B-cell chronic lymphocytic leukemia (B-CLL) is characterized by a resistance toward apoptosis-inducing agents. Nuclear factor-κB (NF-κB)/Rel has been shown to regulate the expression of antiapoptotic genes, such as members of the inhibitor of apoptosis protein (IAP) and tumor necrosis factor receptor-associated factor (TRAF) gene families. Expression and regulation of NF-κB/Rel–dependent inhibitors of apoptosis have not been collectively studied in B-CLL. We examined expression of known NF-κB/Rel–regulated antiapoptotic genes by RNAse protection assay, real-time polymerase chain reaction, and immunoblotting in patients with B-CLL. TRAF1 and to a lesser extent TRAF2 were overexpressed in B-CLL lymphocytes as compared with normal CD19+ B cells. TRAF1 overexpression did not correlate with markers of disease progression or overall survival. Furthermore, we found high constitutive expression of the IAP genes c-IAP-1, c-IAP-2, and XIAP both in normal and B-CLL lymphocytes. Focusing on the regulation of TRAF1, NF-κB/Rel activity in B-CLL nuclear extracts was shown to bind to TRAF1 promoter elements. However, IκB kinase (IKK) activity was not increased in CLL lymphocytes as compared with normal CD19+ B cells. The known IKK inhibitor sulfasalazine did not compromise TRAF1 expression. Thus, although our study revealed a common expression pattern of NF-κB/Rel–regulated inhibitors of apoptosis, our findings indicate an IKK-independent regulation of TRAF1 in B-CLL.
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) is the most common leukemia of the Western hemisphere.1 Although a rare disease in adults younger than 30 years, it reaches an incidence of nearly 50 cases per 100,000 per year in people beyond 70 years of age.2 In spite of the introduction of new drugs such as fludarabine, no progress has been made in prolonging survival of patients. Curative therapy is still limited to a small group of patients eligible for allogeneic stem cell transplantation. The hallmark of B-CLL is a defect in the induction of programmed cell death (PCD).2 Several mechanisms have been implicated in the suppression of apoptosis. High expression levels of the bcl-2 family proteins bcl-2,3,4 bax, and mcl-15 were described. The Fas pathway, which has been shown to be critical in drug-induced cell death,6 was reported to be deficient in B-CLL.7-9 However, the mechanisms underlying the inhibition of apoptosis in B-CLL remain unresolved.
Nuclear factor-κB (NF-κB)/Rel proteins are key regulators of differentiation and cell-specific gene expression in B cells.10 The multisubunit IκB kinase (IKK) is responsible for inducible IκB phosphorylation and is the point of convergence for most NF-κB/Rel activators.11 Induction of NF-κB/Rel activity was demonstrated to block apoptosis induced by tumor necrosis factor alpha (TNFα), cytotoxic drugs, and radiation in different cell types including B cells in vitro and in vivo.12-16 Several genes have been reported to be candidates for conveying NF-κB/Rel–dependent inhibition of apoptosis, such as TRAF1, TRAF2, c-IAP-1, c-IAP-2,17XIAP,18bcl-xL,19bfl-1/A-1,20,21manganese superoxide dismutase (MnSOD),22 andA20.23 24 Expression and regulation of these genes in B-CLL are largely unknown and were the focus of our study.
The tumor necrosis factor receptor-associated factor (TRAF) family of proteins (TRAF1-6) is a group of adaptor molecules involved in the intracellular signal transduction of several members of the tumor necrosis factor receptor (TNFR) family, including TNFR2, CD30, CD40, the Epstein-Barr virus (EBV) encoded latent membrane protein 1 (LMP1) and the lymphotoxin-β receptor (LT-βR).25Several observations prompted us to examine the expression and regulation of TRAF proteins with an emphasis on TRAF1 in B-CLL. Whereas TRAF2 is ubiquituously expressed,25 TRAF1 shows a tissue-specific expression with high levels in lymphoid tissue.26 TRAF1 and TRAF2 are candidates for mediating NF-κB/Rel–mediated inhibition of apoptosis.17 The protective effect of TRAF1 on apoptosis induced by different stimuli has been shown.27,28 Because constitutive NF-κB/Rel activity is observed in B-CLL,29 30 we tested the hypothesis that TRAF1 is up-regulated in B-CLL and that this effect is mediated by IKK-dependent mechanisms.
Patients, materials, and methods
Patients
Peripheral blood samples were obtained from 40 patients with B-CLL treated at our institution from March 1994 to May 1996. All patients met the modified diagnostic National Cancer Institute (NCI) criteria for the diagnosis of B-CLL.31 Cases of mantle cell lymphoma, B-prolymphocytic leukemia (PLL), or chronic lymphocytic leukemia of the T-cell type (T-CLL) were not included. None of the patients had received cytoreductive therapy within 8 weeks prior to blood sampling. The blood samples were subjected to density gradient centrifugation to remove contaminating neutrophils. The share of leukemic B-cells exceeded 90% in all preparations tested. Cells were either directly RNA extracted or cryopreserved in liquid nitrogen for RNA extraction at a later time. Protein extracts for immunoblotting were from frozen samples of a different set of patients (n = 23). Nuclear extracts for electrophoretic mobility shift assays (EMSA) and cell lysates for kinase assays were prepared from freshly obtained patient samples.
B lymphocytes from healthy blood donors
Buffy coats from healthy blood donors (n = 9) were obtained from the local blood bank. After density gradient centrifugation the remaining mononuclear cells were subjected to CD19+selection with a CD19 multisort kit (Miltenyi Biotech, Bergisch-Gladbach, Germany). Preparations were checked for purity by CD19 fluorescence activated cell sorting (FACS) analysis, yielding results of more than or equal to 95% B cells. RNA and protein extractions were done immediately after positive selection.
RNAse protection assays
TRAF1, TRAF2, protein containing C-rich domain associated with RING and TRAF domains (CART), I-TRAF, TRAF5, TRAF6, CD40 receptor–associated factor (CRAF), and TRAF-interacting protein (TRIP) mRNA levels were analyzed with a hAPO-5b multiprobe Riboquant system (Pharmingen, Hamburg, Germany) according to the manufacturer's recommendations. XIAP, survivin, NAIP, c-IAP-1, c-IAP-2, and TRPM2 were examined with a hAPO-5c multiprobe Riboquant system (Pharmingen). A TNFα probe had been added to the hAPO-5c template set by the manufacturer. L32 and glyceraldehyde phosphate dehydrogenase (GAPDH) probes were included as internal controls. Samples were analyzed by electrophoresis on denaturing polyacrylamide gels (6%). Gels were vacuum dried and exposed to Kodak BioMax MR film at −70°C with intensifying screens. Message intensities were quantified by densitometry and a message/L32 message ratio was determined for all samples. Quartiles were calculated for TRAF1/L32 message ratios and used for semiquantitative TRAF1 analysis. The cell lines MHH-cALL2, MHH-cALL3, MHH-cALL4, EHEB, DoHH2, 293, Molt4, NIH929, and RPMI8226 were purchased from DSMZ (Braunschweig, Germany).
Taqman PCR analysis
mRNA expression was evaluated using quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) analysis (TaqMan; PE Applied Biosystems, Norwalk, CT). Reverse transcription of 2 μg total RNA (20 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2, 10 mM dithiothreitol [DTT], 0.5 μM random hexamer primer, 0.5 mM of each dNTP, and 200 U Superscript II RT; Gibco BRL, Rockville, MD) was carried out in duplicate and further processed independently. To exclude DNA contamination in RNA samples, reactions were also carried out without reverse transcriptase. PCRs were performed in duplicate using the primer combinations listed below (Sybr Green PCR Core Reagents; PE Applied Biosystems). Target cDNAs were normalized to the endogenous mRNA levels of the housekeeping gene cyclophyllin for each reaction (ΔCT method). For convenience of presentation the resulting ratios are presented as 10/ΔCT. Thus, high ratios correspond to high transcript levels in relation to amplified cyclophyllin controls. The thermal cycling conditions were 95°C for 2 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute in a thermocycler designed to detect fluorescence changes in real time (ABI PRISM 7700; PE Applied Biosystems). Specific amplification products were confirmed by electrophoresis on an agarose gel resulting in bands with the predicted size. The following primers were used: TRAF1: 5′ GCCCTTCCGGAACAAGGTC 3′; 5′ CGTCAATGGCGTGCTCAC 3′; TRAF2: 5′ GTGGCCACCGGTACTGCTC 3′; 5 ‘CTGCTTTCTAAAATAGAAATGCCTTCTTC 3′; cyclophyllin: 5′ ATGGTCAACCCCACCGTGT 3′, 5′ TCTGCTGTCTTTGGGACCTTGTC 3′; hA20: 5′ AGATCATCCACAAAGCCCTCATC 3′, 5′ AATTGCCGTCACCGTTCGT 3′; hXIAP: 5′ GGTGTTTTCTCAGTAGTTCTTACCAGACA 3′, 5′ ATGCTAAATGGTATCCAGGGTGC 3′; survivin: 5′ GAAACTGGACAGAGAAAGAGCCAA 3′, 5′ GGCACGGCGCACTTTCTT 3′; MnSOD: 5′ TCAATCATAGCATTTTCTGGACAAAC 3′, 5′ GGCTTCCAGCAACTCCCCTT 3′; Bcl-xL: 5′ GAACGGCGGCTGGGAT 3′, 5′ AGCGGTTGAAGCGTTCCTG 3′; Bfl-1/A-1: 5′ ACACAGGAGAATGGATAAGGCAAA 3, 5′ AGTCATCCAGCCAGATTTAGGTTC 3′.
Electrophoretic mobility shift assay
Nuclear protein extracts were prepared according to the procedure described by Dignam et al.32 Nuclear extracts were stored at −70°C. Protein concentrations were determined by a Bradford assay (Bio-Rad, Munich, Germany). A double-stranded oligonucleotide probe corresponding to the high-affinity κB binding site GGGGATTCCC33 was [γ32P]-adenosine triphosphate (ATP)] end labeled by treatment with T4 polynucleotide kinase and purified with G25 Sephadex columns (Roche Molecular Diagnostics, Mannheim, Germany). Approximately 150 000 CPM were added to 6 μg of nuclear protein in the presence of 1 μg of poly(dl-dC) as nonspecific competitor (Pharmacia, Freiburg, Germany). Binding reactions were done in 10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 4% glycerol, for 15 minutes at room temperature. DNA protein complexes were resolved by electrophoresis on a 4% nondenaturing polyacrylamide gel in Tris/glycine/EDTA (ethylenediaminetetraacetic acid) buffer. Gels were vacuum dried and exposed to Kodak BioMax MR film with intensifying screens. For competition experiments unlabeled double-stranded oligonucleotides were added to the binding reactions in 20-fold or 100-fold molar excess, respectively. A double-stranded Oct-1 binding site was used for nonspecific competition. The unlabeled high-affinity κB site GGGGATTCCC and the putative κB binding sites within the human TRAF1 promoter were used for specific competition. The putative κB binding sites found in the TRAF1 promoter are described in detail elsewhere.28
Immune complex kinase assay of the endogenous IKK complex
CLL lymphocytes and normal CD19+ B cells were lysed in Tris lysis buffer (25 mM Tris-HCl pH 8.0, 150 mM NaCl, 50 mM β-glycerophosphate, 1 mM EGTA (ethyleneglycotetraacetic acid), 1 mM EDTA, 10% glycerol, 1% Triton X-100, 5 mM benzamidin, 1 mM phenylmethylsulfonyl fluoride, 0.2 mg/mL leupeptin, 0.4 mg/mL aprotinin, 1 mM DTT). Lysates were clarified by centrifugation. Immunoprecipitation was done with an anti–IKKα antibody (no. sc7218; Santa Cruz Biotechnology, CA) at 4°C for 1 hour. Immunoprecipitates were washed twice in Tris lysis buffer and once in kinase buffer (25 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] pH 7.5, 150 mM NaCl, 25 mM β-glycerolphosphate, 10 mM MgCl2). Kinase reaction was performed in kinase buffer containing 1 mM DTT and 500 ng recombinant GST-IκBα (1-54) or mutated GST-IκBα (1-54, S32A, S36A) in the presence of 5 μCi (0.185 MBq) [γ-32P]ATP for 20 minutes at room temperature. Reaction was stopped by adding boiling Laemmli buffer. Immunoprecipitates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently blotted on a polyvinylidenefluoride (PVDF) membrane. Activity was quantified using a phosphorimager (Fuji, Frankfurt, Germany). Membranes were probed with an antibody against IKKα (Santa Cruz Biotechnology). Immunoprecipitated IKKα was detected using the appropriate secondary reagent and enhanced chemiluminescence (Amersham). The recombinant GST-IκBα (1-54) and mutated GST-IκBα (1-54, S32A, S36A) were kindly provided by C. Weber (University of Ulm, Germany).
Inhibition of NF-κB/Rel and IKK activity
Immunoblotting
Immunoblotting was done following standard protocols. For TRAF1 detection the rabbit antibody sc875 (Santa Cruz Biotechnology) was used. TRAF2 immunoblotting was done with the rabbit antibody sc7187.
Statistical analysis
Comparisons of quantitative TRAF1 and TRAF2 PCR data between healthy subjects and patients with CLL were performed using the Wilcoxon rank sum test. The same method was used for comparisons of clinical parameters between patients with high or low TRAF1 expression. Survival time, measured from the date of blood sampling, was plotted from life tables using Kaplan-Meier estimates. Differences were analyzed by the log-rank test. Statistical calculations were performed using the SAS software package (Version 6.12; SAS, Cary, NC).
Results
IAP gene expression
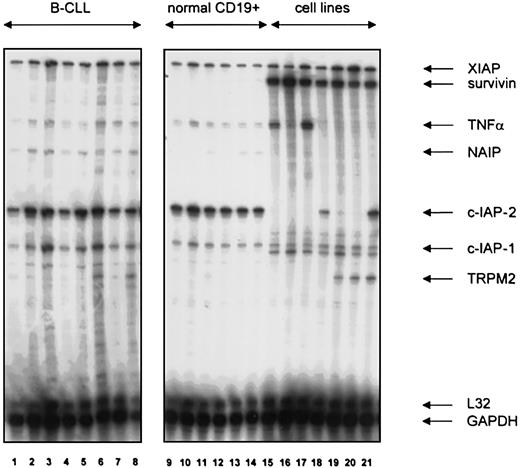
The mRNA expression of the inhibitor of apoptosis (IAP) family genes XIAP, survivin, NAIP, c-IAP-1, and c-IAP-2 was examined by RNase protection assay (RPA) in B-CLL lymphocytes (n = 37), B lymphocytes from healthy donors (n = 6), and lymphoma cell lines. XIAP was expressed at comparable levels in B-CLL, normal B cells, and lymphoma cell lines (Figure 1). Survivin was undetectable in normal B cells and in 36 of 37 B-CLL samples examined. The one positive sample was obtained from an end-stage patient with hyperleukocytosis currently undergoing transformation (data not shown). In contrast, all cell lines showed marked survivin expression. NAIP transcript levels were weak in B-CLL and normal B cells. B-CLL and normal B cells were strongly positive for c-IAP-2, whereas only 2 myeloma lines expressed c-IAP-2. c-IAP-1 transcript levels were comparable in cell lines, B cells, and B-CLL, albeit with a tendency toward higher levels in B-CLL.
IAP gene family expression in normal and neoplastic B cells.
Expression of XIAP, survivin, NAIP, c-IAP-1, c-IAP-2, and TRPM2 mRNA in B lymphocytes from patients with B-CLL (lanes 1-8), healthy blood donors (lanes 9-14), the human B-cell precursor lines MHH-CALL2 (lane 15), MHH-CALL3 (lane 16), MHH-CALL4 (lane 17,) and the human myeloma cell lines LP-1 (lane 18), OPM-2 (lane 19), NCI-H929 (lane 20), and RPMI-8226 (lane 21). A hAPO-5c RNAse protection assay with an added TNFα template provided by the manufacturer was used. A quantity of 10 μg total RNA was used per lane.
IAP gene family expression in normal and neoplastic B cells.
Expression of XIAP, survivin, NAIP, c-IAP-1, c-IAP-2, and TRPM2 mRNA in B lymphocytes from patients with B-CLL (lanes 1-8), healthy blood donors (lanes 9-14), the human B-cell precursor lines MHH-CALL2 (lane 15), MHH-CALL3 (lane 16), MHH-CALL4 (lane 17,) and the human myeloma cell lines LP-1 (lane 18), OPM-2 (lane 19), NCI-H929 (lane 20), and RPMI-8226 (lane 21). A hAPO-5c RNAse protection assay with an added TNFα template provided by the manufacturer was used. A quantity of 10 μg total RNA was used per lane.
TRAF expression
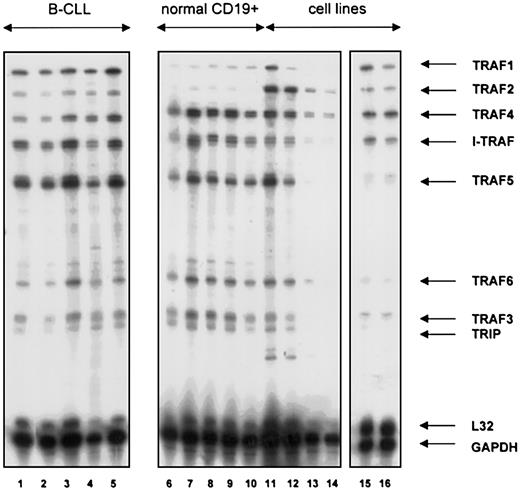
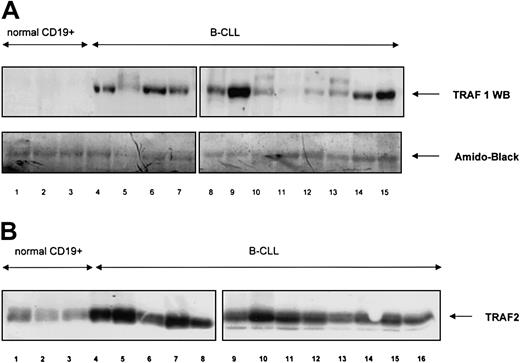
The mRNA expression of TRAF1, TRAF2, TRAF3 (CRAF), TRAF4 (CART), TRAF5, TRAF6, and of the TRAF interacting proteins I-TRAF and TRIP was analyzed by RPA in a similar fashion. TRAF1 mRNA was overexpressed in B-CLL lymphocytes as compared with B cells from healthy volunteers. In 27 patients with B-CLL examined, 25 showed high TRAF1 transcript levels whereas in normal B cells (n = 6) only weak TRAF1 expression was found (Figure 2). Patient characteristics and results are shown in Tables1-3. TRAF2 levels were slightly higher in B-CLL lymphocytes compared with normal B cells. TRAF4 (CART), I-TRAF, and TRAF5 were abundantly expressed both in B-CLL and normal B cell samples. TRAF6, TRAF3 (CRAF), and TRIP showed weak expression without difference between B-CLL lymphocytes and normal B cells. We further examined TRAF1 and TRAF2 protein expression by immunoblotting. As shown in Figure 3A, TRAF1 protein was found in all CLL samples (n = 22), but not in normal CD19+cells (n = 3). Staining with amido-black indicated comparable amounts of protein. Although TRAF2 protein was demonstrated in normal B cells, TRAF2 protein levels were clearly higher in B-CLL (n = 22, Figure 3B).
TRAF gene family expression in normal and neoplastic B cells.
Expression of TRAF1-6, I-TRAF, and TRIP mRNA in B lymphocytes from patients with B-CLL (lanes 1-5), healthy blood donors (lanes 6-10), a tonsil specimen (lane 11), the human B-cell precursor lines MHH-CALL 2 (lane 12) and MHH-CALL4 (lane 13), the human T-cell leukemia line MOLT4 (lane 14), the CD5+, EBV+ B-cell line EHEB (lane 15), and the EBV-Burkitt cell line BJAB (lane 16). A hAPO-5b RNAse protection assay was used for all experiments. A quantity of 10 μg total RNA was used per lane.
TRAF gene family expression in normal and neoplastic B cells.
Expression of TRAF1-6, I-TRAF, and TRIP mRNA in B lymphocytes from patients with B-CLL (lanes 1-5), healthy blood donors (lanes 6-10), a tonsil specimen (lane 11), the human B-cell precursor lines MHH-CALL 2 (lane 12) and MHH-CALL4 (lane 13), the human T-cell leukemia line MOLT4 (lane 14), the CD5+, EBV+ B-cell line EHEB (lane 15), and the EBV-Burkitt cell line BJAB (lane 16). A hAPO-5b RNAse protection assay was used for all experiments. A quantity of 10 μg total RNA was used per lane.
Patient characteristics and quantitative TRAF1 mRNA levels
| Patient no. . | Age . | Prior Tx . | Rai stage . | LDT <12 mo< . | Next Tx . | TRAF1 RPA . | TRAF1 PCR . | |
|---|---|---|---|---|---|---|---|---|
| 1 | 79 | nil | 1 | — | < | ne | +++ | — |
| 2 | 90 | CLB | 4 | < | — | PR | ++ | — |
| 3 | 67 | nil | 2 | < | — | PR | ++++ | — |
| 4 | 59 | ne | 2 | ne | — | PR | Neg | — |
| 5 | 73 | nil | 2 | — | < | PR | + | — |
| 6 | 77 | nil | 2 | — | < | nil | ++ | — |
| 7 | 61 | CLB | 4 | < | — | NC | +++ | — |
| 8 | 67 | nil | 2 | — | < | PR | Neg | — |
| 9 | 60 | nil | 2 | — | < | PR | + | — |
| 10 | 71 | nil | 2 | — | < | PR | +++ | — |
| 11 | 60 | CLB | 1 | — | < | PR | ++ | — |
| 12 | 65 | COP | 2 | — | < | MR | + | — |
| 13 | 59 | nil | 3 | < | — | PR | ++ | — |
| 14 | 52 | nil | 2 | — | < | NC | — | 2.91 |
| 15 | 57 | nil | 4 | < | — | PR | — | 3.50 |
| 16 | 54 | nil | 1 | — | < | mR | — | 1.95 |
| 17 | 57 | CLB | 2 | < | — | mR | — | 2.18 |
| 18 | 61 | nil | 0 | — | < | PR | — | 2.13 |
| 19 | 67 | PRED | 2 | — | < | PR | — | 4.48 |
| 20 | 57 | CLB | 2 | < | — | PR | — | 2.31 |
| 21 | 58 | CLB | 4 | — | < | MR | — | 2.33 |
| 22 | 53 | nil | 2 | — | < | nil | — | 2.45 |
| 23 | 58 | FLUD | 4 | < | — | NC | — | 2.14 |
| 24 | 65 | nil | 2 | < | — | MR | — | 1.61 |
| 25 | 69 | CLB | 4 | — | < | PR | — | 2.22 |
| 26 | 59 | FLUD | 4 | < | — | MR | — | 1.95 |
| 27 | 57 | nil | 2 | < | — | PR | +++ | 2.17 |
| 28 | 61 | CLB | 2 | < | — | PR | +++ | 2.49 |
| 29 | 55 | CLB | 3 | < | — | PR | ++++ | 4.02 |
| 30 | 76 | CLB | 3 | < | — | PR | +++ | 2.81 |
| 31 | 40 | CLB | 4 | < | — | PR | ++++ | 2.30 |
| 32 | 58 | CLB | 4 | < | — | NC | ++++ | 3.13 |
| 33 | 82 | nil | 2 | — | < | ne | + | 1.85 |
| 34 | 57 | CLB | 4 | — | < | PR | + | 1.59 |
| 35 | 75 | nil | 2 | — | < | nil | + | 2.21 |
| 36 | 67 | nil | 2 | — | < | PR | +++ | 2.42 |
| 37 | 58 | CLB | 4 | < | — | PR | ++ | 1.79 |
| 38 | 59 | CLB | 4 | — | < | Pro | +++ | 2.34 |
| 39 | 54 | nil | 2 | — | < | MR | ++ | 2.68 |
| 40 | 57 | FLUD | 4 | < | — | MR | + | 2.82 |
| Patient no. . | Age . | Prior Tx . | Rai stage . | LDT <12 mo< . | Next Tx . | TRAF1 RPA . | TRAF1 PCR . | |
|---|---|---|---|---|---|---|---|---|
| 1 | 79 | nil | 1 | — | < | ne | +++ | — |
| 2 | 90 | CLB | 4 | < | — | PR | ++ | — |
| 3 | 67 | nil | 2 | < | — | PR | ++++ | — |
| 4 | 59 | ne | 2 | ne | — | PR | Neg | — |
| 5 | 73 | nil | 2 | — | < | PR | + | — |
| 6 | 77 | nil | 2 | — | < | nil | ++ | — |
| 7 | 61 | CLB | 4 | < | — | NC | +++ | — |
| 8 | 67 | nil | 2 | — | < | PR | Neg | — |
| 9 | 60 | nil | 2 | — | < | PR | + | — |
| 10 | 71 | nil | 2 | — | < | PR | +++ | — |
| 11 | 60 | CLB | 1 | — | < | PR | ++ | — |
| 12 | 65 | COP | 2 | — | < | MR | + | — |
| 13 | 59 | nil | 3 | < | — | PR | ++ | — |
| 14 | 52 | nil | 2 | — | < | NC | — | 2.91 |
| 15 | 57 | nil | 4 | < | — | PR | — | 3.50 |
| 16 | 54 | nil | 1 | — | < | mR | — | 1.95 |
| 17 | 57 | CLB | 2 | < | — | mR | — | 2.18 |
| 18 | 61 | nil | 0 | — | < | PR | — | 2.13 |
| 19 | 67 | PRED | 2 | — | < | PR | — | 4.48 |
| 20 | 57 | CLB | 2 | < | — | PR | — | 2.31 |
| 21 | 58 | CLB | 4 | — | < | MR | — | 2.33 |
| 22 | 53 | nil | 2 | — | < | nil | — | 2.45 |
| 23 | 58 | FLUD | 4 | < | — | NC | — | 2.14 |
| 24 | 65 | nil | 2 | < | — | MR | — | 1.61 |
| 25 | 69 | CLB | 4 | — | < | PR | — | 2.22 |
| 26 | 59 | FLUD | 4 | < | — | MR | — | 1.95 |
| 27 | 57 | nil | 2 | < | — | PR | +++ | 2.17 |
| 28 | 61 | CLB | 2 | < | — | PR | +++ | 2.49 |
| 29 | 55 | CLB | 3 | < | — | PR | ++++ | 4.02 |
| 30 | 76 | CLB | 3 | < | — | PR | +++ | 2.81 |
| 31 | 40 | CLB | 4 | < | — | PR | ++++ | 2.30 |
| 32 | 58 | CLB | 4 | < | — | NC | ++++ | 3.13 |
| 33 | 82 | nil | 2 | — | < | ne | + | 1.85 |
| 34 | 57 | CLB | 4 | — | < | PR | + | 1.59 |
| 35 | 75 | nil | 2 | — | < | nil | + | 2.21 |
| 36 | 67 | nil | 2 | — | < | PR | +++ | 2.42 |
| 37 | 58 | CLB | 4 | < | — | PR | ++ | 1.79 |
| 38 | 59 | CLB | 4 | — | < | Pro | +++ | 2.34 |
| 39 | 54 | nil | 2 | — | < | MR | ++ | 2.68 |
| 40 | 57 | FLUD | 4 | < | — | MR | + | 2.82 |
Samples were analyzed by either RNase protection assay (RPA), real-time polymerase chain reaction (PCR) (10/ΔCT), or both. Both untreated and pretreated patients were analyzed. Prior treatment (Tx) consisted of prednisone (PRED), standard dose chlorambucil (CLB), fludarabine (FLUD), or cyclophosphamide, vincristine, and prednisone (COP). Lymphocyte doubling times (LDT) are shown as less than 12 months (left column) or longer than 12 months (right column). The response to the next treatment after blood sampling is divided into complete response (CR), partial response (PR), minor response (MR), no change (NC), or progressive disease (Pro).
ne indicates not evaluated; Neg, no expression; —, not done. TRAF1 RPA levels are given from lowest (+) to highest (++++) quartiles as described in “Materials and methods.”
TRAF1 mRNA levels in CD19+ B cells of healthy blood donors
| Donor no. . | TRAF1 RPA . | TRAF1 PCR . |
|---|---|---|
| 1 | + | 1.82 |
| 2 | + | 1.60 |
| 3 | + | 1.95 |
| 4 | + | 1.86 |
| 5 | + | 2.08 |
| 6 | + | 1.99 |
| Donor no. . | TRAF1 RPA . | TRAF1 PCR . |
|---|---|---|
| 1 | + | 1.82 |
| 2 | + | 1.60 |
| 3 | + | 1.95 |
| 4 | + | 1.86 |
| 5 | + | 2.08 |
| 6 | + | 1.99 |
TRAF1 RPA levels are given from lowest (+) to highest (++++) quartiles as described in “Materials and methods.”
TRAF1 mRNA levels in human lymphatic cell lines
| Cell lines . | TRAF1 RPA . | TRAF1 PCR . |
|---|---|---|
| MHH-cALL2 (human B precursor) | Neg | 1.13 |
| MHH-cALL3 (human B precursor) | Neg | 1.19 |
| MHH-cALL4 (human B precursor) | Neg | 1.09 |
| Molt4 (human T-cell leukemia) | Neg | 0.93 |
| EHEB (human chronic B-cell leukemia, EBV+) | ++ | |
| BJAB (human Burkitt lymphoma, EBV−) | + |
| Cell lines . | TRAF1 RPA . | TRAF1 PCR . |
|---|---|---|
| MHH-cALL2 (human B precursor) | Neg | 1.13 |
| MHH-cALL3 (human B precursor) | Neg | 1.19 |
| MHH-cALL4 (human B precursor) | Neg | 1.09 |
| Molt4 (human T-cell leukemia) | Neg | 0.93 |
| EHEB (human chronic B-cell leukemia, EBV+) | ++ | |
| BJAB (human Burkitt lymphoma, EBV−) | + |
TRAF1 RPA levels are given from lowest (+) to highest (++++) quartiles as described in “Materials and methods.” Neg indicates no expression.
TRAF 1 and TRAF2 protein expression in normal and neoplastic B cells.
(A) TRAF1 protein was analyzed in normal CD19+ B cells (lanes 1-3) and B-CLL samples (lanes 4-15). A quantity of 20 μg total protein was used per lane. Amido-black staining indicated similar amounts of protein being loaded per lane. (B) TRAF2 protein was analyzed in normal CD19+ B cells (lanes 1-3) and B-CLL samples (lanes 4-16). A quantity of 20 μg total protein was used per lane.
TRAF 1 and TRAF2 protein expression in normal and neoplastic B cells.
(A) TRAF1 protein was analyzed in normal CD19+ B cells (lanes 1-3) and B-CLL samples (lanes 4-15). A quantity of 20 μg total protein was used per lane. Amido-black staining indicated similar amounts of protein being loaded per lane. (B) TRAF2 protein was analyzed in normal CD19+ B cells (lanes 1-3) and B-CLL samples (lanes 4-16). A quantity of 20 μg total protein was used per lane.
Quantitative analysis of NF-κB–dependent inhibitors of apoptosis by real-time PCR
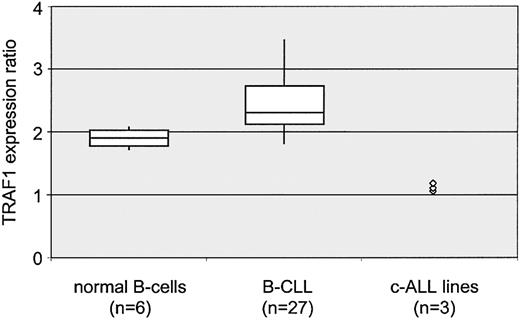
The levels of TRAF1 mRNA were further examined by real-time PCR analysis. TRAF1 transcripts were amplified in B-CLL samples (n = 27), CD19+ lymphocytes from blood donors (n = 6), and pre–B-cell lines (n = 3). Expression of TRAF1 mRNA was compared with levels of cyclophyllin mRNA that were amplified in the same PCR reaction. The TRAF1-cyclophyllin ratios in B-CLL samples correspond to higher TRAF1 expression in B-CLL lymphocytes as compared with normal B cells (Figure 4). The overexpression of TRAF1 mRNA levels in B-CLL lymphocytes compared with normal B cells was statistically significant (P < .05; Table4). The TRAF1 levels were much stronger both in B-CLL lymphocytes and normal B cells as compared with pre–B-cell lines (Figure 4; Tables 1-3). Thus, overexpression of TRAF1 mRNA was confirmed both by RPA and real-time PCR analysis. There were 13 samples analyzed by Taqman PCR only, 13 samples by RPA only, and 14 B-CLL samples examined using both methods (Table 1). TRAF2 mRNA amplification was performed in a similar fashion with simultaneous cyclophyllin amplifications as internal controls. However, the slightly stronger TRAF2 mRNA levels in B-CLL compared with normal B cells shown in Figure 2 were not reconfirmed by PCR analysis (Table 4). mRNA expression of NF-κB–regulated antiapoptotic genes was further analyzed by real-time PCR analysis as shown in Table 4. TRAF1 was the only gene found to be transcriptionally up-regulated in B-CLL compared with normal B cells and common acute lymphoblastic leukemia (c-ALL) cell lines. XIAP and survivin PCR results corresponded to the findings of RPA. MnSOD, A20, and Bfl-1/A-1 were found to have stronger mRNA levels in normal B cells as compared with B-CLL.
Quantitative analysis of TRAF1 mRNA levels.
Boxplot diagram of TRAF1 mRNA levels in B cells from healthy donors (left, n = 6), patients with B-CLL (center, n = 27), and 3 c-ALL cell lines (right). All measurements were done in duplicate by real-time PCR and compared with an internal cyclophillin control amplified in the same reaction. Duplicate experiments also involved independent reverse transcription. Δct ratios were calculated by subtraction of cyclophillin from TRAF1 values. For better illustration, TRAF1 ratios are presented as reciprocal values × 101. High ratios correspond to high TRAF1 mRNA levels. Depicted are the median (horizontal line), first and third quartiles (upper and lower edge of each box), 10% and 90% quantiles (vertical bars). The difference between healthy donors and patients with B-CLL was significant according to the log-rank test (P < .05).
Quantitative analysis of TRAF1 mRNA levels.
Boxplot diagram of TRAF1 mRNA levels in B cells from healthy donors (left, n = 6), patients with B-CLL (center, n = 27), and 3 c-ALL cell lines (right). All measurements were done in duplicate by real-time PCR and compared with an internal cyclophillin control amplified in the same reaction. Duplicate experiments also involved independent reverse transcription. Δct ratios were calculated by subtraction of cyclophillin from TRAF1 values. For better illustration, TRAF1 ratios are presented as reciprocal values × 101. High ratios correspond to high TRAF1 mRNA levels. Depicted are the median (horizontal line), first and third quartiles (upper and lower edge of each box), 10% and 90% quantiles (vertical bars). The difference between healthy donors and patients with B-CLL was significant according to the log-rank test (P < .05).
Transcript levels of NF-κB/Rel-regulated antiapoptotic genes measured by real-time polymerase chain reaction
| Gene . | CD19+ B-cells (n = 6) median . | B-CLL (n = 27) median . | c-ALL lines (n = 3) median . |
|---|---|---|---|
| TRAF1 | 1.92 | 2.31 (P < .05) | 1.13 |
| TRAF2 | 1.54 | 1.52 | 1.64 |
| XIAP | 1.58 | 1.23 | 1.29 |
| Survivin | 1.12 | 0.91 | 3.45 |
| MnSOD | 3.32 (P < .05) | 1.83 | 2.35 |
| A20 | 6.13 (P < .05) | 2.35 | 2.13 |
| Bcl-xL | 1.53 | 1.53 | 2.45 |
| Bfl-1/A-1 | 2.81 (P < .05) | 1.92 | 0.99 |
| Gene . | CD19+ B-cells (n = 6) median . | B-CLL (n = 27) median . | c-ALL lines (n = 3) median . |
|---|---|---|---|
| TRAF1 | 1.92 | 2.31 (P < .05) | 1.13 |
| TRAF2 | 1.54 | 1.52 | 1.64 |
| XIAP | 1.58 | 1.23 | 1.29 |
| Survivin | 1.12 | 0.91 | 3.45 |
| MnSOD | 3.32 (P < .05) | 1.83 | 2.35 |
| A20 | 6.13 (P < .05) | 2.35 | 2.13 |
| Bcl-xL | 1.53 | 1.53 | 2.45 |
| Bfl-1/A-1 | 2.81 (P < .05) | 1.92 | 0.99 |
mRNA expression of NF-κB/Rel-regulated genes with antiapoptotic properties was measured by real-time polymerase chain reaction (PCR). Medians from normal CD19+ cells, B-CLL lymphocytes, and B-precursor cell lines are shown. Values are given as 10/Δct ratios. High ratios correspond to high expression and vice versa. TRAF1 is overexpressed in B-CLL cells compared to normal B cells (P < .05) and B-precursor cell lines. MnSOD, Bfl-1/A-1, and A20 were more strongly expressed in normal B cells (P < .05) than in B-CLL.
Correlation of clinical parameters of B-CLL with TRAF1 expression
Real-time PCR was used to compare the levels of TRAF1 expression in B-CLL patient subgroups. Results were analyzed with regard to Rai stage37 (Rai 0-2 vs Rai 3-4), treatment status (untreated vs treated), lymphocyte doubling time (> 12 months vs < 12 months), and response to next cytotoxic therapy (responders vs nonresponders). None of these analyses gave any evidence for a correlation of these clinical markers with TRAF1 levels. Serum levels of lactate dehydrogenase (LDH) or alkaline phosphatase (AP) also did not correlate with TRAF1 levels (data not shown).
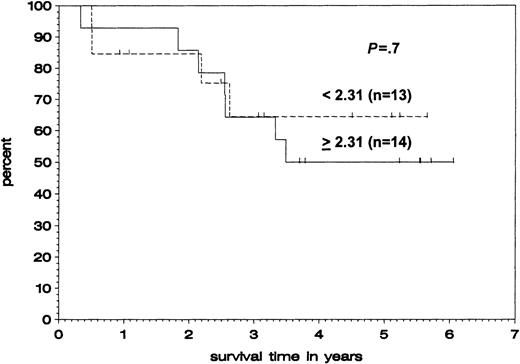
We further examined whether levels of TRAF1 expression correlate with overall survival. Patients were divided into a group with high TRAF1 expression (> median, n = 14) and a group with low TRAF1 expression (< median, n = 13). Although the total number of patients is too small for a statistical log-rank analysis, there was no trend toward a difference in overall survival between the 2 subgroups (Figure 5).
Correlation of TRAF1 mRNA expression and overall survival.
Kaplan-Meier plot of patients with B-CLL with high TRAF1 levels (ratios above or equaling median, solid line) and low TRAF1 levels (ratios below median, broken line), comparing survival from time of blood sampling. The difference between both populations was not significant (P = 0.7).
Correlation of TRAF1 mRNA expression and overall survival.
Kaplan-Meier plot of patients with B-CLL with high TRAF1 levels (ratios above or equaling median, solid line) and low TRAF1 levels (ratios below median, broken line), comparing survival from time of blood sampling. The difference between both populations was not significant (P = 0.7).
Role of autocrine TNFα in TRAF1 regulation
Because TNFα has been implicated as an autocrine growth factor for CLL lymphocytes38,39 and has also been shown to stimulate TRAF1 transcription,27 40 we examined the levels of TNFα mRNA in B-CLL lymphocytes (n = 37) and normal B cells (n = 6; Figure 1). Ratios of TNFα message density to L32 message density were calculated. TNFα mRNA levels were similar in neoplastic and normal B lymphocytes. The median ratio was 0.010 ± 0.003 compared with 0.009 ± 0.004 in B-CLL samples versus normal B cells, respectively, providing no evidence for increased autocrine TNFα production in B-CLL lymphocytes.
NF-κB/Rel binding to TRAF1 promoter sequences in B-CLL
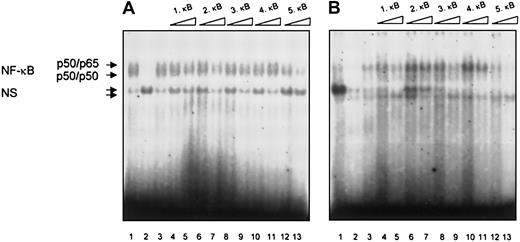
Next we analyzed NF-κB/Rel activity in nuclear extracts from CLL lymphocytes binding to the putative NF-κB/Rel binding sites in the human TRAF1 promoter. The 5κB binding sites found in the human TRAF1 promoter were used for competition analysis.28 Constitutive specific DNA binding was demonstrated in extracts derived from 2 patients. In both patients we observed complete abrogation of NF-κB/Rel binding by competition with the κB site 5 (Figure 6). Competition was observed by the κB sites 1 and 3 in only one patient. The κB sites 1, 3, and 5 have been reported to be functional sites.28
Analysis of NF-
κB/Rel binding to the putative NF-κB/Rel binding sites from the human TRAF1 promoter in B-CLL lymphocytes. Electrophoretic mobility shift assays were performed with nuclear extracts from B cells of 2 patients with B-CLL and the labeled H2TF1 binding site as a probe. Constitutive NF-κB/Rel binding is shown in both patients (lane 1) and was competed by a 20-fold excess of the unlabeled H2TF1 oligonucleotide (lane 2) but not by a 20-fold excess of an unlabeled Oct-1 oligonucleotide (lane 3). Unlabeled double-stranded oligonucleotides spanning the putative NF-κB/Rel binding sites 1-5 of the human TRAF1 promoter region were used as competitors at a 20-fold (lanes 4, 6, 8, 10, 12) or a 100-fold (lanes 5, 7, 9, 11, 13) molar excess. The specific NF-κB/Rel binding complexes consisting of a slower migrating p50/p655 heterodimer and a faster migrating p50 homodimer are indicated by arrows. NS corresponds to nonspecific binding.
Analysis of NF-
κB/Rel binding to the putative NF-κB/Rel binding sites from the human TRAF1 promoter in B-CLL lymphocytes. Electrophoretic mobility shift assays were performed with nuclear extracts from B cells of 2 patients with B-CLL and the labeled H2TF1 binding site as a probe. Constitutive NF-κB/Rel binding is shown in both patients (lane 1) and was competed by a 20-fold excess of the unlabeled H2TF1 oligonucleotide (lane 2) but not by a 20-fold excess of an unlabeled Oct-1 oligonucleotide (lane 3). Unlabeled double-stranded oligonucleotides spanning the putative NF-κB/Rel binding sites 1-5 of the human TRAF1 promoter region were used as competitors at a 20-fold (lanes 4, 6, 8, 10, 12) or a 100-fold (lanes 5, 7, 9, 11, 13) molar excess. The specific NF-κB/Rel binding complexes consisting of a slower migrating p50/p655 heterodimer and a faster migrating p50 homodimer are indicated by arrows. NS corresponds to nonspecific binding.
Influence of NF-κB/Rel inhibitors on NF-κB/Rel DNA binding and TRAF1 transcript levels in B-CLL
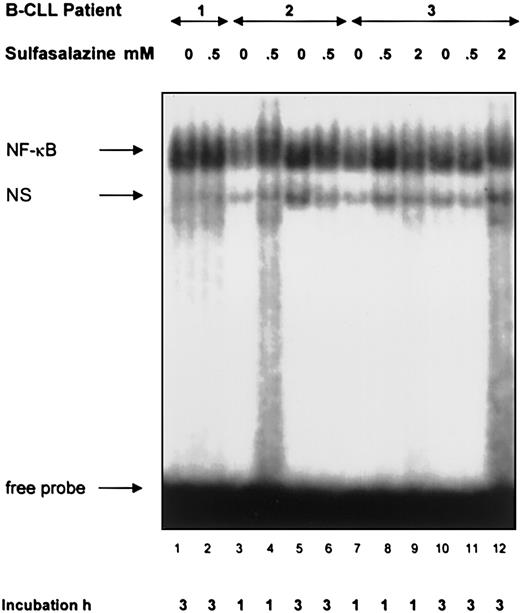
To further examine the influence of constitutive NF-κB/Rel activation on TRAF1 induction we incubated B-CLL lymphocytes from a total of 11 patients with sulfasalazine, an inhibitor of IKK-α and -β.34,41 Concentrations of 0.5 mM and 2 mM were used. Nuclear extracts taken at different time points from 1 hour to 17 hours failed to show any inhibition of NF-κB/Rel activation in the CLL cultures (Figure 7). However, toxic effects were observed at the 2 mM concentration beginning at 3 hours. Incubation with the NF-κB/Rel inhibitor PDTC35did not result in inhibition of NF-κB/Rel–dependent DNA binding, either (data not shown). Next, we analyzed TRAF1 levels in sulfasalazine-treated CLL cultures by real-time PCR analysis. CLL lymphocytes from 4 different patients at different time points and sulfasalazine concentrations were examined. We observed no inhibition of TRAF1 transcript levels (data not shown). Furthermore, we used the proteasome inhibitor lactacystin at 2.5 mM and 10 mM over 6 hours until 36 hours to inhibit NF-κB/Rel activity. However, NF-κB/Rel DNA binding activity was not impaired by lactacystin treatment of B lymphocytes from 2 separate patients (data not shown).
Lack of an inhibitory effect of sulfasalazine on NF-κB/Rel DNA binding in CLL lymphocytes.
Electrophoretic mobility shift assays were performed with nuclear extracts from B cells of patients with B-CLL and the labeled H2TF1 binding site as a probe. B-CLL lymphocytes were incubated with sulfasalazine at the indicated concentrations and times before extraction of nuclear proteins. The specific NF-κB/Rel binding complexes are indicated by an arrow. NS corresponds to nonspecific binding. Extracts from neoplastic lymphocytes of 11 patients incubated for 1 hour to 17 hours consistently failed to indicate an inhibitory effect of sulfasalazine.
Lack of an inhibitory effect of sulfasalazine on NF-κB/Rel DNA binding in CLL lymphocytes.
Electrophoretic mobility shift assays were performed with nuclear extracts from B cells of patients with B-CLL and the labeled H2TF1 binding site as a probe. B-CLL lymphocytes were incubated with sulfasalazine at the indicated concentrations and times before extraction of nuclear proteins. The specific NF-κB/Rel binding complexes are indicated by an arrow. NS corresponds to nonspecific binding. Extracts from neoplastic lymphocytes of 11 patients incubated for 1 hour to 17 hours consistently failed to indicate an inhibitory effect of sulfasalazine.
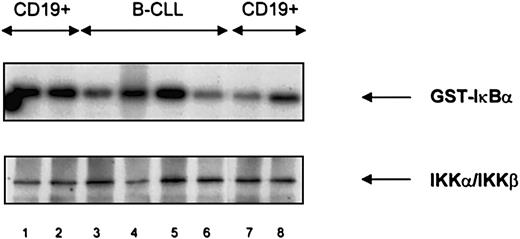
Endogenous IKK-α/IKK-β activity
An immune complex kinase assay showed comparable levels of IKK-α/IKK-β activity both in CD19+ normal B cells and B-CLL lymphocytes (Figure 8). IKK-α/IKK-β did not phosphorylate a substrate mutated at serine residues 32 and 36 (data not shown). Western blot analysis of immunoprecipitates showed a band of approximately 90 kd representing both IKK-α and IKK-β (Figure 8). IKKα/IKK-β levels were similar between CD19+ normal and CLL lymphocytes.
IκB Kinase (IKK) activity and expression in normal B cells and B-CLL lymphocytes.
IKK activity (upper panel) and protein expression (lower panel) were similar in normal CD19+ B cells (lanes 1, 2, 7, 8) and B-CLL lymphocytes (lanes 3-6). An IκB substrate mutated at positions 32 and 36 was not phosphorylated (data not shown).
IκB Kinase (IKK) activity and expression in normal B cells and B-CLL lymphocytes.
IKK activity (upper panel) and protein expression (lower panel) were similar in normal CD19+ B cells (lanes 1, 2, 7, 8) and B-CLL lymphocytes (lanes 3-6). An IκB substrate mutated at positions 32 and 36 was not phosphorylated (data not shown).
Discussion
Our study represents the first comprehensive survey of NF-κB/Rel–regulated inhibitors of apoptosis in B-CLL lymphocytes. We examined expression of the IAP family genes NAIP, c-IAP-1, c-IAP-2, XIAP, and survivin, Bfl-1/A-1, A20, MnSOD, Bcl-xL, TRAF1, and TRAF2 along with other TRAF family genes. We did not find evidence for B-CLL–specific up-regulation of IAP genes. Whereas NAIP levels were low, XIAP, c-IAP-1, and c-IAP-2 were strongly expressed in normal and neoplastic B cells. Survivin was not detected in normal CD19+ B cells and most CLL samples. Survivin expression was found in only one end-stage patient undergoing transformation, which is in line with the recent report of Granziero et al42 and the expression shown in aggressive lymphomas of germinal center origin.43 The mRNA expression of A20, MnSOD, and Bfl-1/A-1 was unexpectedly strong in normal B cells and revealed no indication of up-regulation in B-CLL.
Among the TRAF family genes our interest focused on TRAF1 for several reasons. TRAF1 shows tissue-specific expression with the highest levels in lung, tonsils, and spleen.25,26Additionally, TRAF1 has been shown to be induced by NF-κB/Rel and represents a potential candidate for transducing NF-κB/Rel–mediated inhibition of apoptosis.17,28 Finally, TRAF1 overexpression was demonstrated in Hodgkin and Reed-Sternberg cells44,45 due to NF-κB activation,45 which is the result of constitutive IKK activation.46 Since constitutive NF-κB/Rel activation is also a common feature of B-CLL,29,30 we tested the hypothesis that TRAF1 is up-regulated in B-CLL lymphocytes as a consequence of constitutive IKK activation. Both RNA and protein data clearly confirmed TRAF1 induction in B-CLL compared with normal B cells, which virtually lack TRAF1 expression. TRAF1 mRNA levels did not correlate with clinical parameters of advanced or progressive disease. Also there was no correlation of TRAF1 levels with treatment status, response to therapy, or overall survival. Thus, we consider constitutive TRAF1 expression rather a disease-related than an activity-related feature in B-CLL. In line with our findings, TRAF1 overexpression was shown in a subset of CLL samples in a recent paper by Zapata et al.47
TRAF2 has been reported to be constitutively expressed in various normal and neoplastic tissues including lymphomas.47,48 We observed a modest increase in TRAF2 mRNA levels in B-CLL by RPA which was not confirmed by real-time PCR. However, an increase in TRAF2 protein levels was shown in B-CLL compared with normal B cells. Thus, the induction of TRAF2 protein may be due to additional mechanisms such as protein stabilization, although we did not follow up on this hypothesis. This is the first report on augmented TRAF2 expression in B-CLL. Expression of TRAF3, TRAF4, TRAF5, and TRAF6 mRNAs was similar in B-CLL and normal B lymphocytes. These results are consistent with earlier reports from murine and human studies26 48-50 and do not yield any evidence for aberrant regulation in B-CLL.
What is the mechanism of TRAF1 and TRAF2 induction in B-CLL? TRAF1 and TRAF2 participate in LMP1, TNFR1, TNFR2, CD30, and CD40 signaling.25,26,28,51-55 Because LMP1 expression is absent even in EBV-infected CLL cells,56,57 LMP1 is not a likely candidate for causing the TRAF1 and TRAF2 induction we observed. Transcriptional up-regulation of TRAF1 by TNFα has been demonstrated.28,40 Because we did not detect an increase in TNFα expression in neoplastic versus normal B cells we provide evidence against a TNFα–mediated effect although TNFα production by depleted monocytes cannot be ruled out at this point. CD30 is a member of the TNFR superfamily with pleiotropic effects in lymphocyte regulation.58 CD30 and its ligand are expressed in B-CLL lymphocytes,59 however, the effect of CD30 activation on TRAF1 transcription in B-CLL is not known. The TNFR family protein CD40 is expressed in normal B cells and in B-CLL lymphocytes at comparable levels.59,60 CD40 ligation was shown to induce TRAF1, whereas the effect on TRAF2 transcription may be cell specific.28,48 However, it is currently unclear whether CD40-dependent TRAF1 induction involves NF-κB/Rel or different mechanisms such as the p38 MAPK pathway shown to induce CD40-mediated c-IAP-2 expression.48
What are functional consequences of TRAF1 and TRAF2 expression in B-CLL? TRAF1 was shown to costimulate NF-κB/Rel in concert with TRAF2 after CD30,55 LMP1,53 and TNFα28 stimulation, although these findings are under dispute.40,61-63 Whether NF-κB/Rel activity in B-CLL is modulated by TRAF1 and TRAF2 is currently unclear. There are several reports on antiapoptotic effects of TRAF1 and TRAF2.27,64,65 TNFα-induced apoptosis is inhibited by TRAF117,28 and TRAF217 by inhibition of caspase-8 activity.17 Chemotherapy-induced apoptosis seems to be mediated by up-regulation of Fas ligand and Fas/Fas ligand interaction.6 Fas and TNFα signaling results in cleavage of TRAF1 with the cleaved TRAF1 acting as a dominant-negative inhibitor of NF-κB activation.66,67 Therefore, Fas signaling may amplify the apoptotic response by blockage of NF-κB/Rel–induced inhibitors of apoptosis. The functional integrity of the Fas system in B-CLL has been questioned.7,8 Not only could defects in the Fas/FasL system explain the resistance of B-CLL to cytotoxic drugs but they could also contribute to high TRAF1 transcript levels due to reduced TRAF1 cleavage. The expression pattern of antiapoptotic genes we observed in B-CLL with high TRAF1, TRAF2, c-IAP-1, and c-IAP-2 levels corresponds very well to the scenario reported for NF-κB/Rel–mediated protection from TNFα-induced cytotoxicity.17 Although we demonstrate that the NF-κB/Rel binding activity to TRAF1 promoter elements is functional in B-CLL, our findings argue against a role of NF-κB/Rel in TRAF1 and TRAF2 regulation in B-CLL for 2 reasons: the levels of IKK activity are comparable between normal and CLL lymphocytes; and sulfasalazine, a direct inhibitor of IKK-α and -β,41did not inhibit NF-κB/Rel activation or TRAF1 transcript levels in B-CLL lymphocytes. Therefore, constitutive TRAF1 transcription and NF-κB/Rel activation in B-CLL appears to be independent of IKK activation, which constitutes the point of convergence for most NF-κB/Rel activators.11
Supported by grants from the Deutsche José Carreras Leukämie-Stiftung (DJCLS) and the Medical Faculty of the University of Ulm to G.M.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Gerd Munzert, Boehringer Ingelheim Pharma KG, Birkendorfer Str 65, 88397 Biberach, Germany; e-mail: gerd.munzert@bc.boehringer-ingelheim.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal