B-lineage–committed cells are believed to arise in the liver of mouse embryos at 14 days after coitus (dpc). However, pre-B-specific gene transcripts and DJH gene rearrangements have been detected in earlier, midgestation embryos. We describe here a population of c-kit+AA4.1+CD19+Pax5+cells present in the aorta-gonad-mesonephros (AGM) area and in the livers of 11-dpc mouse embryos. In contrast to multipotent c-kit+AA4.1+CD19−hematopoietic stem cells (HSCs), these c-kit+AA4.1+CD19+ progenitors differentiated only to B-lineage cells in vitro. We propose that mouse embryonic B lymphopoiesis starts earlier than previously thought, at 10 to 11 dpc, both in liver and extra-liver hematopoietic sites. The B-cell differentiation program is not delayed with respect to the emerging lymphohematopoiesis events in the midgestation mouse embryo (8-9 dpc).
Introduction
Embryonic lymphohematopoiesis is a highly dynamic developmental process that takes place in mesoderm-derived locations (yolksac [YS], para-aortic splanchnopleura/aorta-gonad-mesonephros [P-Sp/AGM]), closely after mouse gastrulation (7.5-11 days after coitus [dpc]). A transient, self-limited myeloerythropoiesis first takes place in the early YS from 7.5 dpc to 12 dpc,1 while definitive lymphohematopoiesis arises independently in the intraembryonic P-Sp/AGM (≥ 8.5 dpc), in cell clusters related to the ventral wall of the aorta.2-8These first-arising embryonic hematopoietic stem cells (HSCs) migrate to and colonize fetal hematopoietic organs (liver, spleen, thymus, bone marrow [BM]), where they self-renew and differentiate in inductive microenvironments.9,10 The incipient liver bud is thus seeded by hematopoietic cells at 10 dpc, and becomes the major reservoir of lymphohematopoiesis during late mouse gestation.11 The P-Sp/AGM area expands until 11 dpc and subsequently degenerates with a sharp decline in cell numbers, probably related with the cell export of its HSCs to the circulatory system and homing to the liver.12-14 It has been proposed that midgestation hematopoiesis (8.5-12.5 dpc) is restricted to the generation, self-renewal, and expansion of HSCs, predominantly in the P-Sp/AGM (a putatively pure stem cell organ). In vitro differentiation analyses only revealed multipotent progenitors in embryonic blood, P-Sp/AGM, and liver before 13 dpc.12,15 The first unilineage B-cell–restricted precursors detected by this method were only recovered at 14 dpc in the liver. Programs of lineage differentiation, however, begin to be settled in midgestation embryo progenitors, as happens for the YS myeloerythropoiesis.16Bipotent macrophage/B-cell progenitors (Sca1+AA4.1+B220−) have been reported in the 12.5 dpc liver.17 A fetal hematopoietic precursor with B, T, and macrophage potentials has also been described in the AA4.1+FcγR+ liver cell population at 13 dpc.18 The fetal counterpart of the BM common lymphoid progenitor (CLP; interleukin-7 receptor α [IL-7Rα]+Sca-1lowc-kitlow) has been identified in the 12.5- to 14.5-dpc liver, where it expresses additional nonlymphoid potentials.19,20 On the basis of results from an in vitro multilineage progenitor assay, it has been suggested that lymphoid progenitors are present in early embryo hematopoietic organs, including P-Sp/AGM, blood, and liver, with T progenitors arising much earlier (10-12 dpc) than B progenitors (14-15 dpc).21 The same authors described the so-called earliest B-cell precursor in the 12.5- to 14.5-dpc liver (Lin−CD19−c-kit+IL-7Rα+AA4.1+).22Overall, the prevailing view is that committed B-cell progenitors do not exist in the liver before 14 dpc, 5 days after the emergence of the first definitive HSCs in the P-Sp/AGM. From then on, liver B-cell progenitors expand in a synchronous wave–like pattern, reaching a peak in perinatal stages.23,24 IgM+ B cells have not been detected until 17 dpc.25 By contrast, we and other workers have found evidence of B-lineage–specific gene expressions, DJH rearrangements, and immunoglobulin (Ig) germ line transcripts in P-Sp/AGM and liver before 12.5 dpc.14,26,27Whether these latter findings represent low-level, sporadic transcription of partially accessible gene loci in multipotent HSCs or markers of bona fide B-committed cells is open to debate.28-30
Fetal hematopoietic progenitor populations have been traced with different cell surface receptors. The receptor tyrosine kinase c-kit, besides being present in migrating melanoblasts and germ cells, is also expressed in hematopoietic cell progenitors.31 The AA4.1 molecule, which is involved in cell adhesion,32 is found in all embryonic HSCs,33 and its expression is also up-regulated in BM B-lymphoid progenitors in correlation with the onset of Pax5 transcription, CD19 Ag expression, and loss of pro–T-cell potential.34 IL-7Rα expression represents one of the earliest markers of T/B lymphoid commitment.35 B-cell differentiation is also controlled by a cascade of transcription factors.36 The E2A-encoded (E12 and E47) and early B-cell factor (EBF) proteins regulate the rearrangement and expression of the IgH and Igκ gene loci, and are essential for B-cell development.37,38 The Pax5/BSAP protein is expressed in the earliest B-cell precursors, and has been shown to be determinant for B-cell commitment through the repression of genes acting on the differentiation of alternative cell lineages.39,40 Among others, the B-cell lineage-specific receptor CD19 is a downstream target of Pax5/BSAP activity.41
This wide-ranging information about the emerging embryonic B lymphopoiesis has mostly come from studies with either unpurified and/or mixed cell populations and has frequently been obtained from in vitro differentiation assays of varying efficiency. We decided to search for the earliest-restricted B-cell precursors at the cellular level, by means of genetic and flow cytometry analyses performed on cell populations directly purified ex vivo. In the present paper, we describe a novel population of c-kit+AA4.1+CD19+ cells detected in the 11-dpc P-Sp/AGM and liver, which selectively expresses pre-B-specific genes. In contrast to multipotent c-kit+AA4.1+CD19− cells purified from the very same sites, CD19+ cells exclusively differentiate in vitro to B-lineage cells. They are absent from Pax5-deficient mouse embryos, a finding that confirms their B-cell restriction. Our data show that B lymphopoiesis starts 3 days earlier than previously believed in liver and extra-liver sites of mouse embryo.
Materials and methods
Mice and embryo cell preparations
BALB/c (CD45.2), C57BL/6 (CD45.2), C57BL/6-CD45.1, and heterozygous C57BL/6-Pax5+/− mice were maintained under pathogen-free conditions in the animal facilities of the ISCIII and the CBMSO. Timed pregnancies were determined by the observation of the vaginal plug in the early morning after overnight mating (gestation day: 0 dpc). Single-cell suspensions from P-Sp (8-9 dpc), AGM, liver, blood, and YS (10-13 dpc) were prepared as previously described.14 Viable cells were counted by trypan blue exclusion.
Monoclonal antibodies and flow cytometry
The following monoclonal antibodies (mAbs) used for staining were purified from hybridoma culture supernatants on protein G columns and labeled with fluorescein isothiocyanate (FITC) or biotin by standard procedures, or with Cy5 with a labeling kit (Amersham Pharmacia Biotech, Pisataway, NJ), according to the manufacturer instructions: anti-B220/CD45R (RA3-6B2), CD19 (1D3), AA4.1, Sca-1/Ly-6A/E, HSA/CD24 (J11d), CD43 (S7), CD16/CD32 (2.4G2), Mac-1/CD11b (M1/70), Thy-1/CD90 (T-24), CD4 (GK1.5), αβ-TCR (H57.597), and CD3ε (145-2C11). All other reagents were obtained from BD Pharmingen (San Diego, CA) as purified, biotinylated, FITC-, or phycoerythrin (PE)–coupled antibodies: c-kit/CD117 (2B8), CD34 (RAM34), IL-7Rα/CD127 (B12-1), TER-119/Ly-76, CD45 (30-F11), CD45.2 (104), CD8 (53-6.7), and rat immunoglobulin isotype controls. Embryonic cell suspensions were prepared in cold fluorescence activated cell sorting (FACS) medium (phosphate buffered saline [PBS], 5% fetal calf serum [FCS], 0.1% sodium azide) and incubated in the presence of anti-CD16/CD32 mAb to block nonspecific Fc-dependent mAb bindings (except when the CD16/CD32 expressions were analyzed). After washing, cells were incubated with optimal concentrations of purified, biotin-, FITC-, and/ or Cy5-labeled mAbs. PE-conjugated streptavidin (Southern Biotechnology Associates, Birmingham, AL) revealed the biotinylated mAbs in a third incubation. Cell debris and dead cells were excluded by light scatter parameters and propidium iodide gating. Specific mAb signals were defined against the fluorescence provided by isotype-matched irrelevant mAbs. Cytoplasmic μ heavy chain (μHC) detection was performed as described42 after surface staining with PE-labeled anti-CD19 and Cy5-labeled anti-IgM mAbs. Cells were then permeabilized43 and stained with a μHC-specific FITC goat anti–mouse IgM or FITC goat anti–hamster Ig (Southern Biotechnology Associates). Forward, side scatter, and 3 fluorescent parameters were collected on at least 4 × 104 gated cells per sample. Flow cytometric analysis were performed on a dye laser–equipped FACSCalibur with CELLQuest program (Becton Dickinson, Mountain View, CA). Cells were sorted with an Altra Hypersort system EPICS (Coulter Electronics, Hialeah, FL) under sterile conditions. Reanalysis of sorted cells showed more than 95% purity for CD19+ populations. After the initial detection studies, most analyses and purifications of embryonic cells were done in pooled 11-dpc livers, due to the limited cell numbers recovered earlier in midgestation embryos.
Polymerase chain reaction analyses
Total RNA was extracted, and cDNA prepared and amplified, with Taq-DNA Polymerase (Sigma Chemical, St Louis, MO) for rag-2, VpreB, and λ5 (2 μL cDNA/reverse transcriptase–polymerase chain reaction [RT-PCR]), and for β-actin (1 μL cDNA) as described.42 FastStart Taq-Polymerase (Roche Diagnostics GmbH, Mannhein, Germany) was used for Pax5 (4 μL cDNA), CD19, EBF, and Ikaros (2 μL cDNA) amplifications. Each cycle was repeated 25 times for β-actin, 35 times for VpreB and λ5, and 40 times for the other PCRs. Primers and annealing conditions have been described for rag-2,44 VpreB, λ5,42 Pax5,45CD19,37 EBF, Ikaros,21 and β-actin.45 Amplification products (one-fifth each) were separated electrophoretically on 2% agarose gels, transferred, and hybridized. Rag-2, VpreB, and λ5 cloned probes, oligonucleotides 5′-ACTCACCCTATGCCATTGTGC-3′ (EBF-in), 5′-ATGTGGATATTGTGGCCGGA-3′ (Ikaros-in), 5′-CCAGTACGGGAATGTGCTCT-3′ (CD19-in) and those described for Pax5 and β-actin45 were used as32P-labeled probes. PCR signal intensities were quantified by densitometry on a FLA-3000 detector (Fuji, Tokyo, Japan) using the AIDA software (Raytest, Stauhenhardt, Germany). Relative message levels were calculated by referring the gene-specific signal intensity values to the value obtained for β-actin in the same cDNA. HC locus transcripts (VDJCμ) were revealed by 2 rounds of amplification (35 cycles each), by using described primers43,46(VH7183, VHJ558, and Cμ1), and a newly designed for Cμ2 (5′-GGGAAATGGTGCTGGGCAGGAA-3′). For transcript expressions from individual Pax5−/− embryo livers, 2 μL of the first PCR products were subjected to 35 cycles of amplification with internal primers for rag-2, VpreB, and CD19 (rag-2-3′ in: 5′-GGTTCAGGGACATCTCCTTACTAAG-3′; VpreB in: 5′-CCCACGGCACACTAATACACA-3′; CD19: 5′-TTGAGTGGAGCTGAGGAGCT-3′ and the above-described CD19-in). The nested PCR (60°C, annealing temperature) products were visualized on ethidium bromide–stained gels. Pax5 genotypes41 were determined by PCR assays on 11-dpc embryo tail DNA preparations, using α-actin44 to verify DNA content. Control experiments with sequential numbers of cycles confirmed the linear range of the amplification process for the cDNA inputs. As additional controls for the differential sensitivity of VpreB and λ5 RT-PCR assays, amplifications were performed on cDNA samples from serial dilutions of the 70Z/3 cell line.
Lymphoid, myeloid, and erythroid potentials from sorted embryonic cells
B-lymphoid progenitor frequencies were calculated by seeding dilutions of 50 to 500 sorted cells per well in 96-well plates bearing confluent layers of Mitomycin C (Sigma)–treated ST2 stromal cells (5 × 103 cells/well; 7-10 days of culture). The medium used was Iscove modified Dulbecco medium (IMDM) (BioWhittaker Europe, Verviers, Belgium), supplemented with 10% IL-7–conditioned medium (IL-7 sup), as previously described.42 The stromal cell cultures were refreshed every 96 hours. The B-lymphoid, myeloid, and erythroid potentials were evaluated in parallel by using a modification of described methods.12 In brief, sorted cells (100-500 cells/well, 96-well plates) were seeded on an ST2 stromal cell monolayer, and grown in IL-7 sup (10%), IL-11 (20 ng/mL; Pepro Tech, Rocky Hill, NJ), and stem cell factor (SCF) (20 ng/mL; R&D Systems, Abingdon, United Kingdom) for 7 days. The growing cell clones were then split into 3 aliquots that were set up in (1) SCF alone or in combination with granulocyte-macrophage colony-stimulating factor (GM-CSF) (2 ng/mL; Pepro Tech) (myeloid conditions), (2) ST2 plus 1% methylcellulose, SCF, and erythropoietin (EPO, 2 U/mL) (erythroid conditions), and (3) ST2 plus IL-7 sup (B-lymphoid conditions). After an additional culture period of 10 to 14 days, wells harboring growing cells were scored and collected. Additional cultures of sorted populations were made with the Methocult GFM3434 system (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with recombinant IL-3, IL-6, SCF, and EPO to optimize the recovery of erythroid/myeloid precursors, as described16 (0.5-5 × 103cells/well; 24-well plates). After a period of 7 to 9 days, colonies were scored on the basis of their morphology, and individually picked for flow cytometric studies. Myeloid lineage colonies were also identified by May-Grünwald-Giemsa staining of cytospin preparations.
Fetal thymic organ cultures
Fetal thymic organ cultures (FTOCs) were performed as described.47 Briefly, 15-dpc thymic lobes were obtained from C57BL/6-CD45.1 mouse embryos and cultured on polycarbonate filters, floating on IMDM containing 2-deoxyguanosin (1.35 mM; Sigma) for 6 days. After washing the lobes, they were separated into individual Terasaki plate wells (Sarstedt, Australia). Unfractionated 11-dpc liver cells, and sorted c-kit+AA4.1+CD19− and c-kit+AA4.1+CD19+ populations recovered from 11-dpc livers of C57BL/6 (CD45.2) mouse embryos were expanded on ST2 cells with IL-7, IL-11, and SCF (500 cells/well) for 3 days. Cells from each well were added to the thymic lobes in the Terasaki plates (250 initial cells/lobe) and were placed in hanging drop IMDM/IL-7 cultures during the first 48 hours of cultures maintained for 12 additional days. Cells were then pooled from several thymic lobes (n = 5-10 lobes for each cell population) and were stained for flow cytometry. The anti-CD45.2 mAb identified cells of donor origin (C57BL/6).
Results
Lymphoid-specific gene expression in 9- to 13-dpc mouse embryos
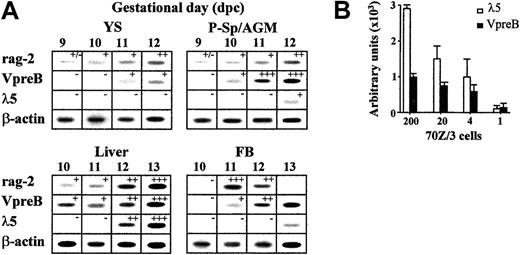
Lymphohematopoietic sites of the early BALB/c mouse embryo (YS, P-Sp/AGM, liver, blood) were microdissected, cells were mechanically dissociated, and mRNA levels of rag-2, VpreB,λ5, and β-actin genes were evaluated in semiquantitative RT-PCR assays. A summary of the results obtained is shown in Figure 1A. While all these transcripts were undetectable in 8-dpc mouse samples (n = 8 for P-Sp and n = 10 for YS; data not shown), recombinase-encoding rag-2 gene expression was frequently found in 9-dpc P-Sp and YS (3 of 8 and 4 of 9 independent samples, respectively). This result is consistent with previous findings about the simultaneous rag-1 gene transcription at these sites.14 VpreB transcripts were found one day later in P-Sp/AGM and liver, and their levels increased during the subsequent days of gestation. The other surrogate light chain (SLC) component, the λ5 gene, was not up-regulated until 12 dpc in the AGM and liver, 2 days later than the VpreB gene, suggesting the existence of VpreB+λ5− B-cell progenitors at the earliest ontogenic stages. The asynchronous expressions of the SLC-encoding VpreB and λ5 genes were not due to different sensitivity thresholds of the PCR assays, because these revealed positive VpreB/λ5 transcripts up to the single-cell level in titrated dilutions of the SLC+ 70Z/3 pre-B-cell line. These signals were in fact stronger for λ5 gene transcripts (Figure 1B). All of these genes were markedly up-regulated in later liver samples (rag-2, VpreB, and λ5 transcript levels increased 5-, 18-, and 10-fold, respectively, from 12 to 15 dpc; data not shown), while they progressively reduced in AGM and YS, probably in relation to the exhaustion of the local lymphohematopoietic sustaining potential.
Expression of lymphoid lineage-specific genes in midgestation (9-13 dpc) mouse embryos.
(A) Gene transcript levels of rag-2, VpreB, λ5, and β-actin in sequential samples of embryo lymphohematopoietic progenitor–containing areas (YS, P-Sp/AGM, liver, and blood), as detected by semiquantitative RT-PCR. After hybridization and densitometric quantification, the means of the relative message levels obtained for rag-2, VpreB, and λ5 were referred to those calculated in simultaneous 13-dpc liver PCR amplifications (as a maximum; n = 6-10 independent samples per organ per gestational day). These results are expressed by the +/− symbols in the figure, which represent −, < 1%; +/−, 1%-10%; +, 10%-40%; ++, 41%-70%; +++, 71%-100%. (B) Sensitivity threshold of λ5- and VpreB-specific RT-PCRs calculated in a titration of the 70Z/3 pre–B-cell line up to the single-cell level. Densitometric quantification of the specific signals obtained in 3 independent experiments; means ± SD are shown.
Expression of lymphoid lineage-specific genes in midgestation (9-13 dpc) mouse embryos.
(A) Gene transcript levels of rag-2, VpreB, λ5, and β-actin in sequential samples of embryo lymphohematopoietic progenitor–containing areas (YS, P-Sp/AGM, liver, and blood), as detected by semiquantitative RT-PCR. After hybridization and densitometric quantification, the means of the relative message levels obtained for rag-2, VpreB, and λ5 were referred to those calculated in simultaneous 13-dpc liver PCR amplifications (as a maximum; n = 6-10 independent samples per organ per gestational day). These results are expressed by the +/− symbols in the figure, which represent −, < 1%; +/−, 1%-10%; +, 10%-40%; ++, 41%-70%; +++, 71%-100%. (B) Sensitivity threshold of λ5- and VpreB-specific RT-PCRs calculated in a titration of the 70Z/3 pre–B-cell line up to the single-cell level. Densitometric quantification of the specific signals obtained in 3 independent experiments; means ± SD are shown.
A population of c-kit+AA4.1+CD19+ cells is present in 11- to 13-dpc mouse embryos
To establish whether the earliest B-lymphoid–specific gene expressions were due to low-level gene transcriptions in multipotent HSCs or if they corresponded to bona fide B-lineage–restricted precursors, we attempted to identify directly the putative B-lineage cells with specific cell surface markers and flow cytometry. We examined the appearance of the B-lineage–specific CD19 Ag, as well as the expression of B220, IL-7Rα, AA4.1, CD43, and other surface receptors in mouse embryos. As shown in Figure2A, a small population of c-kit+CD19+ cells was clearly observed in both AGM and liver cell suspensions of mouse embryos at 11 dpc. Tiny CD19+ cell subsets were also detected in isolated samples of blood (Figure 2A) and YS (data not shown). The fact that CD19+ cells represent a higher fraction of c-kit+ cells in AGM than in liver (5.9% and 1.2%, respectively) is due to the presence of other nonlymphohematopoietic c-kit+ progenitors in the latter organ, since CD19+ cells constitute 10% to 11% of AA4.1+hematopoietic precursors in both locations. When the percentages of these CD19+ cells were referred to the absolute cell recoveries obtained in the embryo lymphohematopoietic organs, we observed that 11-dpc AGM and liver harbored 97.8 ± 58 and 480 ± 100 CD19+ cells per organ, respectively (Figure2B; n = 6 for AGM, and 17 for liver; mean ± SD). The number of AA4.1+ cells increased exponentially in the liver between 11 and 13 dpc, but not in the simultaneous AGM, where they declined after 12 dpc. CD19+ cells expanded in the liver during the same period and, to a lesser extent, in the AGM. CD19+cells were also detected in 11-dpc livers of C57BL/6 embryos (275 ± 45 CD19+ cells per liver; n = 4). The CD19 immunofluorescent findings were confirmed and extended by CD19-specific RT-PCR assays (Figure 2C). At 11 dpc, high levels of CD19 gene transcripts were always found in both AGM and liver, and weaker signals were also present in YS and blood. In contrast, CD19 transcripts were absent from all 8-dpc samples, and they began to appear in 9-dpc (2 of 4 P-Sp and 1 of 5 YS) and 10-dpc (3 of 4 P-Sp and 3 of 5 YS) mouse embryos (data not shown).
CD19+ cells in the midgestation mouse embryo.
(A) Dot plots obtained after 2-color FACS analyses with biotinylated anti-B220 and anti-CD19 mAbs versus FITC anti–c-kit mAb in 11- to 12-dpc AGM-, liver-, and blood-derived cell suspensions. After Fc blocking and PI exclusion of dead cells, specific mAb signals were defined against those of isotype-matched, irrelevant mAbs. The windows shown include cells positive for established markers, and the numbers above the graphs are means ± SD of 5 independent FACS analyses/sample. (B) Absolute cell recoveries in 11- to 13-dpc AGM (▵) and FL (●). Total cells, AA4.1+, and CD19+ cells/organ are shown, the latter 2 obtained from their relative numbers in FACS analyses and absolute cell numbers/organ. n = 5-7 independent analyses; means ± SD are shown. (C) CD19 gene transcripts detected in the displayed lymphohematopoietic sites at 8 and 11 dpc. The data are presented as in Figure 1A. ST2 stromal cells were used as negative control. n = 4 independent samples. (D) Surface phenotype of 11-dpc CD19+liver cells. The 2-color FACS analyses were done with biotinylated anti-CD19 mAb versus the FITC mAbs specific for the shown receptors. White and shaded histograms represent negative and specific signals, respectively, obtained from gated CD19+cells.
CD19+ cells in the midgestation mouse embryo.
(A) Dot plots obtained after 2-color FACS analyses with biotinylated anti-B220 and anti-CD19 mAbs versus FITC anti–c-kit mAb in 11- to 12-dpc AGM-, liver-, and blood-derived cell suspensions. After Fc blocking and PI exclusion of dead cells, specific mAb signals were defined against those of isotype-matched, irrelevant mAbs. The windows shown include cells positive for established markers, and the numbers above the graphs are means ± SD of 5 independent FACS analyses/sample. (B) Absolute cell recoveries in 11- to 13-dpc AGM (▵) and FL (●). Total cells, AA4.1+, and CD19+ cells/organ are shown, the latter 2 obtained from their relative numbers in FACS analyses and absolute cell numbers/organ. n = 5-7 independent analyses; means ± SD are shown. (C) CD19 gene transcripts detected in the displayed lymphohematopoietic sites at 8 and 11 dpc. The data are presented as in Figure 1A. ST2 stromal cells were used as negative control. n = 4 independent samples. (D) Surface phenotype of 11-dpc CD19+liver cells. The 2-color FACS analyses were done with biotinylated anti-CD19 mAb versus the FITC mAbs specific for the shown receptors. White and shaded histograms represent negative and specific signals, respectively, obtained from gated CD19+cells.
A more detailed characterization of 11-dpc liver c-kit+CD19+ cells was performed (Figure 2D). The majority of the cell population was positive for the AA4.1 and CD34 Ags, markers representative of fetal HSCs33 and very early progenitors.48 Levels of Sca-1 Ag, which is up-regulated at this point in mouse ontogeny,14 were not expressed by the c-kit+CD19+ cell population. The IL-7Rα chain, which traces the earliest stages of lymphoid restriction,35 is present in up to 70% of the CD19+ cells. The CD45 common leukocyte Ag was also positive in the c-kit+CD19+ cell population. Unexpectedly, these CD19+ cells were negative for the B220 Ag (CD45R isoform). Other progenitor cell markers such as CD24, CD43, and CD16/CD32 were present in subsets of the c-kit+CD19+ cell population. Cell surface receptors typical of non–B-hematopoietic lineages, such as CD11b, CD4, Gr-1, and TER-119, as well as Ags of more mature B-cell stages (CD23, surface IgM) were absent from the embryonic c-kit+CD19+ cell population (data not shown). These findings suggest that this novel population of midgestation c-kit+AA4.1+CD19+B220−cells may represent intermediate and/or very early pro-B progenitors.
Selective expression of lymphoid-specific genes in purified CD19+ cells from 11-dpc embryonic liver
We next investigated whether the earliest detected lymphoid gene expressions in the embryo (see the first paragraph of “Results”) were restricted to the newly revealed population of CD19+cells and/or to other embryonic hematopoietic cells. The following cell subsets were purified by electronic sorting from 11- and 13-dpc livers after 3-color stainings (c-kit, AA4.1, CD19) (Figure3A): (R1) c-kit+AA4.1− cells, which excluded any CD19+ cell, and represented 54 ± 6% of total liver cells at 11 dpc (mean ± SD; n = 5), (R2) c-kit+AA4.1+CD19− cells (6 ± 1%; n = 5), and (R3) c-kit+AA4.1+CD19+ cells (0.67 ± 0.3%; n = 10), and their mRNAs were studied in RT-PCR assays. The remaining c-kit−AA4.1−cells were not analyzed because we knew from previous studies that this population was constituted by TER-119+ erythroid cells and hepatocyte progenitors, and that it lacked any other lymphohematopoietic progenitor potentials (S.M., M.L.G., M.A.R.M., unpublished data, June 2002). Pre-B–specific rag-2 and VpreB transcripts were selectively detected in the 11-dpc liver CD19+ (R3) cell population, while the λ5 gene expression was delayed up to 13 dpc in the same population (Figure 3B, top diagram). We also tested for the expression of transcription factors involved in B-lineage commitment, such as Ikaros (expressed by both T- and B-cell progenitors),49 EBF (upstream regulatory gene ofPax5), and Pax5 (essential for B-cell commitment) in 11-dpc liver c-kit+CD19− cells (R1 + R2) and in CD19+ cells (R3) (Figure 3B, bottom diagram). The 3 genes(Ikaros, EBF, and Pax5) were highly expressed in CD19+ (R3) liver cells. Gene transcripts for Ikaros and EBF (but not Pax5) were also present among c-kit+CD19− cells. We conclude that the expression of pro/pre-B-specific genes observed in midgestation mouse embryos (VpreB, λ5, rag-2, Pax5) is restricted to the population of c-kit+AA4.1+CD19+ cells. Finally, VDJCμ transcripts were negative at 11 dpc, both in the sorted populations and in unfractionated liver samples (Figure 3B), as well as cytoplasmic μHC (Figure 3C).
Lymphoid-specific gene expressions in purified 11- and 13-dpc liver cell populations from normal and Pax5-deficient mice.
(A) The 3-color mAb stainings were done with FITC anti–c-kit, biotinylated anti-AA4.1 and Cy5 anti-CD19 mAbs. The selected cell populations shown in the figure were electronically gated and positively sorted. There were 2 sorting windows set up for c-kit+AA4.1− (R1) and c-kit+AA4.1+ cell populations, as shown in the representative dot plot of the figure. These cell populations were separated on the basis of CD19 Ag signals in c-kit+AA4.1+CD19− (R2) and c-kit+AA4.1+CD19+ (R3) cells (right histograms). The R1 cell population lacked CD19+ cells. (B) Gene expression obtained in purified 11-dpc (R1, R2, R3, R1+R2) and 13-dpc (R3) cell preparations, unseparated 11-, 13-, and 19-dpc liver cells, and adult thymocytes (Thy). The data are representative of 3 independent analyses/sample, and are displayed as in Figure 1. First and second round VDJCμ (VH7183 and VHJ558) PCR reactions (rows 1 and 2, respectively) and β-actin are shown at the right as ethidium bromide gels. (C) Cytoplasmic μHC staining (cμHC) of 11-dpc liver and BM cells. CD19+ surface IgM− (sIgM−) BM cells and CD19+11-dpc liver cells were electronically gated (left histograms) and analyzed for cμHC (right histograms). White and shaded right histograms represent negative and specific signals, respectively. (D) Detection of rag-2, VpreB, and CD19 gene transcripts in representative samples of isolated 11-dpc liver cells from wild-type (+/+), heterozygous (+/−), and homozygous Pax5-deficient (−/−) mouse embryos. The Pax5 genetic background of each embryo was identified with a genomic PCR. Bottom and top bands correspond to the wild-type and mutant Pax5 genes, respectively.41 α-Actin was used as a positive control of genomic DNA content. Rag-2, VpreB, and CD19 gene expressions were obtained with a nested RT-PCR, as described in “Materials and methods.”
Lymphoid-specific gene expressions in purified 11- and 13-dpc liver cell populations from normal and Pax5-deficient mice.
(A) The 3-color mAb stainings were done with FITC anti–c-kit, biotinylated anti-AA4.1 and Cy5 anti-CD19 mAbs. The selected cell populations shown in the figure were electronically gated and positively sorted. There were 2 sorting windows set up for c-kit+AA4.1− (R1) and c-kit+AA4.1+ cell populations, as shown in the representative dot plot of the figure. These cell populations were separated on the basis of CD19 Ag signals in c-kit+AA4.1+CD19− (R2) and c-kit+AA4.1+CD19+ (R3) cells (right histograms). The R1 cell population lacked CD19+ cells. (B) Gene expression obtained in purified 11-dpc (R1, R2, R3, R1+R2) and 13-dpc (R3) cell preparations, unseparated 11-, 13-, and 19-dpc liver cells, and adult thymocytes (Thy). The data are representative of 3 independent analyses/sample, and are displayed as in Figure 1. First and second round VDJCμ (VH7183 and VHJ558) PCR reactions (rows 1 and 2, respectively) and β-actin are shown at the right as ethidium bromide gels. (C) Cytoplasmic μHC staining (cμHC) of 11-dpc liver and BM cells. CD19+ surface IgM− (sIgM−) BM cells and CD19+11-dpc liver cells were electronically gated (left histograms) and analyzed for cμHC (right histograms). White and shaded right histograms represent negative and specific signals, respectively. (D) Detection of rag-2, VpreB, and CD19 gene transcripts in representative samples of isolated 11-dpc liver cells from wild-type (+/+), heterozygous (+/−), and homozygous Pax5-deficient (−/−) mouse embryos. The Pax5 genetic background of each embryo was identified with a genomic PCR. Bottom and top bands correspond to the wild-type and mutant Pax5 genes, respectively.41 α-Actin was used as a positive control of genomic DNA content. Rag-2, VpreB, and CD19 gene expressions were obtained with a nested RT-PCR, as described in “Materials and methods.”
We also analyzed CD19, rag-2, and VpreB mRNA expression in 11-dpc livers from Pax5-deficient mice, which are devoid of B-lineage cells, as an additional proof of the B-cell–restricted character of the embryonic c-kit+AA4.1+CD19+ cells (Figure 3D). Isolated livers from Pax5−/− embryos were completely negative for VpreB and CD19 transcripts (n = 7 individual livers), while rag-2 was infrequently detected (1 of 5 samples). Pax5+/− and Pax5+/+ mice expressed all these transcripts at similar frequencies (n = 20 and n = 22, respectively). The Pax5 dependence of midgestation embryo c-kit+AA4.1+CD19+ cells reproduces the findings observed in late fetal liver and adult BM,39 41 and confirms the B-lineage character of these cell progenitors.
11-dpc c-kit+AA4.1+CD19+ liver cells are restricted to B-lineage differentiation
We analyzed the lymphohematopoietic potentials of c-kit+AA4.1+CD19+ (R3) cells in comparison to those of simultaneous c-kit+AA4.1+CD19− (R2) cells from 11-dpc livers. Both cell populations were established in low-cell density, 2-step stromal cell cultures, and in the Methocult system, as described in “Materials and methods.” Representative findings of these experiments are shown in Figure 4. Sorted 11-dpc c-kit+AA4.1+CD19−(R2) liver cells differentiated to populations of (1) small, round CD19+CD90−CD11b−TER-119−cells (n = 200 wells; 27% positives), and (2) big, granular, macrophagelike CD19−CD11b+ cells (n = 200 wells; 43% positives) in both B-lymphoid and myeloid conditions, respectively. The selective addition of GM-CSF to the myeloid cultures increased their proportion of Gr-1+ granulocytes with respect to CD11b+ macrophages (data not shown). R2 cells also gave rise to small, anucleated, CD19−TER-119+ erythrocytes in EPO-methylcellulose cultures. They also generated several types of colonies when seeded in the Methocult system (500-1000 erythroid, 50 to 400 myeloid/granulocyte, and 50 to 200 erythro/myeloid colonies per 105 cells; n = 4). In contrast, although well expanded in the first culture step (ST2 + IL-7, SCF, IL-11), the c-kit+AA4.1+CD19+ (R3) starting cultures rapidly died when secondarily transferred to conditions in the absence of IL-7. Purified 11-dpc liver c-kit+AA4.1+CD19+ (R3) cells only expanded successfully in defined B-cell conditions (ST2 + IL-7) that allowed the establishment of 45% of the initially obtained cultures (n = 150 wells). The frequencies of B-cell precursors obtained in limiting dilution experiments performed as described42 were 1/500 and 1/160 for the R2 and the R3 cell populations, respectively. The B220 Ag (which was absent in vivo) was partially up-regulated in B-cell cultures of multipotent R2 cells, and became completely positive in those derived from R3 cells. B-lineage cell lines that acquired the stage of LPS responsiveness and matured to surface IgM+ cells were derived from both R2 and R3 cell populations (data not shown). We conclude that, in contrast to multipotent embryonic c-kit+AA4.1+CD19− (R2) cells, c-kit+AA4.1+CD19+ (R3) cells represent progenitors functionally committed to the B-lymphoid lineage and are devoid of any other myeloerythroid potentials.
In vitro differentiation potentials of c-kit+AA4.1+CD19− (R2) and c-kit+AA4.1+CD19+ (R3) cells purified from 11-dpc livers.
Purified R2 and R3 cells were first expanded in vitro (ST2 + IL-7, IL-11, SCF) and then transferred to cultures under B-lymphoid (ST2 + IL-7), myeloid (SCF ± GM-CSF), and erythroid conditions (methlycellulose plus EPO, SCF). Positive wells were counted, photomicrographed, and analyzed by FACS at the end of the culture periods. Left column, light field photomicrographs of the cultures under B-lymphoid (× 20) and erythroid (× 4) conditions, and May-Grünwald-Giemsa photomicrographs from cytospin preparations of the myeloid cultures (× 20). The 2-color FACS analyses were performed as in Figure 2A, by using FITC anti-Thy1, anti-B220 and anti-CD11b, biotinylated anti-TER119, and Cy5 anti-CD19 mAbs.
In vitro differentiation potentials of c-kit+AA4.1+CD19− (R2) and c-kit+AA4.1+CD19+ (R3) cells purified from 11-dpc livers.
Purified R2 and R3 cells were first expanded in vitro (ST2 + IL-7, IL-11, SCF) and then transferred to cultures under B-lymphoid (ST2 + IL-7), myeloid (SCF ± GM-CSF), and erythroid conditions (methlycellulose plus EPO, SCF). Positive wells were counted, photomicrographed, and analyzed by FACS at the end of the culture periods. Left column, light field photomicrographs of the cultures under B-lymphoid (× 20) and erythroid (× 4) conditions, and May-Grünwald-Giemsa photomicrographs from cytospin preparations of the myeloid cultures (× 20). The 2-color FACS analyses were performed as in Figure 2A, by using FITC anti-Thy1, anti-B220 and anti-CD11b, biotinylated anti-TER119, and Cy5 anti-CD19 mAbs.
Embryonic c-kit+AA4.1+CD19+liver cells lack T-cell potential
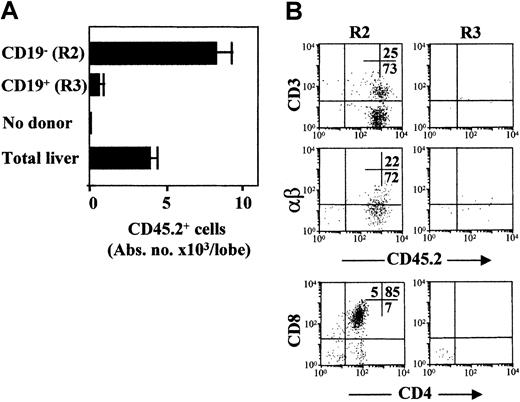
We also examined whether the embryonic CD19+ cell progenitors showed any T-cell differentiation potential by seeding them in FTOC. Purified 11-dpc liver R2 and R3 cells obtained from C57BL/6 (CD45.2+) mouse embryos were expanded for 3 days in vitro (ST2 + IL-7, IL-11, SCF) and then were seeded on 15-dpc 2-deoxyguanosin–treated thymic epithelia from C57BL/6-CD45.1+ mouse embryos. After 2 weeks, the FTOCs were dispersed and donor-derived CD45.2+ cells were analyzed by flow cytometry. Sorted R2 cells expanded in vitro and efficiently matured throughout the stages of CD4+CD8+, CD4+, and CD8+ thymocytes, up to CD3+αβ+cells. However, only occasional donor-derived CD45.2+ cells were seen in FTOC established with R3 cells (Figure5). FTOC initiated with total cells from 11-dpc liver embryos were also reconstituted (Figure 5). We conclude that the embryonic c-kit+AA4.1+CD19+ (R3) cell progenitors are devoid of T-cell precursor potential, and thus are selectively restricted to differentiation along the B-cell lineage.
In vitro T-cell potential of 11-dpc liver cell populations established in FTOC.
(A) Purified R2 and R3 cells and total liver cells were first expanded in vitro (ST2 + IL-7, IL-11, SCF) and then seeded on thymic lobes (250 initial cells/lobe). Absolute cell numbers of CD45.2+donor-derived cells/lobe are shown. Data are representative of 3 independent cultures and are expressed as means ± SD of 4-8 lobes/bar. (B) Shown are 2-color FACS dot plots of thymocyte populations recovered at the end of FTOC assays seeded with purified R2 and R3 cell populations. FITC anti-CD3ε and anti–αβ-TCR, PE anti-CD8, Cy5 anti-CD4, and biotinylated anti-CD45.2 mAbs were used. Stainings and FACS analyses were as described in Figure 2A. The numbers inside the plots show the cell percentages in the corresponding quadrants.
In vitro T-cell potential of 11-dpc liver cell populations established in FTOC.
(A) Purified R2 and R3 cells and total liver cells were first expanded in vitro (ST2 + IL-7, IL-11, SCF) and then seeded on thymic lobes (250 initial cells/lobe). Absolute cell numbers of CD45.2+donor-derived cells/lobe are shown. Data are representative of 3 independent cultures and are expressed as means ± SD of 4-8 lobes/bar. (B) Shown are 2-color FACS dot plots of thymocyte populations recovered at the end of FTOC assays seeded with purified R2 and R3 cell populations. FITC anti-CD3ε and anti–αβ-TCR, PE anti-CD8, Cy5 anti-CD4, and biotinylated anti-CD45.2 mAbs were used. Stainings and FACS analyses were as described in Figure 2A. The numbers inside the plots show the cell percentages in the corresponding quadrants.
Discussion
Primitive myeloerythropoiesis and definitive lymphohematopoiesis emerge early in the postgastrulation mouse embryo (7-8 dpc) in mesoderm-derived sites such as the YS and the P-Sp/AGM.1-3,5,6,50 From these primary sites, multipotent progenitors home to secondary hematopoietic organs (liver, spleen, BM, thymus).10 While the first definitive HSCs are found in the 8- to 9-dpc P-Sp,3,6 and erythromyeloid programs of differentiation are rapidly established in the YS,16 it was thought that restricted B-lymphopoiesis did not appear until 13 to 14 dpc, and that it occurred exclusively in the liver.12,15 These (and additional data related to other hematopoietic differentiation pathways) led to the proposal of P-Sp/AGM as being a pure organ of hematopoiesis where multipotent progenitors self-renewed and expanded, while the B-lineage differentiation program was initiated later in fetal liver.15,30 The findings of low-level lymphoid lineage–specific gene transcripts and of DJH rearrangements in early extra-liver sites14,26,27 were considered to be the result of partially accessible gene loci in “primed,” but not yet committed, multipotent progenitors.30 The above scenario leaves an extended developmental window (9 to 13 dpc) where HSCs would only proliferate but not up-regulate differentiation programs, at least for the B-lymphoid lineage.
The present work has isolated and characterized a novel population of c-kit+AA4.1+CD19+ cells emerging at 10 to 11 dpc in the AGM and liver, and to a lesser extent, in the YS. They represent a few hundred cells per organ between 11 and 13 dpc, whose numbers increase during this period, before the later synchronous and exponential expansion of B-cell progenitors that takes place in the liver after 13 dpc.23,24 It is of particular note that the B-lineage–specific gene transcripts previously detected in hematopoietic sites of postgastrulation mouse embryos (rag-1 and -2, VpreB, λ5) cosegregated with the purified c-kit+AA4.1+CD19+ cell population. In vitro differentiation cultures in B-lymphoid, myeloid, erythroid, and T-cell conditions demonstrated that, while purified c-kit+AA4.1+CD19− liver cells matured throughout all these hematopoietic pathways, simultaneous c-kit+AA4.1+CD19+ cells only differentiated successfully to B-lineage cells. Pax5, crucial for B-lineage commitment,39,41 was consistently expressed in 11-dpc liver c-kit+AA4.1+CD19+cells, as well as in 11-dpc AGM (data not shown), while it was detected in some samples (1 of 5) from multipotent c-kit+AA4.1+CD19− cells. This observation is consistent with the simultaneous up-regulation of Pax5 and CD19, and loss of pro–T-cell potentials in AA4.1+ (in contrast to AA4.1−) cells observed in the earliest ontogenical stages of BM B-cell differentiation.22 34Also, the analysis of Pax5-deficient 11-dpc mouse embryos showed that, while Pax5+/+ and Pax5+/− embryos expressed CD19 transcripts, Pax5−/− embryos lacked VpreB and CD19 B-lineage–specific transcript expression. Taken together, these findings lead us to propose that mouse embryo B lymphopoiesis arises at 10 to 11 dpc, immediately after the starting events of definitive lymphohematopoiesis that happen in the P-Sp/AGM region. Although the embryonic liver microenvironment greatly promotes B-cell growth and differentiation programs (specifically after 13 dpc), these processes are not at all limited to this hematopoietic organ in midgestation mouse embryos.
We consider that the studied population of committed B-lineage c-kit+AA4.1+CD19+ progenitors has probably escaped detection before now, due to (1) the unusual features of these B-lineage cells (Figure 2D), (2) the heterogeneous experimental approaches that employed either total or variously purified cell populations in which the AA4.1+c-kit+CD19+ cells might represent a minor subset, and (3) the indirect conclusions from in vitro culture assays of variable efficiency. Although we know that midgestation B-cell progenitors are highly sensitive to IL-7 signals and that they can be very efficiently established in vitro on ST2 + IL-7 cell cultures,42 it may be that only a subset of the embryonic CD19+IL-7Rα+ cells isolated ex vivo can develop under stromal cells plus IL-7 culture conditions or in the multilineage progenitor assay.21 In fact, the cloning efficiency of the positively sorted c-kit+AA4.1+CD19+ cells was 1/160 in the B-lymphoid conditions used in this work, a finding that leads to an estimate of absolute numbers of B-lymphoid progenitors per 11-dpc liver that is not very different from those suggested by others in in vitro protocols.21 Also, the limited number of clones (13 analyzed) from in vitro–established 11-dpc AA4.1+ cells in the report of Delassus and Cumano12 made the detection of any B-committed cell unlikely, when considering the small percentages of CD19+ cells within the AA4.1+ population. Furthermore, our findings extend those reported by Andersson's group, which observed 1 × 103 CD19+ cells in 13-dpc liver.24 The c-kit+AA4.1+CD19+ progenitors characterized here probably account for a fraction of the earliest-detected DJH rearrangements14,26 27 and Ig germ line transcripts (M.L.G., M.A.R.M., unpublished work, 2000). However, neither VDJCμ transcripts nor cytoplasmic μHC were observed in c-kit+AA4.1+CD19+cells, findings that support their pro-B-cell differentiation stage.
The 11-dpc mouse embryo c-kit+AA4.1+CD19+B220−cells continue to be detected until birth in liver, as also happens in adult BM.51 In fact, the first CD19+B220+ cells were not detected until 14 dpc in liver (Egawa et al22; P.G., unpublished data, June 2002), which explains why several groups did not detect this cell population.12,52 However, Velardi and Cooper53 already described the presence of 300 cytoplasmic μ+B220− cells per liver at 13 dpc. Also, the absence of λ5 transcripts in the 11-dpc AGM was considered to be proof of inexistent in situ B lymphopoiesis.30 The current study confirms and extends the findings of previous work,4,14 showing that the transcription of the SLC-encoding λ5 gene (but not that ofVpreB) is delayed until 12 dpc, at which time it is restricted to the c-kit+AA4.1+CD19+cells. The functional significance of the VpreB+λ5− expression pattern in the earliest B-lineage progenitors is unclear, and deserves analysis at the protein level with the recently developed mAbs.54 These latter results recall the λ5-independent pathway of B-cell differentiation described in adult BM.55 If λ5 plays a role in the SLC-dependent proliferation of B-cell precursors, its absence in embryonic CD19+ progenitors could contribute to explain the limited expansion rates of these cells in the liver before 13 dpc. The lack of λ5 and B220 expression in the c-kit+AA4.1+CD19+ cells is probably related to (absent?) microenvironment-induced signals, as both genes were rapidly up-regulated once the ex vivo cells had been established in vitro (Figure 4).42
Although lacking the CD45R/B220 molecule, embryonic CD19+cells were positive for the CD45 common leukocyte antigen. Other phenotypical traits of the CD19+ cells also indicate their immature stage (AA4.1, IL-7Rα, CD43, HSA/CD24, FcγR/CD16/CD32), activation status (CD34),56 and exclusive B-lineage character (Sca-1−CD90−CD11b−Gr-1−TER-119−). The B-lineage restriction of the cell population is confirmed by the lack of other hematopoietic differentiation potentials (myeloid, erythroid, and T-lymphoid), as well as of natural killer and dendritic cell differentiations revealed in recent experiments (data not shown). The embryonic CD19+ cells differed phenotypically from various recently described oligolineage progenitors. In contrast to the CLP,20,57 the embryonic B-committed cells described here lacked T-cell potential and were c-kit+ and CD19+. The divergent ontogenical dynamics of T- and B-cell generation (greater numbers of T progenitors than of B-cell progenitors at 12 dpc versus predominant B progenitors at 14-15 dpc)21also suggested that the embryo equivalent of CLP might be an elusive event in early ontogeny. Bipotential B-macrophage progenitors have been described in 12-dpc fetal liver17 and, very recently, in adult BM.51 Unlike in the population detected here, the former cells expressed Sca-1 and the latter were c-kitlow. The bipotential BM CD19+ cells were also B220−, as is the case for the embryonic CD19+cell population described here. It is worth noting that the BM CD19+B220− cells also contained unilineage B-committed cells.51 Finally, the c-kit+AA4.1+CD19+ cell progenitors differ from the Lin−CD19−c-kit+IL-7Rα+AA4.1+B-cell precursors revealed in the 12.5-dpc liver in their CD19 expression22; in the same report, the first B220+CD19+ cells did not become apparent in the liver until 14.5 dpc.
In conclusion, the findings of this study support the idea that the developmental process of embryonic B lymphopoiesis begins at 10 to 11 dpc with a population of fully B-committed c-kit+AA4.1+CD19+B220−cell progenitors. This new cell population places B-lineage differentiation at least 3 days earlier than has been previously believed, near to the time in ontogeny when the first HSCs arise. Although it seems clear that late fetal liver (after 13 dpc) represents an inductive microenvironment for B-lineage expansion and differentiation, embryonic B lymphopoiesis also evolves in other lymphohematopoietic sites such as P-Sp/AGM and YS.
We are grateful to M. Busslinger and C. Martı́nez-Alonso for critical reading of the manuscript, to M. Busslinger for providing us the Pax5+/− mice, A. Cumano for the AA4.1 and Sca-1 mAbs, A. Rolink for the IL-7–transfected 3T3 cells, and G. Guenechea for his advice on Methocult system assays. We also thank P. Mason for editorial assistance, P. Ferrero for technical support, and C. Moreno for help with the sorting procedures.
Prepublished online as Blood First Edition Paper, July 18, 2002; DOI 10.1182/blood-2002-03-0809.
Supported by grants from the Comunidad Autónoma de Madrid (CAM) (08.3/0009/1997), the Ministerio de Ciencia y Tecnologı́a (MCyT) (PM 99-0104), and ISCIII (01/34). The CBMSO is partially funded by the Fundpción Ramón Areces. B.d.A., P.G., and S.M. are supported by MCyT-Ramón y Cajal Program, ISCIII, and MCyT, respectively.
B.d.A. and P.G. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
M. L. Gaspar, Centro Nacional de Microbiologı́a, Instituto de Salud Carlos III, Ctra Majadahonda-Pozuelo km 2, Majadahonda 28220, Spain; e-mail:mlgaspar@isciii.es.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal