Table 3). No antibody was detected in patients in group D, but no immunization challenge could be documented.
Identification of antibodies in sera from individuals 1 to 12
| . | Origin of immunization . | Serum reactivity . | Titer . | Antibodies in serum . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Common RBCs . | Rhnull RBCs . | D- RBCs . | RH:−18 RBCs . | ceMO RBCs . | Other RBCs . | After absorption on DccEE RBCs . | ddccee RBCs . | DCCee RBCs . | DccEE RBCs . | |||
| Group A | ||||||||||||
| 1 | 2 pregnancies transfusion | + | 0 | 0 | nt | + | 9 + | anti-e–like | 256 | 128 | 256 | anti-RH18, anti-E |
| 2 | 2 pregnancies | + | 0 | + | nt | nt | anti-e–like | 256 | 64 | 64 | anti-RH18, anti-D | |
| 3 | 3 pregnancies | + | 0 | + | 0 | nt | 5 0 | anti-e–like | 16 | 32 | 64 | anti-RH18, anti-D |
| 4 | 3 pregnancies | + | 0 | + | 0 | nt | anti-e–like | 8 | 4 | 4 | anti-RH18, anti-D | |
| 5 | 3 pregnancies | + | 0 | 0 | 0 | + | 3 + | anti-e–like, anti-C | 32 | 256 | 4 | anti-RH18, anti-C |
| 6 | 2 pregnancies | + | 0 | 0 | nt | + | anti-e–like | 64 | 16 | 64 | anti-RH18, anti-E | |
| 7 | 1 pregnancy transfusion | + | 0 | 0 | 0 | + | anti-e–like, anti-C | 256 | 256 | 256 | anti-RH18, anti-C, anti-Fya, anti-S | |
| 8 | transfusion | + | 0 | 0 | 0 | nt | anti-e–like | 2000 | 512 | 512 | anti-RH18 | |
| Group B | ||||||||||||
| 9 | 2 pregnancies transfusion | + | 0 | + | + | nt | anti-Ce–like | 32 | 8000 | 8000 | anti-RH34, anti-D | |
| 10 | transfusion | + | 0 | 0 | + | + | anti-Ce–like | 64 | 128 | 16 | anti-RH34 | |
| Group C | ||||||||||||
| 11 | 2 pregnancies | 0/+ | 0 | 0 | nt | 0 | 9 + | nt | 2 | 1 | 0 | anti-e, ce |
| 12 | 4 pregnancies | 0/+ | 0 | 0 | + | + | 9 + | nt | 16 | 64 | 0 | anti-e, Ce |
| . | Origin of immunization . | Serum reactivity . | Titer . | Antibodies in serum . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Common RBCs . | Rhnull RBCs . | D- RBCs . | RH:−18 RBCs . | ceMO RBCs . | Other RBCs . | After absorption on DccEE RBCs . | ddccee RBCs . | DCCee RBCs . | DccEE RBCs . | |||
| Group A | ||||||||||||
| 1 | 2 pregnancies transfusion | + | 0 | 0 | nt | + | 9 + | anti-e–like | 256 | 128 | 256 | anti-RH18, anti-E |
| 2 | 2 pregnancies | + | 0 | + | nt | nt | anti-e–like | 256 | 64 | 64 | anti-RH18, anti-D | |
| 3 | 3 pregnancies | + | 0 | + | 0 | nt | 5 0 | anti-e–like | 16 | 32 | 64 | anti-RH18, anti-D |
| 4 | 3 pregnancies | + | 0 | + | 0 | nt | anti-e–like | 8 | 4 | 4 | anti-RH18, anti-D | |
| 5 | 3 pregnancies | + | 0 | 0 | 0 | + | 3 + | anti-e–like, anti-C | 32 | 256 | 4 | anti-RH18, anti-C |
| 6 | 2 pregnancies | + | 0 | 0 | nt | + | anti-e–like | 64 | 16 | 64 | anti-RH18, anti-E | |
| 7 | 1 pregnancy transfusion | + | 0 | 0 | 0 | + | anti-e–like, anti-C | 256 | 256 | 256 | anti-RH18, anti-C, anti-Fya, anti-S | |
| 8 | transfusion | + | 0 | 0 | 0 | nt | anti-e–like | 2000 | 512 | 512 | anti-RH18 | |
| Group B | ||||||||||||
| 9 | 2 pregnancies transfusion | + | 0 | + | + | nt | anti-Ce–like | 32 | 8000 | 8000 | anti-RH34, anti-D | |
| 10 | transfusion | + | 0 | 0 | + | + | anti-Ce–like | 64 | 128 | 16 | anti-RH34 | |
| Group C | ||||||||||||
| 11 | 2 pregnancies | 0/+ | 0 | 0 | nt | 0 | 9 + | nt | 2 | 1 | 0 | anti-e, ce |
| 12 | 4 pregnancies | 0/+ | 0 | 0 | + | + | 9 + | nt | 16 | 64 | 0 | anti-e, Ce |
In this table, samples of individuals 1–12 are divided into 3 groups (A, B, C) according to similar results of Rh antibody characterization. Group D individuals (13-17) were not immunized. Immunization challenge is indicated for each individual. Serum reactivity was studied on common RBCs. When sera did not react with all common RBCs (sera 11-12: 0/+), adsorption studies were not performed. For the other sera, after adsorption on DccEE RBCs, the specificity found is followed by “like” when the individual expressed the specificity of the antibody. All sera were tested on Rhnull, D- - and titration performed on ddccee, DCCee, and DccEE RBCs. RH:−18 from the SCARF were tested with some sera. ceMO RBCs (typed Dccee) were not tested when an anti-D was present in the serum. Some sera also were tested with RBCs from these study (“Other RBCs”): serum 3 was tested with RBCs 5; serum 5 with RBCs 3; sera 1, 11, and 12 with RBCs 9 (defined as RH:−34 in this study). The last column indicates all the antibodies found in each serum.
nt indicates not tested.
Rh antigen reactivity and corresponding molecular backgrounds
| . | Rh phenotype with routine reagents . | Rh antibodies in serum . | Anti-e MoAbs . | Anti–low and –high-frequency antigens . | Molecular background . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MS16 . | MS21 . | MS63 . | MS62 . | MS69 . | MS70 . | RH10/20 . | RH32 . | RH50 . | RH19 . | RH46 . | RHD . | RHCE . | |||
| Controls | |||||||||||||||
| ceMO | Dccee; weak e | 0 | w | w | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | + | D | ceMO/ceMO |
| RN | DCCee; weak C, weak e | 0 | w | 0 | 0 | 0 | w | 0 | 0 | + | 0 | + | 0 | D | RN/RN |
| C48 | Dccee | 0 | + | + | + | + | w | + | 0 | 0 | 0 | + | + | D | ce(48)/ce(48) |
| Group A | |||||||||||||||
| 1 | Dccee weak D | anti-RH18, anti-E | + | + | w | + | + | + | + | 0 | 0 | 0 | + | DAR | ceAR/ceEK |
| 2 | Dccee weak D | anti-RH18, anti-D | + | + | w | 0/w | + | + | + | 0 | 0 | 0 | + | DAR | ceAR/ceEK |
| 3 | Dccee weak D | anti-RH18, anti-D | + | + | + | + | + | + | 0 | 0 | 0 | 0 | + | DAR | ceEK/ceEK |
| 4 | Dccee weak D | anti-RH18, anti-D | + | + | 0 | 0 | w | + | + | 0 | 0 | 0 | + | DAR | ceAR/ceAR |
| 5 | Dccee weak D | anti-RH18, anti-C | + | + | 0 | 0 | w | + | + | 0 | 0 | 0 | + | DAR | ceAR/ceAR |
| 6 | Dccee | anti-RH18, anti-E | + | + | + | + | + | + | 0 | 0 | 0 | 0 | + | DAR/D(667) | ceEK/ceBI |
| Group B | |||||||||||||||
| 9 | ddCcee | anti-RH34, anti-D | + | + | + | + | + | 0 | + | 0 | 0 | + | + | D-Ce(3-8)-D | ces(1006)/ces (1006) |
| 10 | ddCcee | anti-RH34 | + | + | + | + | + | 0 | + | 0 | 0 | + | + | D-Ce(3-8)-D | ces(1006)/ces (1006) |
| Group C | |||||||||||||||
| 11 | DCcee weak C, weak e | anti-e, anti-Ce | + | w | 0 | 0 | w | 0 | 0 | + | 0 | + | + | D | RN/ceMO |
| 12 | DccEe weak e | anti-e, anti-Ce | 0 | 0 | + | + | 0 | 0 | w | 0 | 0 | w | + | D/D(674) | cE/ces(340) |
| Group D | |||||||||||||||
| 13 | Dccee weak e | 0 | w | w | 0 | 0 | 0 | 0 | 0 | 0 | + | 0 | + | D/DIVa | ceMO/ce-D(5)-ce |
| 14 | DCcee weak C weak e | 0 | + | + | 0 | w | 0 | 0 | w | + | 0 | + | + | D | RN/ces(748) |
| 15 | DCcee weak C weak c weak e | 0 | w | 0 | 0 | 0 | w | 0 | 0 | w | 0 | w | 0 | D | RN/ce-D(4–9)-ce |
| 16 | DCcee weak C weak e | 0 | + | w | 0 | 0 | w | 0 | 0 | + | 0 | + | + | D | RN/ceMO |
| 17 | DccEe weak e | 0 | 0 | 0 | + | + | 0 | 0 | w | 0 | 0 | w | + | D | cE/ces(340) |
| . | Rh phenotype with routine reagents . | Rh antibodies in serum . | Anti-e MoAbs . | Anti–low and –high-frequency antigens . | Molecular background . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MS16 . | MS21 . | MS63 . | MS62 . | MS69 . | MS70 . | RH10/20 . | RH32 . | RH50 . | RH19 . | RH46 . | RHD . | RHCE . | |||
| Controls | |||||||||||||||
| ceMO | Dccee; weak e | 0 | w | w | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | + | D | ceMO/ceMO |
| RN | DCCee; weak C, weak e | 0 | w | 0 | 0 | 0 | w | 0 | 0 | + | 0 | + | 0 | D | RN/RN |
| C48 | Dccee | 0 | + | + | + | + | w | + | 0 | 0 | 0 | + | + | D | ce(48)/ce(48) |
| Group A | |||||||||||||||
| 1 | Dccee weak D | anti-RH18, anti-E | + | + | w | + | + | + | + | 0 | 0 | 0 | + | DAR | ceAR/ceEK |
| 2 | Dccee weak D | anti-RH18, anti-D | + | + | w | 0/w | + | + | + | 0 | 0 | 0 | + | DAR | ceAR/ceEK |
| 3 | Dccee weak D | anti-RH18, anti-D | + | + | + | + | + | + | 0 | 0 | 0 | 0 | + | DAR | ceEK/ceEK |
| 4 | Dccee weak D | anti-RH18, anti-D | + | + | 0 | 0 | w | + | + | 0 | 0 | 0 | + | DAR | ceAR/ceAR |
| 5 | Dccee weak D | anti-RH18, anti-C | + | + | 0 | 0 | w | + | + | 0 | 0 | 0 | + | DAR | ceAR/ceAR |
| 6 | Dccee | anti-RH18, anti-E | + | + | + | + | + | + | 0 | 0 | 0 | 0 | + | DAR/D(667) | ceEK/ceBI |
| Group B | |||||||||||||||
| 9 | ddCcee | anti-RH34, anti-D | + | + | + | + | + | 0 | + | 0 | 0 | + | + | D-Ce(3-8)-D | ces(1006)/ces (1006) |
| 10 | ddCcee | anti-RH34 | + | + | + | + | + | 0 | + | 0 | 0 | + | + | D-Ce(3-8)-D | ces(1006)/ces (1006) |
| Group C | |||||||||||||||
| 11 | DCcee weak C, weak e | anti-e, anti-Ce | + | w | 0 | 0 | w | 0 | 0 | + | 0 | + | + | D | RN/ceMO |
| 12 | DccEe weak e | anti-e, anti-Ce | 0 | 0 | + | + | 0 | 0 | w | 0 | 0 | w | + | D/D(674) | cE/ces(340) |
| Group D | |||||||||||||||
| 13 | Dccee weak e | 0 | w | w | 0 | 0 | 0 | 0 | 0 | 0 | + | 0 | + | D/DIVa | ceMO/ce-D(5)-ce |
| 14 | DCcee weak C weak e | 0 | + | + | 0 | w | 0 | 0 | w | + | 0 | + | + | D | RN/ces(748) |
| 15 | DCcee weak C weak c weak e | 0 | w | 0 | 0 | 0 | w | 0 | 0 | w | 0 | w | 0 | D | RN/ce-D(4–9)-ce |
| 16 | DCcee weak C weak e | 0 | + | w | 0 | 0 | w | 0 | 0 | + | 0 | + | + | D | RN/ceMO |
| 17 | DccEe weak e | 0 | 0 | 0 | + | + | 0 | 0 | w | 0 | 0 | w | + | D | cE/ces(340) |
Results are represented by “+” for a normal positive reaction, “w” for a weak positive reaction, and “0” for a negative reaction. “0/w” stands for reactions that may vary depending of the batch used. RH49 (STEM) low-frequency antigen expressed on some RH:−18 RBCs was not tested (serum was not available in the laboratory).24 The molecular background indicates the homozygous or heterozygous status of the RHD gene only when it could be determined by family study or by the presence of 2RHD genes at sequencing analysis. For the RHCEalleles, the major mutation is indicated in parentheses, except for group 1, in which mutations are multiple. cesstands for other alleles with at least mutation at 733. Patients 7 and 8 (DAR-ceAR/DAR-ceAR) do not appear in this Table, as recent transfusion made Rh typing not meaningful.
RH46 antigen expression has been tested with polyclonal and monoclonal reagents.
Group A sera (samples 1 to 8) reacted with all common RBCs. Sera 1, 5, 6, 7, and 8 were negative with Rhnull and D- - RBCs, indicating antibodies recognizing common RHCE gene products. Sera 2, 3, and 4 were negative with Rhnull RBCs but positive with D- - RBCs, indicating that sera contained antibodies recognizing the common RHCE gene products as well as anti-D. After adsorption of sera on Rhe-negative RBCs (DccEE), in all cases, the remaining antibody was an anti-e–like antibody reacting with all normal Rhe-positive RBCs. These data met criteria defined by Shapiro to identify anti-RH18.7 Anti-RH18 specificity was confirmed for sera 3, 4, 5, 7, and 8 that could be tested negative with known RH:−18 RBCs from the SCARF. After adsorption on DccEE RBCs, anti-C also was found in sera 5 and 7. Adsorption of sera on other RBCs (data not shown) indicated that anti-E also was present in sera 1 and 6 and anti-Fya plus anti-S in serum 7. The titer reflected for each serum the specificity of the antibodies. For serum 8, containing only anti-RH18, the titer was higher on ddccee RBCs. Sera 1 to 6 did not agglutinate their own RBCs, indicating that all the antibodies identified were alloantibodies. The auto-control and phenotype analysis were not meaningful for samples of individuals 7 and 8, who were transfused before the tests.
In this group, RBCs were typed Dccee. Samples 1 to 6 had a normal reactivity with anti-Rhe routine reagents and were negative with the anti-RH19 reagent, but exhibited different patterns with anti-e MoAbs and anti-RH10/20 serum.
Compatibility testing between RBCs and sera from samples 3 and 5 showed that serum from sample 5 agglutinated RBCs from sample 3, whereas serum from sample 3 failed to react with RBCs from sample 5. When serum from sample 5 was adsorbed on RBCs from sample 3 and then eluted, the reactivity was anti-e. Variants 1 to 5 exhibited the specific profile for the DAR partial phenotype when tested with a panel of anti-D MoAbs (data not shown).25 This result was consistent with the anti-D found in sera 2 to 4.
Group B sera (samples 9 and 10) also reacted with all RBCs tested, except those from Rhnull. Serum 9 reacted with D- - RBCs, indicating that an anti-D was associated with the antibody against the common products of the RHCE gene, whereas serum 10 did not react with D- -, eliminating the presence of an anti-D. When sera were absorbed on Rhe-negative RBCs (DccEE), the remaining reactivity was anti-Ce. As defined by Shapiro,7 these results suggest that sera 9 and 10 both contained anti-RH34 antibody. No RH:−34 RBCs from the SCARF were available to confirm this hypothesis. Titers reflected the antibodies determined in sera. Sample 10, which contained only anti-RH34, had a higher titer with DCCee RBCs. Furthermore, group B RBCs, typed ddCcee, had the same serological profile with anti-e MoAbs and reacted with anti-RH10/20 and anti-RH19.
In group C, individual 11 serum contained anti-e and anti-ce. Antibodies reacted with RBCs from sample 9 (considered as RH:−34) but failed to react with ceMO variant. Individual 11 RBCs, typed DCcee with weak C and weak e, were positive for anti-RH19, did not react with some anti-e MoAbs, and expressed RH32 and RH46 (antithetical for RH32).
Individual 12's serum contained anti-e and anti-Ce that reacted with RH:−18 RBCs and RBCs from sample 9. Individual 12's RBCs, typed DccEe with weak e, also were negative with some anti-e MoAbs and reacted weakly with the anti-RH10/20 serum and anti-RH19.
In group D (samples 13 to 17; Table 3), distinct serological patterns were found. Sample 13, typed Dccee with weak e, was similar to ceMO in terms of Rhe reactivity and RH19 reactivity (RH:−19), but RBCs from sample 13 also expressed the RH50 antigen. Sample 14, typed DCcee with weak C and weak e, reacted with anti-RH10/20 (weak), anti-RH32, anti-RH46, and anti-RH19. Anti-e MoAbs defined a new reactivity profile for this RBC sample. RBCs from sample 15 typed RH:32,-46 exhibited an unusual DCcee phenotype with weak C, weak c, and weak e antigens. RH:32,-46 RBCs are classically DCCee with weak C and weak e antigens. Reactivity was weak with anti-RH19. RBCs from samples 16 and 17 reacted as samples 11 and 12 from group C, respectively.
cDNA sequence analysis
Complete sequence of RHD and RHCEtranscripts were analyzed for samples 1, 3 to 6, 9 to 12, 14, and 15. Known transcripts as well as new transcripts were identified (Table4, Figure1). For individual 1, 3 different transcripts were found: (1) the DAR allele,25carrying 602C>G (Thr201Arg) in exon 4, 667T>G (Phe223Val) in exon 5, and 1025T>C (Ile342Thr) in exon 7; (2) the ceARallele25 carrying 48G>C (Trp16Cys) in exon 1; 712A>G (Met238Val), 733C>G (Leu245Val), 787A>G (Arg263Gly), and 800T>A (Met267Lys) in exon 5 and 916A>G (Ile306Val) in exon 6; (3) the new ceEK allele, carrying 48G>C (Trp16Cys) in exon 1; 712A>G (Met238Val), 787A>G (Arg263Gly), and 800T>A (Met267Lys) in exon 5. In sample 3, we found the DAR and theceEK alleles. In sample 4, we found the DAR and the ceAR alleles. Individuals 3 and 4 were homozygous forceEK and ceAR, respectively, as shown by the absence of double peak when direct sequencing of PCR products was performed. In individual 6, 4 different transcripts were characterized: (1) DAR; (2) the new D(667) allele carrying 667T>G in exon 5 (Phe223Val); (3) ceEK; (4) the newceBI allele, carrying 48G>C in exon 1 (Trp16Cys); 712A>G in exon 5 (Met238Val); 818C>T in exon 6 (Ala273Val), and 1132C>G in exon 8 (Leu378Val). For individuals 1, 3, and 4, transcript analysis could not determine if DAR was in single or double dose. Analysis of transcripts from individuals 9 and 10 showed the known(C)ces haplotype (cesstands for a ce allele that produced the RH20 antigen and is associated with the 733C>G mutation).16 As described by Faas et al,16(C)ces is composed of 2 altered genes: (1) a hybrid D-Ce-D transcript in which exons 1 and 2, the first 3 polymorphic positions of exon 3 (361, 380, and 383), and exons 9 and 10 derived from the RHD gene. The hybrid transcript carries 4 extra nucleotide substitutions: 186G>T in exon 2 (Leu62Phe), 410C>T in exon 3 (Ala137Val), 733C>G in exon 5 (Leu245Val), and 1006G>T in exon 7 (Gly336Cys); (2) and aces transcript carrying 733C>G in exon 5 but also 1006G>T (Gly336Cys) in exon 7. Individuals 9 and 10 were homozygous for (C)ces as shown by the absence of double peak when direct sequencing of PCR products was performed.
Observed RH haplotypes with alteredRHCE alleles in black individuals
| RHD-RHCE haplotype . | Name of haplotype . | References . | Comments . |
|---|---|---|---|
| DAR-ceAR | 25, this report (samples 1, 4, 5, 7, 8) | — | |
| no D-ceAR | 25 | — | |
| DAR-ceEK | This report (sample 3) | — | |
| no D-ceEK | This report (sample 1) | — | |
| D-Ce-D(4)-Ce | RN | 6 | — |
| D-Ce-D(part of 3,4)-Ce | RN | 6 | — |
| D-Ce(3-8)-D-ces(1006) | rs or (C)ces | 16, 17, this report (samples 9, 10) | Extra mutations can be present in the hybrid D-Ce-D gene (186G>T exon 2, 410C>T exon 3, 733C>G exon 5, 1006G>T exon 7). |
| D-ceMO | 18 | — | |
| D-ces | 16, 17, this report (samples I-1, II-2, II-3, II-4, II-5 in Figure2) | — | |
| D-ce(48) | 19, 20, 21 | — | |
| no D-ce-D(5)-ce | RoHar | 26 | RoHarwas described in whites. In this report (sample 13),ce-D(5)-ce is in linkage with D orDIVa. |
| RHD-RHCE haplotype . | Name of haplotype . | References . | Comments . |
|---|---|---|---|
| DAR-ceAR | 25, this report (samples 1, 4, 5, 7, 8) | — | |
| no D-ceAR | 25 | — | |
| DAR-ceEK | This report (sample 3) | — | |
| no D-ceEK | This report (sample 1) | — | |
| D-Ce-D(4)-Ce | RN | 6 | — |
| D-Ce-D(part of 3,4)-Ce | RN | 6 | — |
| D-Ce(3-8)-D-ces(1006) | rs or (C)ces | 16, 17, this report (samples 9, 10) | Extra mutations can be present in the hybrid D-Ce-D gene (186G>T exon 2, 410C>T exon 3, 733C>G exon 5, 1006G>T exon 7). |
| D-ceMO | 18 | — | |
| D-ces | 16, 17, this report (samples I-1, II-2, II-3, II-4, II-5 in Figure2) | — | |
| D-ce(48) | 19, 20, 21 | — | |
| no D-ce-D(5)-ce | RoHar | 26 | RoHarwas described in whites. In this report (sample 13),ce-D(5)-ce is in linkage with D orDIVa. |
— indicates no comment.
Rare RHCE alleles in black individuals producing loss of immunogenic epitopes and/or weak e antigens.
Open boxes illustrate the RHCE exons. Black lines or solid boxes within the RHCE gene represent replacement ofRHCE nucleotides by the counterpart of the RHDgene. Bold lines stand for mutations at nonpolymorphic sites. Variants represented were described in this study or by other authors (references indicated). In variants associated with the loss of immunogenic high-incidence antigens, there are schematically 3 categories: (1) the RH:−18 phenotype, with 3 possible RHCEalleles, (2) the RH:−34 phenotype, characterized by a haplotype with aD-CE(3-8)-D hybrid gene plus 4 extra mutations, encoding some RhC epitopes, in linkage with aces allele represented in this figure carrying an extra mutation at 1006 ((C)ces), and (3) the RH:32,-46 phenotype, with 2 CE-D-CE hybrid alleles. Another group of variants corresponds to altered ce alleles with the loss of Rhe immunogenic epitopes, like the ceMOallele and a ceS allele with an extra mutation in 340 [ces(340)]. The last group of variants includes other ce alleles associated with decreased Rhe [ce(C48), ces, ces(697), ces(748), ce-D(5)-ce]. No immunization has been shown, so far, against lacking Rhe epitopes. The frequent cytosine at position 48 in blacks may be carried or not in the differentRHCE alleles. The RHD gene (normal or altered), which cosegregates with these alleles, is indicated in Table 4 when known either because this report or other authors could demonstrate the associated RHD gene.
Rare RHCE alleles in black individuals producing loss of immunogenic epitopes and/or weak e antigens.
Open boxes illustrate the RHCE exons. Black lines or solid boxes within the RHCE gene represent replacement ofRHCE nucleotides by the counterpart of the RHDgene. Bold lines stand for mutations at nonpolymorphic sites. Variants represented were described in this study or by other authors (references indicated). In variants associated with the loss of immunogenic high-incidence antigens, there are schematically 3 categories: (1) the RH:−18 phenotype, with 3 possible RHCEalleles, (2) the RH:−34 phenotype, characterized by a haplotype with aD-CE(3-8)-D hybrid gene plus 4 extra mutations, encoding some RhC epitopes, in linkage with aces allele represented in this figure carrying an extra mutation at 1006 ((C)ces), and (3) the RH:32,-46 phenotype, with 2 CE-D-CE hybrid alleles. Another group of variants corresponds to altered ce alleles with the loss of Rhe immunogenic epitopes, like the ceMOallele and a ceS allele with an extra mutation in 340 [ces(340)]. The last group of variants includes other ce alleles associated with decreased Rhe [ce(C48), ces, ces(697), ces(748), ce-D(5)-ce]. No immunization has been shown, so far, against lacking Rhe epitopes. The frequent cytosine at position 48 in blacks may be carried or not in the differentRHCE alleles. The RHD gene (normal or altered), which cosegregates with these alleles, is indicated in Table 4 when known either because this report or other authors could demonstrate the associated RHD gene.
Transcript analysis of sample 11 identified theRN and ceMO alleles (ceMOcarries C48 in exon 1 and T667 in exon 5)6 18 with a normal RHD transcript. In sample 12, a normal cEallele was associated with a new ces allele referred to as ces(340) and carrying 733C>G in exon 5 but also 340C>T in exon 3 (Arg114Trp). A newD(674) transcript carrying 674C>T in exon 5 (Ser225Phe) was also found for individual 12. Individual 14 carried a normalRHD gene, a hybrid Ce-D(4)-Ce allele characteristic of an RN haplotype, and a newces allele referred to asces(748) with 733C>G and 748G>A (Val250Met) in exon 5. Individual 15 carried a normal RHDgene, one RN allele, and one hybridce-D(4-9)-ce allele.
Genomic DNA analysis
The transcript analysis was confirmed on genomic DNA (except for C48 in exon 1, very frequent in black individuals).
RHD and RHCE exons 4 to 5, RHCE exon 6, and RHD exon 7 were sequenced from individuals 2, 5, and 7. Individual 2 carried DAR, ceAR, and ceEKalleles, whereas individuals 5 and 7 carried DAR andceAR alleles. Individual 8 genotype(DAR-ceAR/DAR-ceAR) was deduced from genomic analysis of the parents, both who carried a DAR allele plus a normalRHD gene and one ceAR allele plus a normalce allele.
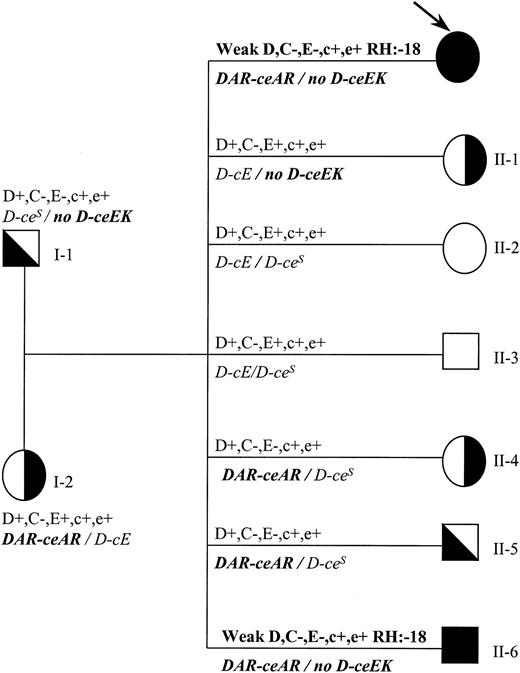
Family study (Figure 2) of individual 1 (Rh phenotype and genomic sequencing) showed that DAR was in single dose and also that individual 1 inherited a DAR-ceARhaplotype from the mother and a noD-ceEK haplotype from the father.
Family tree indicating genotypes and phenotypes for individual 1.
Rh phenotype and genotype (italics) are shown. Propositus is indicated by an arrow. DAR is in linkage with ceAR(indicated as DAR-ceAR), whereas ceEK is not associated with any RHD gene (indicated as no D-ceEK). Propositus and individual II-6 have the same molecular background and the same rare phenotype (RH:−18).
Family tree indicating genotypes and phenotypes for individual 1.
Rh phenotype and genotype (italics) are shown. Propositus is indicated by an arrow. DAR is in linkage with ceAR(indicated as DAR-ceAR), whereas ceEK is not associated with any RHD gene (indicated as no D-ceEK). Propositus and individual II-6 have the same molecular background and the same rare phenotype (RH:−18).
RHCE exon 3 and exons 4 to 5 were sequenced from samples 13, 16, and 17. Individual 13 carried a ceMO allele and one hybrid ce-D(5)-ce allele. Individual 16 (with a serological profile similar to that of individual 11) carried oneRN allele associated to a ceMOallele. Individual 17 (with a serological profile similar to that of individual 12) carried a normal cE allele and aces(340) allele. Sequencing of RHDexons 3 to 7 (in which the more frequent mutations in partial D are localized) was normal for samples 16 and 17. For sample 13, we found a normal RHD gene next to an RHD gene carrying mutations previously described in DIVa: 455A>C and 1048G>C mutations. RHD exon 2 was not sequenced, but expression of RH30 confirmed the hypothesis of a DIVa gene for individual 13 (data not shown).
Screening of SCD patients for rare RHalleles
Since the carriers of some rare RH alleles may develop antibodies following transfusion or pregnancy, the frequency of such alleles was estimated in a population of 146 SCD patients. For the detection of ceEK, ceAR, and ceBIalleles, the presence of the common G712 mutation was determined by a specific PCR assay. Among 6 patients carrying G712, sequencing ofRHCE exons 4, 5, and 6 PCR products showed that 4 and 2 patients were found heterozygous for ceAR andceEK, respectively. Three patients carried ceMOheterozygously, as they carried both the T667 mutation and the wild-type G667 mutation. To identify the (C)ceshaplotype and the ces(340) allele, a first step was performed by screening the G733 mutation inRHCE exon 5. Among the 75 patients positive for the G733 mutation, one also carried T340 heterozygously in exon 3 [ces(340) allele], and 10 carried both T1006 and a hybrid exon 3 [(C)ces]. Among patients carrying the (C)ces haplotype, one was homozygous, as shown by absence of normal RHD exon 3, wild-type G1006 mutation, and wild-type C733 mutation. From these findings, we calculated the incidence of the rare alleles and haplotypes in our population of 146 SCD patients as follows:ceEK: 1.4%; ceAR: 2.7%; ceMO: 2%;(C)ces: 7.5%; andces(340): 0.7%.
Discussion
This report shows that a variety of RHCE alleles are present in Afro-Caribbean black individuals, among which 4 are new alleles: ceEK, ceBI, ces(340), and ces(748). Some of the alleles found in this ethnic group are associated with the loss of immunogenic epitopes, and there is a risk of immunization if exposition to normal Rh antigens occurs. Some of the antibodies produced are clinically significant. Therefore, for efficient transfusion purposes, a complete characterization of the variants is required. Serological diagnosis of these variants is difficult, especially when different combinations of altered alleles occur. The serological and molecular data described here led us to propose a procedure to detect these variants within a population of SCD patients and black blood donors, to ensure patient transfusion safety.
Clinical issues must be addressed for patients homozygous forceEK, ceAR, ceBI,(C)ces, ceMO, andces(340) or for those who are composite heterozygous, as we showed that they can produce anti-Rh antibodies. Individuals homozygous for ceAR within a DAR-ceAR haplotype can produce a clinically significant anti-RH18, as illustrated by the fatalities of individuals 7 and 8 following incompatible transfusion. In both cases, these SCD patients were first transfused with DccEE RBCs because of the presence of an antibody that reacted mainly to Rhe-positive RBCs, in addition to anti-C for patient 7. As the transfusion was inefficient, a further transfusion with DccEE blood units was decided upon, despite complete incompatibility. Hemolysis and death occurred rapidly for both patients. Patient 7 was a pregnant woman who received transfusions after a cesarean delivery. The serum obtained after the last transfusion contained anti-RH18, anti-C, anti-Fya, and anti-S. The DAT was strongly positive (IgG and complement). The anti-RH18 (serum titer: 256 on ddccee RBCs) was eluted from the RBCs. The newborn typed (DCcee) was healthy despite a strongly positive DAT (IgG) and elution of the anti-RH18. Patient 8 received transfusions for acute anemia. After the last transfusion, the serum contained only anti-RH18 (titer 2000 on ddccee RBCs) and was eluted from the RBCs. No anti-D was found in the sera of the 2 patients despite their partial D phenotype (DAR) and transfusion of D-positive RBCs. No rare RH:−18 blood was available. These 2 cases emphasize both the clinical relevance of anti-RH18 in transfusions in SCD patients and the difficulty identifying this antibody, as already discussed by Issitt,9 since it may resemble an auto–anti-Rhe produced by Rhe-positive individuals at the beginning of the immunization. Individuals 4 and 5, who produced anti-RH18 through pregnancies, had the same molecular background as patients 7 and 8(DAR-ceAR/DAR-ceAR).
The other Afro-Caribbean individuals who produced anti-RH18 in group A shared a similar Dccee phenotype but exhibited different reactivity profiles with the anti-e MoAbs and the anti-RH10/20 serum. TheRHCE genes were as follows: ceAR/ceEK(individuals 1 and 2), ceEK/ceEK (individual 3), andceEK/ceBI (individual 6). The RH haplotype was precisely defined for individual 1, for whom the family study showed one DAR-ceAR haplotype and one noD-ceEK haplotype (noD stands for no RHD gene). The ceEKallele also can cosegregate with DAR, as individual 3, who is homozygous for ceEK, carried at least one DARallele. Since the 3 RH:−18 polypeptides encoded by ceEK,ceAR, and ceBI are distinct in terms of amino-acid substitutions (Figure 1), it was of interest to determine if the anti-RH18 produced by individuals carrying these different alleles had exactly the same specificity. Serum of individual 3(ceEK/ceEK) was compatible with RBCs of individual 5(ceAR/ceAR), whereas serum of individual 5 was not compatible with RBCs of individual 3. An antibody with the anti-e specificity could be eluted from RBCs from sample 3 after adsorption of serum from sample 5. These results suggest that serum from sample 3 is defined as anti-RH18 and that serum from sample 5 had a broader specificity and contained probably the same anti-RH18 plus some anti-Rhe epitopes. No conclusion can be drawn for the ceBIallele, as no immunized homozygous individual is known. These findings add one more angle of complexity to the definition of RH:−18 individuals, as molecular background has to be taken into account to reach serological compatibility. Also, as shown in Table 2, ceMO RBCs cannot be transfused to RH:−18 individuals producing anti-RH18 despite their common RH:−19 status (Table 3). It has to be understood that anti-RH19 defines some but not all epitopes of the RH18 antigen because anti-RH19 is obtained from an anti-RH18 serum adsorbed on DccEE RBCs. Therefore, anti-RH19 is always negative with RH:−18 RBCs (Table 3) and also with some variants of the e antigen such as ceMO, which lacks probably some but not all epitopes lacking in RH:−18 RBCs.
Anti-RH18 was eluted from RBCs from the newborn of individual 2(ceAR/ceEK), but no hemolysis was detected. No antibody was eluted from RBCs of the newborn of individual 3 (ceEK/ceEK). For the other newborns, only clinical data were available. They all were healthy. As opposed to the findings of another author,10 we did not find anti-RH18 involvement in any hemolytic disease of the newborn.
Clinical issues also are to be taken into account for the other alleles described in this study, as the carriers may develop anti-Rh antibodies.
Patients 9 and 10, homozygous for the(C)ces haplotype, produced an anti-RH34. Patient 9 was a pregnant woman. Despite the high titer of the antibodies (RH34 and anti-D), the newborn was healthy, but no biologic data (newborn phenotype, DAT) were available. Patient 10 was a young SCD patient who received only 3 transfusions (phenotype of blood units unknown) before production of the antibody. For this patient, it was strongly advised that no more transfusions occur, as the anti-RH34 titer was 128 on DCCee RBCs.
Among the immunized patients, individual 11 carried theceMO allele next to a RN allele. We showed already that ceMO encoded a weak Rhe antigen18 and bring evidence here that it may encode a partial Rhe antigen, as individual 11 produced anti-e and anti-ce antibodies. We showed previously that variants homozygous forceMO or with a ceMO/cE genotype were typed RH:−19.18 Individual 11 is typed RH:19 because the associated RN allele encodes a polypeptide expressing RH19 (Table 3). Titers of antibodies were low, but we think that one should be cautious if transfusion occurs. Then, long-term cryopreservation of RBCs for patients 11 and 16, who carried the sameceMO/RN genotype, would be strongly recommended. The ceMO allele also was found in individual 13. For instance, individual 13 carried a ceMO allele but expressed the RH50 antigen, consistent with the presence of a hybridce-D(5)-ce allele.26 27 Although no antibody is currently present in the serum of individual 13, there is a risk of immunization through transfusion or pregnancy, because the loss of Rhe epitopes caused by the ceMO polypeptide is not compensated by the polypeptide encoded by ce-D(5)-ce (lack of epitopes detected with anti-e MoAbs). As individual 13 is typed RH:−19, it can be deduced that the ce-D(5)-ce allele produced a polypeptide lacking epitopes recognized by the anti-RH19 reagent.
The ces(340) allele was found in individual 12, who produced allo–anti-e and allo–anti-Ce antibodies (through pregnancies). Her serum was not compatible with RH:−18 and RH:−34 RBCs (sample 9). Since this new allele was next to a normalcE allele, we postulated thatces(340) encodes a partial e antigen that has to be taken into account when transfusion occurs. The nonimmunized patient 17 exhibited the same pattern of reactivity and similar molecular background. For individuals 12 and 17, common DccEE RBCs could be transfused, but transfusion problems will probably arise for patients carrying homozygous ces(340) or association ofces(340) with another allele that encodes partial e antigen. For such patients, cryopreservation of RBCs will be recommended. It must be pointed out that 340T is the same polymorphism that occurs in weak D type 17 and in weak C produced by theCeMA allele described recently.27 28
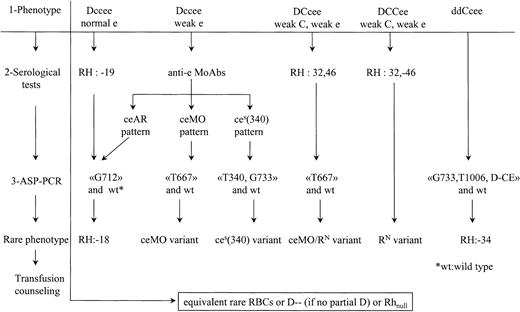
The evidence that ceEK, ceAR, ceBI,(C)ces, ceMO, andces(340) but also RN may induce partial phenotypes with risk of immunization in blacks led us to evaluate the frequency of these alleles in a population of black SCD patients. Incidence of the corresponding rare phenotypes has been deduced from frequency of the alleles: RH:−18: 1/600; RH:−34: 1/190; partial e (ceMO): 1/2370; partial e[ces(340)]: 1/21 000. NoRN allele was found. Since there are approximately 4000 SCD patients in France, we think that it is relevant to detect these rare phenotypes in patients and donors. The procedure shown in Figure 3 is based on RBC serological reactivity and PCR assays to detect individuals who carry a molecular background associated with a risk of immunization, such as in groups A to C. In all cases, the partial D status also has to be taken into account for transfusion safety.
Process to distinguish rare RhCE phenotypes associated with a risk of immunization.
Three steps are proposed: (1) routine phenotype, (2) serological studies with anti-e MoAbs or human sera, and (3) ASP-PCR to detect mutations correlated to the serological profile as shown in this study. Wild-type ASP-PCR is also performed to determine zygosity. The ceAR phenotype can be detected in the category of Dccee phenotypes with normal Rhe or with depressed Rhe, depending of the routine reagents used. For DCcee phenotypes with decreased expression of C and e antigens, expression of RH32 and specific mutation of theceMO allele indicates the composite heterozygousceMO/RN genotype. When there is a decreased expression of both C and e antigens within a DCCee phenotype, serological testing is sufficient to detect RH:32,-46. For the ddCcee phenotype, few serological alterations are exhibited in the RH:−34 individuals. Therefore, an ASP-PCR to detect the altered(C)ces haplotype would be straightforward. In all cases, rare blood has to be provided if transfusion is needed. The case of DccEe with depressed Rhe has not been considered because rather common DccEE units can be transfused to avoid anti-e immunization or immunohemolytic reaction when anti-e is already produced.
Process to distinguish rare RhCE phenotypes associated with a risk of immunization.
Three steps are proposed: (1) routine phenotype, (2) serological studies with anti-e MoAbs or human sera, and (3) ASP-PCR to detect mutations correlated to the serological profile as shown in this study. Wild-type ASP-PCR is also performed to determine zygosity. The ceAR phenotype can be detected in the category of Dccee phenotypes with normal Rhe or with depressed Rhe, depending of the routine reagents used. For DCcee phenotypes with decreased expression of C and e antigens, expression of RH32 and specific mutation of theceMO allele indicates the composite heterozygousceMO/RN genotype. When there is a decreased expression of both C and e antigens within a DCCee phenotype, serological testing is sufficient to detect RH:32,-46. For the ddCcee phenotype, few serological alterations are exhibited in the RH:−34 individuals. Therefore, an ASP-PCR to detect the altered(C)ces haplotype would be straightforward. In all cases, rare blood has to be provided if transfusion is needed. The case of DccEe with depressed Rhe has not been considered because rather common DccEE units can be transfused to avoid anti-e immunization or immunohemolytic reaction when anti-e is already produced.
In group D (samples 13 to 17), patients were nonimmunized. Clinical issues have been discussed above for patients 16 and 17, as they were similar in terms of phenotype and genotype as patients 11 and 12, respectively, but also for patient 13, who carried ceMO next to a hybrid ce-D(5)-ce allele. Two other composite heterozygous individuals have been found. Individual 15 carried oneRN haplotype associated to ace-D(4-9)-ce allele, which probably encodes only a few epitopes of both c and e antigens and no RH46 antigen. It can be assumed that the absence of RH46 antigen makes this phenotype clinically significant for transfusion. Individual 14 carried a newces(748) allele next to anRN allele. Alterations found in individual 14 have not been associated, so far, to the production of Rh antibodies. Therefore, transfusion counseling is even more complex because the risk is unknown.
In conclusion, we characterized rare Rh phenotypes found in black individuals as we determined the serological reactivity and the molecular background of RH:−18 and RH:−34 and of several partial e and weak e phenotypes. This is a step toward a better definition of the numerous Rh variants in the black population. We associated some of these phenotypes to the presence of alloantibodies. We underlined the clinical significance in transfusion practice of the anti-RH18 produced by black individuals carrying the DAR-ceAR haplotype at the homozygous state. These data should lead to screening studies of black blood donors to obtain rare blood units for cryopreservation. These programs should improve transfusion safety, especially in the field of sickle cell disease and hemolytic disease of the newborn.
We are grateful to Martine Verdier, MD, Jérôme Babinet, MD, Danièle Vanahaeke, MD, and Lucienne Mannessier, MD (Etablissement Français du Sang, France) for referring to our laboratory the donor samples. We also acknowledge the SCARF contribution to this work.
Prepublished online as Blood First Edition Paper, August 1, 2002; DOI 10.1182/blood-2002-01-0229.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
France Noizat-Pirenne, 20 rue Bouvier, 75522 Paris Cedex 11 France; e-mail: pirenne@ints.fr.

![Fig. 1. Rare RHCE alleles in black individuals producing loss of immunogenic epitopes and/or weak e antigens. / Open boxes illustrate the RHCE exons. Black lines or solid boxes within the RHCE gene represent replacement ofRHCE nucleotides by the counterpart of the RHDgene. Bold lines stand for mutations at nonpolymorphic sites. Variants represented were described in this study or by other authors (references indicated). In variants associated with the loss of immunogenic high-incidence antigens, there are schematically 3 categories: (1) the RH:−18 phenotype, with 3 possible RHCEalleles, (2) the RH:−34 phenotype, characterized by a haplotype with aD-CE(3-8)-D hybrid gene plus 4 extra mutations, encoding some RhC epitopes, in linkage with aces allele represented in this figure carrying an extra mutation at 1006 ((C)ces), and (3) the RH:32,-46 phenotype, with 2 CE-D-CE hybrid alleles. Another group of variants corresponds to altered ce alleles with the loss of Rhe immunogenic epitopes, like the ceMOallele and a ceS allele with an extra mutation in 340 [ces(340)]. The last group of variants includes other ce alleles associated with decreased Rhe [ce(C48), ces, ces(697), ces(748), ce-D(5)-ce]. No immunization has been shown, so far, against lacking Rhe epitopes. The frequent cytosine at position 48 in blacks may be carried or not in the differentRHCE alleles. The RHD gene (normal or altered), which cosegregates with these alleles, is indicated in Table 4 when known either because this report or other authors could demonstrate the associated RHD gene.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/12/10.1182_blood-2002-01-0229/5/m_h82323472001.jpeg?Expires=1769103246&Signature=Hm1VZty3U2hA93h6PKC-0GO0XPS48smVy4fCvlUIXmfWdZ1yB3W893RatWTN713iRFNAw6kC6CJYXi0Wsc-cuG5rwNnV8oGPfUi7gMY6mYTKexba8vhoADfQDEViiJJfUsfwY9TesIBDxXzDesSlujXSinBLINExQ9yo4-T0w0F5X1KoyI1DXHNLW2AvWRlX-6sW-JGbucQfVbONImsW0VJC8xq1GOa0L00~DLGy2XE27t7xMQXssGj7lapTY5bqgGiZf-Al0qb6BS3hldUSc1Al-bPqki6Y~wr1TMJLQfMxmds1r4FF00OTqYdPQlqCKaGBJSqYENg3IhzZSbE5WQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal