Abstract
Mutations at the Nramp1 gene cause susceptibility to infections with intracellular pathogens. In human blood, polymorphonuclear (PMN) leukocytes are the most abundant site ofNRAMP1 messenger RNA (mRNA) expression, suggesting that NRAMP1 plays an important role in the activity of these cells. By Northern blot analysis, NRAMP1 mRNA was only detected in most mature neutrophils from bone marrow (band and segmented cells). A high-affinity polyclonal rabbit antihuman NRAMP1 antibody directed against the amino terminus of the protein was produced and used to study cellular and subcellular localization of the protein in primary human neutrophils. Subcellular fractionation of granule populations together with immunoblotting studies with granule-specific markers indicate that NRAMP1 expression is primarily in tertiary granules. These granules are positive for the matrix enzyme gelatinase and the membrane subunit of the vacuolar H+/ATPase and can be recruited for exocytosis by treatment of neutrophils with phorbol myristate acetate. Immunogold studies by cryoelectron microscopy with primary neutrophils confirm that a majority (75%) of NRAMP1-positive granules are also positive for gelatinase, but they also suggest further heterogeneity in this granule population. Presence of NRAMP1 in tertiary granules is in agreement with the late-stage appearance ofNRAMP1 mRNA during neutrophil maturation in bone marrow. Finally, immunofluorescence studies of Candida albicans–containing phagosomes formed in neutrophils indicate that NRAMP1 is recruited from tertiary granules to the phagosomal membrane on phagocytosis, supporting a role for NRAMP1 in the antimicrobial defenses of human neutrophils.
Introduction
In the mouse, naturally occurring1 or experimentally introduced mutations2 at theNramp1 gene cause susceptibility to infection with several unrelated intracellular parasites such as Salmonella,Leishmania, and Mycobacterium (reviewed by Skamene et al3). These mutations impair the ability of macrophages to restrict intracellular replication of these microbes, suggesting an important role of the Nramp1 protein in bacteriostatic or bactericidal defense mechanisms of these cells.4Nramp1 messenger RNA (mRNA) is expressed in primary mouse macrophages and in J774 and RAW297 macrophage cell lines, where its expression can be strongly induced by exposure to interferon-γ and bacterial lipopolysaccharide.5 Sequence analysis indicates that Nramp1 is an integral membrane protein composed of 12 transmembrane domains, which define a superfamily of divalent cation transporters that has been highly conserved during evolution from bacteria to humans (reviewed by Cellier et al6 and Forbes and Gros7). Nramp1 protein is expressed in the membrane fractions of mouse macrophages,8 and subcellular localization studies by immunofluorescence indicate that it is present in the Lamp-1–positive compartment of these cells.9,10Double-immunofluorescence experiments together with biochemical studies in purified vesicles indicate that on phagocytosis Nramp1 is rapidly recruited to the membrane of phagosomes containing inert particles such as latex beads9 and zymosan11 or live bacteria such as Salmonella andYersinia.4 Although the mechanism of action of Nramp1 at the phagosomal membrane remains debated (reviewed by Forbes and Gros7), recent studies show that it functions as a divalent cation transporter.11-13 In particular, microfluorescence-imaging experiments in live macrophages allowed to internalize zymosan particles labeled with metal-sensitive fluorescent dyes show that Nramp1 mediates active, pH-dependent efflux of divalent cations (Mn2+) from the luminal space of the phagolysosome.11 It has been proposed that removal of divalent cations essential for microbial survival and replication may account for the pleiotropic effect of Nramp1 mutations on different intracellular pathogens.7
In humans, population-based association studies and linkage analyses in multigeneration pedigrees have demonstrated an association between polymorphic variants at NRAMP1 and susceptibility to tuberculosis and leprosy in endemic areas of disease and in the outbreak situation.14-17 Additional reports have also suggested association of NRAMP1 polymorphisms with inflammatory diseases in humans such as rheumatoid arthritis18-20 and Crohn disease.21Interestingly, functional polymorphisms have been identified in the promoter of the NRAMP1 gene in the form of sequence variations in a Z-DNA–forming dinucleotide repeat.22These polymorphisms define a 4-allele system that shows significant functional properties in transcriptional activity with reporter genes.22 Searle and Blackwell22proposed a hypothesis in which chronic activation of theNRAMP1 promoter in allele 3 of this polymorphism is associated with autoimmune disease susceptibility, while showing a protective effect against infectious diseases. The reverse would be true for the low-expressing allele 2 at NRAMP1.
Human NRAMP1 mRNA is expressed in lungs, spleen, and liver but is highest in peripheral blood leukocytes.23,24 There, polymorphonuclear leukocytes represent by far the major site ofNRAMP1 mRNA expression followed to a lesser degree by monocytes. Migration of monocytes to tissues (alveolar macrophages) or maturation in vitro is associated with increased NRAMP1expression compared with blood monocytes.24 Studies in the promyelocytic leukemia cell line HL-60 indicate that differentiation toward either the monocytic pathway (with vitamin D3 and phorbol ester) or the granulocyte pathway (dimethylformamide, dimethyl sulfoxide [DMSO]) is concomitant to strong induction ofNRAMP1 mRNA expression.24 Together, these results have suggested that NRAMP1 may play an important role in the antimicrobial response or inflammatory process mediated by polymorphonuclear (PMN) leukocytes.
Both PMN leukocytes and macrophages are capable of engulfing and destroying microorganisms by the action of toxic radicals, ions, and proteolytic enzymes produced by these cells. Neutrophils contain different types of granules that can be recruited for release by exocytosis or for fusion to phagosomes containing ingested particles.25 These granules are divided into 3 or 4 subtypes, depending on the purification method used to isolate them. The biochemical properties and content of bactericidal species of these granules have been extensively characterized in human neutrophils (reviewed in Borregaard et al26). Simple separation by sedimentation on density gradients yields 4 major fractions: a cytosolic soluble fraction, a PM/secretory vesicle fraction (positive for alkaline phosphatase and nicotinamide adenine dinucleotide phosphate [NADPH] oxidase), a specific granule fraction (secondary and tertiary granules, positive for lactoferrin and vitamin B12 binding protein), and the azurophil granules (positive for myeloperoxidase and β-glucuronidase).27 In addition, recruitment of particular granules for exocytosis after stimulation follows established kinetics.28
The role of the NRAMP1 protein in the antimicrobial defenses of the neutrophil remains unknown. However, the previous localization of the murine Nramp1 protein to the lysosomal compartment of macrophages9 suggests that it may also be present in one or more granule populations in neutrophils. To address this issue, we have generated a polyclonal antibody against the human NRAMP1 protein and used this reagent to study cellular and subcellular localization of NRAMP1 protein in human neutrophils.
Materials and methods
Cell isolation procedures
Bone marrow (BM) cells were obtained (15 mL) by aspiration from the posterior superior iliac crest from healthy volunteers as described.29 Procedures were approved by the institutional review board at the University of Copenhagen, and informed consent was provided according to the Declaration of Helsinki. Following sedimentation (15-20 minutes), the clear supernatant containing leukocytes was removed, and the cells were recovered by centrifugation (200g for 10 minutes at room temperature). The pelleted cells were resuspended in phosphate-buffered saline (PBS) and placed over a 2-layer Percoll gradient of densities 1.065 and 1.080 g/mL. Following centrifugation (1000g for 20 minutes at 4°C), 3 cell bands can be recovered: “band 1” containing band and segmented cells; “band 2”containing myelocytes and metamyelocytes; and “band 3” containing myeloblasts and promyelocytes.29The distribution of the neutrophils precursors in the 3 bands was assessed by differential counting of cytospin preparations. Mature neutrophils were isolated from peripheral blood anticoagulated in acid-citrate dextrose. Erythrocytes were sedimented by centrifugation on 2% Dextran T-500, and the leukocyte-rich supernatant was pelleted and resuspended in saline for subsequent centrifugation on Lymphoprep (Nyegaard, Oslo, Norway) at 400g for 30 minutes to separate polymorphonuclear cells from platelets and mononuclear cells.30
Cell culture
Chinese hamster ovary (CHO) LR73, the promyelocytic leukemia–derived HL-60, and the macrophagelike histiocytic lymphoma U937 cells were cultured as previously described.23,31HL-60 cells were induced to differentiate into granulocyte by the presence of 1.25% DMSO in the culture medium for 6 days. Differentiation was monitored by using flow cytometry to follow surface expression of CD11b. CHO and U937 cells were transfected with a humanNRAMP1 complementary DNA (cDNA) modified by the in-frame addition of a c-Myc tag at the carboxy terminus of the protein. Crude membrane fractions were prepared from control CHO and U937 cells and from CHO-NRAMP1–c-Myc and U937-NRAMP1–c-Myc as previously described.31
RNA isolation and Northern blotting
Total cellular RNA was extracted from the different BM cell populations by a single-step procedure.32 TheNRAMP1 hybridization probe was a 1.2-kilobase (kb) cDNA fragment corresponding to positions −120 to +1120 of the published sequence.23 Control hybridization probe for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was produced and used exactly as described.29 The membrane was washed for 2 × 15 minutes at 60°C in 2 × sodium chloride/sodium citrate (SSC; 1 × SSC is 150 mM NaCl, 15 mM sodium citrate, pH 7.0) 0.5% sodium dodecyl sulfate (SDS) followed by 2 × 15 minutes in 0.2 × SSC, 0.5% SDS. The membrane was stripped by boiling in 0.1% SDS before rehybridization.
Subcellular fractionation
Neutrophils were resuspended in Krebs-Ringer phosphate (130 mM NaCl, 5 mM KCl, 1.27 mM MgSO4, 0.95 mM CaCl2, 5 mM glucose, 10 mM NaH2PO4/Na2HPO4, pH 7.4) at 3 × 107 cells/mL and divided into 2 equal portions. One portion was kept on ice as control, the other was stimulated at 37°C for 15 minutes with 2 μg/mL phorbol myristate acetate (PMA). The stimulation was stopped by adding 2 volumes of ice-cold buffer. Control and stimulated cells were sedimented at 200g for 10 minutes and resuspended in ice-cold buffer before subcellular fractionation. Individual granule fractions were purified from resting and PMA-stimulated neutrophils exactly as previously described.33 Briefly, neutrophils were subjected to nitrogen cavitation followed by centrifugation to remove nuclei and unbroken cells. The resulting postnuclear supernatant was applied on top of a discontinuous, 3-layer Percoll gradient (1.050/1.09/1.12 g/mL) and centrifuged at 37 000g for 30 minutes at 4°C. Gradients were aspirated from the bottom through a peristaltic pump attached to a fraction collector set to deliver 1 mL in each fraction. Fractions 1 to 6, 7 to 12, 13 to 18, and 19 to 24 were pooled in 4 distinct populations. Percoll was removed by centrifugation, and the biologic material was resuspended in 1 mL PBS. Aliquots of each sample were assayed for the presence of marker proteins corresponding to individual compartments (indicated in parenthesis): myeloperoxidase (α band/azurophils or primary granules), lactoferrin (β1 band/specific or secondary granules), gelatinase (β2 band/gelatinase or tertiary granules), and albumin (γ band; secretory vesicles).28
Production and purification of NRAMP1 antibodies
A glutathione-S-transferase (GST) fusion protein was constructed by subcloning in plasmid vector pGEX (Pharmacia, Montreal, QC, Canada) a NRAMP1 N-terminal cDNA sequence (nucleotides 1 to 177, corresponding to amino acids 1 to 59; GST-59NT) obtained by PCR amplification from full-lengthNRAMP1 cDNA template using oligonucleotides 5HF (5′TCGGATCCT CAA TGA CAG GTG AC-3′) and 5HR (5′-TAGAATTCG CAG GCT GAA GGT G-3′). The PCR product was digested with BglII and EcoRI (restriction enzymes sites are underlined in the oligonucleotide sequence), prior to subcloning into pGEX-2 digested with BamHI and EcoRI to create the in-frame GST fusion. The fusion proteins were produced in Escherichia coli as previously described.31 Approximately 0.4 to 0.8 mg GST-59NT protein was injected in New Zealand white male rabbits as previously described.8 The polyclonal antiserum was then purified by an affinity chromatography protocol against the same NRAMP1 peptide segment (1-59 aas) fused to dihydrofolate reductase (DHFR), as previously described.31
Immunofluorescence and immunocytochemistry
Cells were fixed with Bouin solution for 20 minutes and permeabilized (or not) with Triton-X100 (0.1% in PBS) for 10 minutes. Cells were incubated at 20°C in blocking solution (bovine serum albumin [BSA] 1%, 10% heat-inactivated goat serum for CHO cells or BSA 1%, 20% heat-inactivated donkey serum for HL-60) for 1 hour, prior to incubation with primary antibodies for 2 hours at 20°C. Dilution of primary antibodies in blocking solution was as follows: anti-NRAMP1 antibody (rabbit polyclonal), 1/50 to 1/75 or nonimmune serum, 1/50 to 1/75. After washing with PBS-0.5% Tween 20, secondary antibodies were incubated for 1 hour at room temperature. Dilution of secondary antibodies was as follows: goat Texas red–antirabbit 1/400 and donkey Alexa Fluor488–antirabbit, 1/400 (Molecular Probes). Cells were examined by using a Nikon optical fluorescence microscope with a ×60 to ×100 objective (×600 to ×1000 magnification) or by using a confocal microscope. For immunocytochemistry, after permeabilization with Triton X-100, CHO cells were incubated in (1%) H2O2 in PBS for 10 minutes to inhibit endogenous peroxidase activity. Primary antibody incubation was followed by incubation with a swine secondary antirabbit immunoglobulin G (IgG; 1/100; DAKO) for 1 hour at room temperature and subsequently with a rabbit peroxidase antiperoxidase (1/100; DAKO). Specific staining was revealed by using 3′-diaminobenzidine tetrahydrochloride in solution. Finally, cells were counterstained with 0.1% methylene blue in PBS, dehydrated, and mounted with Permount.
Phagocytosis
DMSO-differentiated HL-60 cells were seeded in Poly-D-Lysine–coated glass slides (Biocoat; BD-Falcon) and further incubated for 1 day to promote adherence. Heat-inactivatedCandida albicans was opsonized for 15 minutes at 25°C with rabbit complement (Biotest). Cells were washed twice with PBS, and the culture medium was replaced by phagocytosis buffer (Hanks balanced salt solution with Ca++ and Mg2+ [Sigma], 0.2% BSA [Fisher]) containing opsonized C albicans A at a cell-to–C albicans ratio of 1:10. Incubation was at 37°C for 30 minutes. Slides were washed and processed for light microscopy or epi-immunofluorescence.
Immunoblotting
Crude membrane preparations from control CHO/U937 cells (5 μg) and aliquots (25 μg) of enriched granule fractions were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 10% acrylamide gels followed by transfer to a polyvinylidene diflouride (PVDF) membrane (16 hours, 4°C).31 Primary antibodies were used at the following dilution: rabbit antihuman NRAMP1, 1/250; mouse anti–c-Myc (9E10) 1/100; rabbit antigelatinase, 1/2500; rabbit antimyeloperoxidase, 1/2500, rabbit antihuman serum albumin, 1/2500; rabbit antilactoferrin, 1/2500; and rabbit anti-δ subunit (39 kd) of the mammalian V-type H+-ATPase, 1/400.
Electron microscopy
Neutrophils from peripheral blood were fixed for 24 hours in 4% paraformaldehyde in 0.1 M PHEM (240 mM PIPES, 100 mM HEPES, 8 mM MgCl2, 40 mM EGTA, pH 6.9) and then processed for ultrathin cryosectioning, as previously described.34Cryosections (45 nm) were cut at −125°C with the use of diamond knives (Drukker, Cuijk, The Netherlands) in an ultracryomicrotome (Leica Aktiengesellschaft, Vienna, Austria) and transferred with a mixture of sucrose and methylcellulose onto formvar-coated copper grids.35 The grids were placed on 35-mm Petri dishes containing 2% gelatin. For immunolabeling, the sections were incubated with rabbit antihuman NRAMP-1 (1/25) for 45 minutes, followed by a 30-minute incubation with 10-nm protein A–conjugated colloidal gold (EM Lab, Utrecht University). For double immunolabeling with rabbit anti–NRAMP-1 and rabbit antigelatinase (1/300), the procedure described by Slot et al36 was followed with 15- and 10-nm protein A–conjugated colloidal gold probes. After immunolabeling, the cryosections were embedded in a mixture of methylcellulose and uranyl acetate and examined with a Philips CM 10 electron microscope (Eindhoven, The Netherlands).
Results
NRAMP1 mRNA expression in developing polymorphonuclear leukocytes
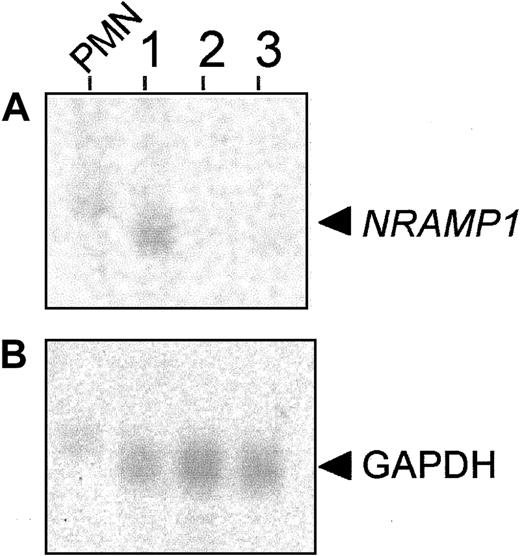
The development of neutrophils from stem cell precursors involves different maturational steps, including the acquisition of specific granules.25 The mRNAs encoding proteins present in different types of granules display a unique and sometimes diagnostic pattern of expression during the maturation process of neutrophils. For example, mRNA for myeloperoxidase, a protein present in primary/azurophyl granules, appears very early in neutrophil precursors and is largely absent from mature cells, whereas the mRNAs for gelatinase (tertiary granules) and for CD35 and fMLP-R (secretory vesicles) appear late during differentiation and remain present in mature cells.29 Concordance of temporal expression profiles between the mRNA for a novel granule protein and that previously established for mRNAs of matrix or membrane proteins of unique granule populations can provide initial clues on the site of expression of this novel protein. With the use of a discontinuous Percoll gradient protocol, BM cells of the neutrophil lineage can be fractionated into 3 cell populations consisting of early immature (primarily myeloblasts and promyelocytes found in band 3), late immature (primarily myelocytes and metamyelocytes found in band 2), and mature neutrophils (primarily band and segmented neutrophils found in band 1).29 Total RNA was prepared from these BM populations as well as from circulating PMN leukocytes, separated by gel electrophoresis and probed with a human NRAMP1 cDNA fragment (Figure 1A). NRAMP1mRNA expression was detected in circulating PMN leukocytes and also in the most mature neutrophil precursors corresponding to band 1 (Figure1A). The apparent faster electrophoretic mobility of NRAMP1mRNA in the sample of band 1 cells compared with mature PMN leukocytes is due to a gel artifact that also affects migration ofGAPDH control mRNA in the same samples. However, noNRAMP1 expression was detectable in RNA preparations from band 2 and band 3 cells, suggesting that NRAMP1 is not expressed in less mature cells (myeloblasts, promyelocytes, myelocytes, and metamyelocytes). These results suggest that NRAMP1 mRNA expression occurs late in neutrophil maturation, with expression persisting in circulating PMN leukocytes. Previously, band 1 cells were shown to express mRNAs corresponding to markers of tertiary granules and secretory vesicles (gelatinase, fMLP-R, CD35), suggesting that the NRAMP1 protein may be expressed in these granule populations.
NRAMP1 expression in mature granulocytes from peripheral blood (PMN leukocytes) and 3 populations of neutrophil precursors from BM.
Total RNA was extracted from (1) band 1 cells, band cells/segmented cells; (2) band 2 cells, myelocytes/metamyelocytes; and (3) band 3 cells, myeloblasts/promyelocytes and from PMN leukocytes. After separation by electrophoresis in denaturing formaldehyde gels (10 μg/lane), RNA was blotted to a hybridization membrane. The blot was probed with cDNA probes corresponding to NRAMP1 gene (A) andGAPDH gene (B) as a positive control.
NRAMP1 expression in mature granulocytes from peripheral blood (PMN leukocytes) and 3 populations of neutrophil precursors from BM.
Total RNA was extracted from (1) band 1 cells, band cells/segmented cells; (2) band 2 cells, myelocytes/metamyelocytes; and (3) band 3 cells, myeloblasts/promyelocytes and from PMN leukocytes. After separation by electrophoresis in denaturing formaldehyde gels (10 μg/lane), RNA was blotted to a hybridization membrane. The blot was probed with cDNA probes corresponding to NRAMP1 gene (A) andGAPDH gene (B) as a positive control.
Production and characterization of rabbit antihuman NRAMP1 antisera
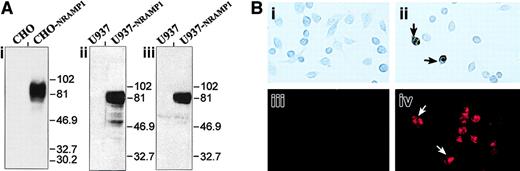
To further study expression and subcellular localization of the NRAMP1 protein in human neutrophils, a specific antiserum raised against the first 59 amino acid residues of NRAMP1 was developed and purified by affinity.31 The specificity of the resulting antiserum was assessed by immunoblotting (Figure2) by using membrane fractions from CHO cells or human U937 cells transfected or not with a humanNRAMP1 cDNA epitope tagged with a c-Myc antigenic peptide inserted at the C-terminus of the protein. The affinity-purified rabbit anti-NRAMP1 polyclonal antiserum specifically detected a protein species of apparent molecular mass 80 to 100 kd in the NRAMP1-transfected cells that was absent from the membrane fractions of control cells (Figure 2i-ii). NRAMP1 migrated as a broad immunoreactive species of size considerably larger than that predicted from the cDNA sequence (61 kd), suggesting that the protein is extensively modified by posttranslational modification in CHO and U937 cells, possibly by N-linked glycosylation, as previously reported for mouse Nramp1.8 The NRAMP1 protein detected by this polyclonal antiserum displayed electrophoretic mobility characteristics identical to that observed if a mouse anti–c-Myc monoclonal antibody was used to probe the immunoblot (Figure 2Aiii), confirming the specificity of the anti-NRAMP1 antiserum. Finally, the antihuman NRAMP1 antibody produced could also recognize the NRAMP1 protein by immunohistochemistry (Figure 2Bi-ii) and by immunofluorescence (Figure 2Biii-iv) in transfected CHO cells.
Detection of the NRAMP1 c-Myc–tagged proteins in CHO and U937 cells.
(A) Western blot analysis with crude membrane extracts from untransfected CHO and U937 cells and from CHO-NRAMP1 and U937-NRAMP1 transfectants was separated by SDS-PAGE on a 10% acrylamide gel followed by transfer to PVDF membranes. Immunodetection was with affinity-purified anti-NRAMP1 (i-ii) or with anti–c-Myc (iii) antibodies. The positions and sizes (in kilodaltons) of molecular weight markers are shown. (B) Untransfected CHO cells (i,iii) and CHO-NRAMP1 (ii,iv) were processed for immunochemistry (i-ii) or immunofluorescence (iii-iv) with our affinity-purified anti-NRAMP1 antibody. Arrows indicate some NRAMP1-positive cells.
Detection of the NRAMP1 c-Myc–tagged proteins in CHO and U937 cells.
(A) Western blot analysis with crude membrane extracts from untransfected CHO and U937 cells and from CHO-NRAMP1 and U937-NRAMP1 transfectants was separated by SDS-PAGE on a 10% acrylamide gel followed by transfer to PVDF membranes. Immunodetection was with affinity-purified anti-NRAMP1 (i-ii) or with anti–c-Myc (iii) antibodies. The positions and sizes (in kilodaltons) of molecular weight markers are shown. (B) Untransfected CHO cells (i,iii) and CHO-NRAMP1 (ii,iv) were processed for immunochemistry (i-ii) or immunofluorescence (iii-iv) with our affinity-purified anti-NRAMP1 antibody. Arrows indicate some NRAMP1-positive cells.
Detection of NRAMP1 in human neutrophil granules by immunoblotting
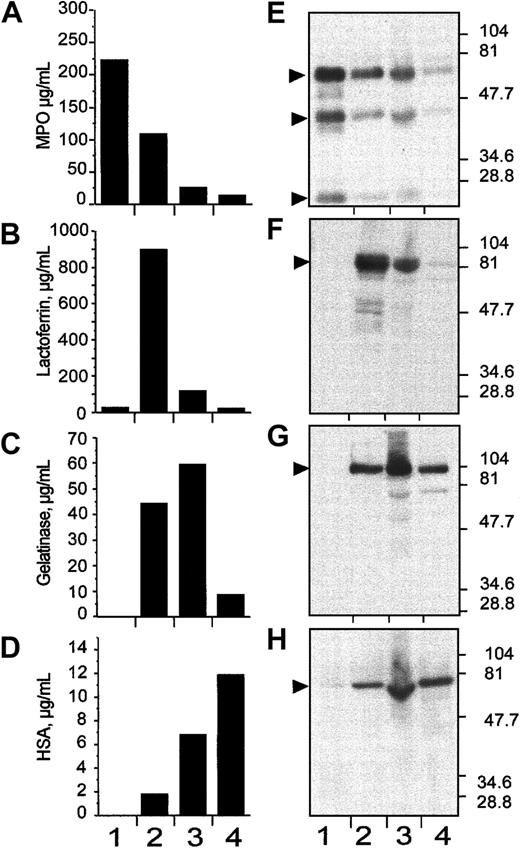
Granule populations were prepared from human neutrophils by separation on a 3-step discontinuous Percoll gradient.33After centrifugation, 4 bands are formed at the interface of the gradient steps; these correspond (from the bottom) to the α-band, which contains primarily azurophil granules, the β1-band enriched for the specific or secondary granules, the β2-band enriched for the gelatinase-positive tertiary granules, and the γ-band that contains secretory vesicles and plasma membranes.33 One-milliliter fractions were collected from the bottom of the tube and the fractions 1 to 6, 7 to 12, 13 to 18, and 19 to 24 were pooled in 4 populations designated fractions: 1, 2, 3, and 4 (Figure3). Biologic material from these pools (1-4) were recovered by centrifugation and were assayed for the presence of granule-specific markers by either enzyme-linked immunoassay (ELISA; Figure 3A-D) and by immunoblotting after SDS-PAGE (Figure 3E-H), using specific antibodies. The soluble matrix marker protein myeloperoxidase was used as a marker for the α-band,37 lactoferrin for the β1-band,37gelatinase for the β2-band, and human serum albumin (HSA) for the γ-band.38 Results from both analyses were consistent and, despite some degree of overlap, showed clear enrichment of primary (α-band), secondary (β1-band), tertiary (β2-band), and secretory vesicles (γ-band) in the 1-, 2-, 3-, and 4-pooled fractions, respectively (Figure 3).
Subcellular fractionation of nitrogen-cavitated neutrophils on 3-layer Percoll gradient.
After nitrogen cavitation, 10 mL postnuclear supernatant was applied on top of a 3-layer Percoll gradient. After centrifugation, the gradients were fractionated by aspiration from the bottom of the tube in 1-mL fractions. Fractions 1 to 6, 7 to 12, 13 to 18, and 19 to 24 were pooled in 4 distinct populations named 1, 2, 3, and 4, respectively. These pooled fractions were assayed for myeloperoxidase (A,E), lactoferrin (B,F), gelatinase (C,G), and albumin (D,H) by immunologic assays (ELISA; A-D) and by immunoblotting (E-H). In both markers assay, the pooled fractions 1, 2, 3, and 4 were identified as α-, β1-, β2-, and γ-band, respectively. In E to H, the positions and sizes of the molecular weight markers are shown.
Subcellular fractionation of nitrogen-cavitated neutrophils on 3-layer Percoll gradient.
After nitrogen cavitation, 10 mL postnuclear supernatant was applied on top of a 3-layer Percoll gradient. After centrifugation, the gradients were fractionated by aspiration from the bottom of the tube in 1-mL fractions. Fractions 1 to 6, 7 to 12, 13 to 18, and 19 to 24 were pooled in 4 distinct populations named 1, 2, 3, and 4, respectively. These pooled fractions were assayed for myeloperoxidase (A,E), lactoferrin (B,F), gelatinase (C,G), and albumin (D,H) by immunologic assays (ELISA; A-D) and by immunoblotting (E-H). In both markers assay, the pooled fractions 1, 2, 3, and 4 were identified as α-, β1-, β2-, and γ-band, respectively. In E to H, the positions and sizes of the molecular weight markers are shown.
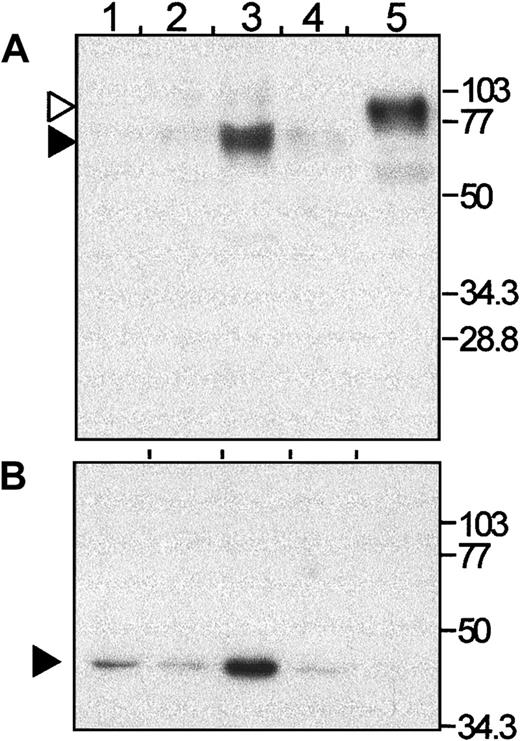
The presence of NRAMP1 protein was then investigated in these fractions by immunoblotting (Figure 4). Interestingly, NRAMP1 was found expressed mostly in the β2-fraction, with a small amount of immunoreactive protein detected in the β1- and the γ-fractions (Figures 4 and 5). The anti-NRAMP1 antiserum detected a single protein species in the β2-fraction of apparent electrophoretic mobility 70 to 75 kd, which migrated faster than the immunoreactive NRAMP1–c-Myc–tagged protein expressed in U937 transfected cells (80-90 kd), which was used as a positive control in these experiments. The nature of this difference remains unknown but may reflect different posttranslational modification of NRAMP1 in the 2 cells types. These results indicate that NRAMP1 is primarily expressed in the tertiary granule population. Reprobing the same immunoblot with a monoclonal antibody directed against the 39-kd δ subunit of the vacuolar H+/ATPase (Figure 4B), strongly suggests that both NRAMP1 and V-ATPase are coexpressed in tertiary granules of human neutrophils.
Detection of NRAMP1 in human neutrophils granules by Western blotting.
An immunoblot of purified fractions (lane 1, α; lane 2, β1; lane 3, β2; and lane 4, γ; 25 μg of each fraction per lane) was probed with an antibody against NRAMP1 (NT) (A). After stripping, the membrane was reprobed with an antibody against the 39-kd subunit of the VATPase (B). As a positive control for the anti-NRAMP1 antibody, 5μg membrane proteins from U937 transfectants expressing the c-Myc–tagged NRAMP1 was included (lane 5). The positions and sizes (in kilodaltons) of the molecular weight markers are shown.
Detection of NRAMP1 in human neutrophils granules by Western blotting.
An immunoblot of purified fractions (lane 1, α; lane 2, β1; lane 3, β2; and lane 4, γ; 25 μg of each fraction per lane) was probed with an antibody against NRAMP1 (NT) (A). After stripping, the membrane was reprobed with an antibody against the 39-kd subunit of the VATPase (B). As a positive control for the anti-NRAMP1 antibody, 5μg membrane proteins from U937 transfectants expressing the c-Myc–tagged NRAMP1 was included (lane 5). The positions and sizes (in kilodaltons) of the molecular weight markers are shown.
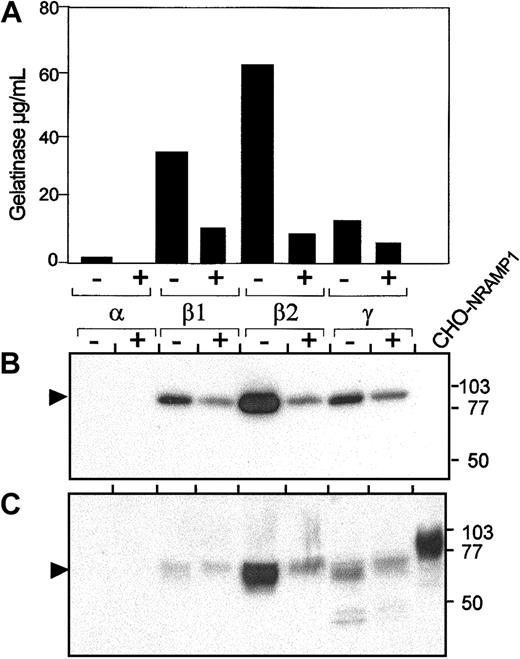
Expression of NRAMP1 in human neutrophils granules after PMA treatment.
Prior to disruption, cells were either kept on ice (control; lane −) or stimulated for 15 minutes at 37°C with 2μg/mL PMA (lane +). After washing and disruption of the cells, the postnuclear supernatants were centrifuged on a 3-layer Percoll gradients, fractionated, and pooled in 4 samples 1, 2, 3, and 4, as described in Figure 3. Pooled fractions 1, 2, 3, and 4 were assayed by ELISA for myeloperoxidase, lactoferrin, albumin (not shown), and gelatinase (A) and identified as α-, β1-, β2-, and γ-bands. The same fractions (25 μg) were separated by SDS-PAGE on a 10% acrylamide gel followed by transfer to PVDF membrane (B,C). Immunoblotting was performed with antibodies raised against gelatinase (B) and NRAMP1 (C). The positions and sizes (in kilodaltons) of the molecular weight markers are shown.
Expression of NRAMP1 in human neutrophils granules after PMA treatment.
Prior to disruption, cells were either kept on ice (control; lane −) or stimulated for 15 minutes at 37°C with 2μg/mL PMA (lane +). After washing and disruption of the cells, the postnuclear supernatants were centrifuged on a 3-layer Percoll gradients, fractionated, and pooled in 4 samples 1, 2, 3, and 4, as described in Figure 3. Pooled fractions 1, 2, 3, and 4 were assayed by ELISA for myeloperoxidase, lactoferrin, albumin (not shown), and gelatinase (A) and identified as α-, β1-, β2-, and γ-bands. The same fractions (25 μg) were separated by SDS-PAGE on a 10% acrylamide gel followed by transfer to PVDF membrane (B,C). Immunoblotting was performed with antibodies raised against gelatinase (B) and NRAMP1 (C). The positions and sizes (in kilodaltons) of the molecular weight markers are shown.
Effect of PMA treatment on NRAMP1-positive neutrophil granules
Recruitment of different granule populations for exocytosis can be induced by stimulation with compounds such as PMA or fMLP and follows established kinetics. This recruitment can be used to confirm the identity of an NRAMP1-positive granule population and to determine if NRAMP1 can be delivered to the pm after fusion of this granule population. In these experiments, neutrophils were treated with 10−6 M PMA, a treatment that has been previously established to induce recruitment of secretory vesicles and tertiary and secondary but not primary granules.28 After nitrogen cavitation and granule separation on Percoll gradient, fractions were collected as 4 pools, as described in Figures 3 and 4. Such fractions were then analyzed by ELISA and immunoblotting for the presence of gelatinase (Figure 5A-B, respectively) and for other granule-specific matrix markers (data not shown). Immunoblotting data indicate that PMA treatment of neutrophils resulted in disappearance of approximately 80% of gelatinase reactivity, demonstrating tertiary granule recruitment to the plasma membrane for exocytosis (Figure 5A-B). Analysis of the same fractions with the anti-NRAMP1 antiserum (Figure5C) indicates that PMA treatment of neutrophils results in drastic reduction of level of NRAMP1 protein in the β2 granule fraction in a manner and proportion very similar to that observed for the matrix marker gelatinase. These results support the contention that NRAMP1 is expressed in the membrane of PMA-recruitable, gelatinase-containing tertiary granules. Interestingly, disappearance of NRAMP1 from the tertiary granules was not accompanied by a concomitant appearance of NRAMP1 in the plasma membrane/secretary vesicles-containing γ fraction (Figure 5A), as has been previously observed for other membrane proteins.39 These results suggest the possibility that NRAMP1 may not be stable at the plasma membrane.
Electron microscopy detection of NRAMP1 in human neutrophils
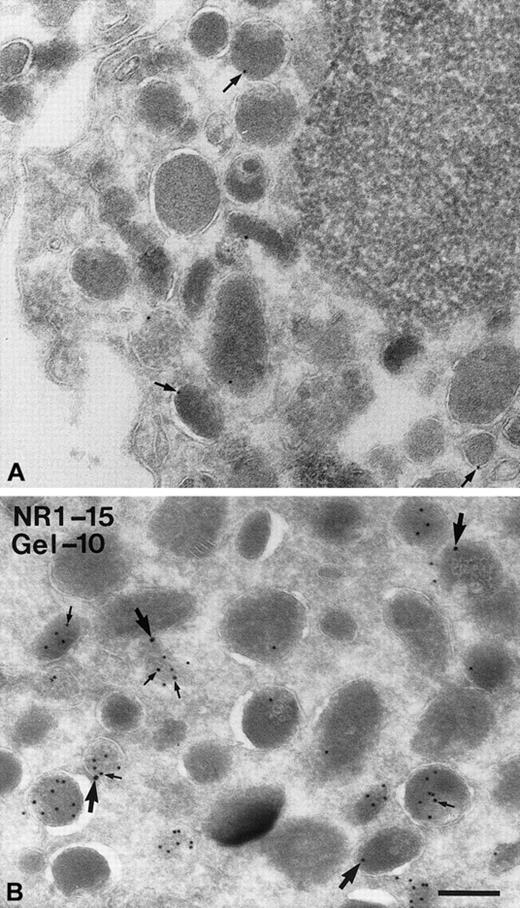
The subcellular localization of NRAMP1 was also examined by immunoelectron microscopy. For this examination, human neutrophils were harvested and 45-nm thin cryosections were incubated with the rabbit anti-NRAMP1 polyclonal antiserum (Figure6A), and specific immune complexes were revealed with protein A–conjugated colloidal gold (10-nm particles). Although few gold particles were detected, immunoreactive NRAMP1 was found associated with granules in neutrophils (arrows in Figure 6A) but was absent from the cytosol or from the nucleus and mitochondria. Interestingly, gold particles were found at the membrane but not in the matrix, in agreement with the fact that NRAMP1 is an integral membrane protein, and thus possibly expressed in the membrane of neutrophil granules. As results in Figures 1, 4, and 5 strongly suggested that NRAMP1 may localize to tertiary granules, additional double-labeling experiments were carried out. In this case, NRAMP1 was labeled with 15-nm gold particles, and gelatinase was revealed with a specific antibody coupled to 10-nm gold particles. In these experiments, several granules could be seen heavily labeled by the antigelatinase antibody (Figure 6B; small arrows). Labeling appeared to be in the lumen of the granule in agreement with the known matrix location of this protein in tertiary granules. Although a large proportion of granules were positive for gelatinase only (96 of 115, ∼83%; see “Discussion”), the majority (14 of 19, ∼75%) of NRAMP1-positive granules were also positive for gelatinase (Figure 6B; large arrows), in agreement with the proposal that NRAMP1 is present in gelatinase granules. Interestingly, a subpopulation of NRAMP1-positive granules (5 of 19; ∼25%) were negative for gelatinase (Figure 6B), suggesting that NRAMP1 can also be present in gelatinase-negative granules.
Detection of NRAMP1 and gelatinase in human neutrophils granules by immunoelectron microscopy.
PMN leukocytes were fixed, cryosectioned, and labeled with anti-NRAMP1 and protein A–gold (10 nm; A) or double-labeled with antigelatinase and protein A–gold (10 nm) followed by labeling with anti-NRAMP1 and protein A–gold (15 nm; B). (A) Labeling on the membranes of some granules (arrows) is shown. (B) A population of granules is highly labeled for gelatinase on the matrix (small arrows), and labeling for NRAMP1 (large arrows) is shown on the membrane of some granules containing gelatinase and some negative for gelatinase. Bar = 200 nm.
Detection of NRAMP1 and gelatinase in human neutrophils granules by immunoelectron microscopy.
PMN leukocytes were fixed, cryosectioned, and labeled with anti-NRAMP1 and protein A–gold (10 nm; A) or double-labeled with antigelatinase and protein A–gold (10 nm) followed by labeling with anti-NRAMP1 and protein A–gold (15 nm; B). (A) Labeling on the membranes of some granules (arrows) is shown. (B) A population of granules is highly labeled for gelatinase on the matrix (small arrows), and labeling for NRAMP1 (large arrows) is shown on the membrane of some granules containing gelatinase and some negative for gelatinase. Bar = 200 nm.
Recruitment of NRAMP1 to phagosomes in HL-60–derived granulocytes
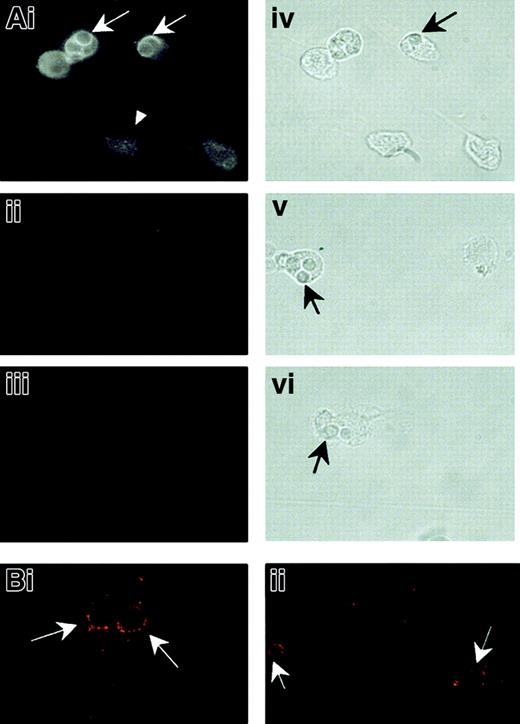
Localization of NRAMP1 protein to tertiary granules in primary human neutrophils suggests that NRAMP1 may be recruited to the membrane of phagosomes, after phagocytosis of microbial particles by these cells. This possibility was tested by using HL-60 cells differentiated in vitro into neutrophils following treatment with DMSO. We have previously shown that this treatment results in robust induction ofNRAMP1 mRNA24 as well as other markers of the granulocytic lineage. HL-60–derived neutrophils were allowed to ingest heat-killed, opsonized C albicans cells for 30 minutes at 37°C. Cells were then permeabilized and analyzed by immunofluorescence for reactivity with anti-NRAMP1 antiserum (Figure7). In intact, particle-free, differentiated HL-60 cells, NRAMP1 staining appeared intracellular and punctate, with no obvious labeling of the plasma membrane (Figure 7Ai; arrowheads). Staining was specific and absent when cells were incubated with either normal preimmune serum (Figure 7Aii) or secondary antibody alone (Figure 7Aiii). This staining is similar to that previously observed for the mouse Nramp1 protein expressed in primary macrophages9 or for the human NRAMP1 protein overexpressed in transfected CHO cells (Figure 2). In individual cells having engulfed C albicans, NRAMP1 staining appeared concentrated at the periphery of the internalized particles (Figure 7Ai; arrows). This staining could result from active recruitment of NRAMP1 to the phagosomal membrane or could be an optical artifact caused by displacement of unfused NRAMP1-positive vesicles by the internalized particle. To distinguish between the 2 possibilities, C albicans phagosomes formed in HL-60–derived neutrophils were analyzed for NRAMP1 reactivity by confocal microscopy (Figure 7Bi-ii). This analysis revealed NRAMP1 staining concentrated at the periphery of individual phagosomes, with 80% (67 of 83 scored) of phagosomes scoring positive. This finding strongly suggests active NRAMP1 recruitment at that site following fusion with tertiary granules.
NRAMP1 association withC albicans–containing phagosomes in HL-60 cells differentiated into granulocytes.
After treatment for 5 days with DMSO, HL-60 cells were allowed to ingest heat-killed C albicans for 30 minutes at 37°C. The cells were then washed free of uningested particles, fixed, and permeabilized before indirect immunofluorescence examined by using a Nikon optical fluorescence microscope (A) or a confocal microscope (B). Detection was performed with an affinity-purified rabbit anti-NRAMP1 antibody (Ai, Bi, and Bii), nonimmune serum (Aii), or no primary antibody (Aiii) on the cells. Immunofluorescence micrographs (Ai-Aiii) and the corresponding bright field images (Aiv-Avi) are shown. Arrowhead shows an intact, particle-free, differentiated HL-60 cell. Arrows point to ingested C albicans.
NRAMP1 association withC albicans–containing phagosomes in HL-60 cells differentiated into granulocytes.
After treatment for 5 days with DMSO, HL-60 cells were allowed to ingest heat-killed C albicans for 30 minutes at 37°C. The cells were then washed free of uningested particles, fixed, and permeabilized before indirect immunofluorescence examined by using a Nikon optical fluorescence microscope (A) or a confocal microscope (B). Detection was performed with an affinity-purified rabbit anti-NRAMP1 antibody (Ai, Bi, and Bii), nonimmune serum (Aii), or no primary antibody (Aiii) on the cells. Immunofluorescence micrographs (Ai-Aiii) and the corresponding bright field images (Aiv-Avi) are shown. Arrowhead shows an intact, particle-free, differentiated HL-60 cell. Arrows point to ingested C albicans.
Discussion
Neutrophils are polymorphonuclear leukocytes that play a primordial role in the first line of defense against bacterial infections. Neutrophils possess a large number of bacteriostatic and bactericidal enzymes, one of the most important being the production of superoxide anions via NADPH oxidase. However, continuous production of such oxygen radicals by neutrophils can cause inflammatory reactions and is seen in autoimmune disorders.40 We have previously reported very high levels of NRAMP1 mRNA expression in human neutrophils, either in primary cells or in HL-60 cells induced to differentiate along the granulocytic pathway.24 The fact that neutrophils (1) play a key role in antimicrobial defenses and in inflammatory reaction and (2) express high-level NRAMP1 mRNA and that NRAMP1 polymorphisms are associated with susceptibility to infections and autoimmune diseases in humans suggest that the NRAMP1 may play an important role in neutrophil function.
In the present study, we have used a rabbit antihuman NRAMP1 antibody to demonstrate robust NRAMP1 protein expression in human neutrophils. Subcellular fractionation studies indicate that NRAMP1 protein copurifies with a granule population but shows very little if any expression in the plasma membrane fraction of these cells. The NRAMP1-positive granule population is positive for the matrix marker gelatinase and is also enriched for the 39-kd δ subunit of the V-type H+-ATPase (Figure 3). Interestingly, results in Figures 4and 5 suggest that NRAMP1 may be more “specific” for β2 granules than the classic gelatinase matrix marker. At least 2 factors may have contributed to this observation: The first is the sensitivity of the immunoblotting technique used, including a possible threshold effect of the anti-NRAMP1 antiserum with no detectable signal below a certain level (as opposed to the more abundant matrix marker gelatinase). The second is that NRAMP1 may be more susceptible than gelatinase to proteolytic degradation when present in either the β1 or the γ fractions. The presence of NRAMP1 protein expression in gelatinase-positive granules is paralleled by the similar kinetics of mRNA expression of the 2 corresponding genes during neutrophil ontogeny (Figure 1). Finally, immunoelectron microscopy analysis on cryosections of primary cells show that NRAMP1 indeed localizes to a subset of granules, where it appears to be present in their membrane, in agreement with known structural features of the protein.41For double-labeling studies, a total of 115 positive granules were scored for gelatinase (G) and/or NRAMP1 reactivity (N). Of 110 G+ granules, only 14 (∼13%) were G+/N+. We believe that this low percentage of doubly positive granules is an underestimate and is due at least in part to the low activity of the immunopurified antihuman NRAMP1 antiserum in electron microscopy experiments. This belief is based on the observation that all N+ granules contained a single gold particle, thus the possibility of a threshold effect, with a number of N+ granules possibly going undetected. By opposition, gelatinase is quite abundant in these granules, and most G+ granules harbored multiple grains. Nevertheless, of the total 19 N+ granules recorded, 14 (∼75%) were G+/N+, whereas 5 (∼25%) were G−/N+, confirming an important colocalization of the 2 markers in tertiary granules. The G−/N+ granules are likely to be truly G−/N+ but could also be G+/N+, with gelatinase going undetected. Despite limitations associated with detection, these results suggest possible heterogeneity in tertiary granules in general, and in N+ granules in particular.
Tertiary granules can be recruited to fuse with phagosomes into which they deliver enzymes that are important for the destruction of ingested bacteria.25 Their matrix is characterized by the presence of the protease gelatinase, which is related to collagenase and heparanase present in other granules of these cells. They also contain cytochrome b, which consists of the heme-containing, membrane-bound gp91-phox and p22-phox subunits that ultimately form the superoxide-producing complex NADPH oxidase. Therefore, recruitment of tertiary granules to phagosomes formed in neutrophils after ingestion of microbial pathogens would be expected to deliver the NRAMP1 protein to the membrane of such phagosomes. This proposition was verified by immunofluorescence and confocal microscopy on HL-60 cells differentiated into granulocytes and that clearly show association of NRAMP1 to C albicans–containing phagosome. This situation is reminiscent to that seen in mononuclear phagocytes such as monocytes and macrophages in which the mouse Nramp1 protein localizes to Lamp-1–positive lysosomes in resting cells and can be quickly recruited to the membrane of phagosomes containing either inert particles or live bacteria.4,9,10 These findings also suggest that NRAMP1 may have the same biochemical activity and play the same functional role in neutrophils and macrophages. In macrophages, Nramp1 functions as a divalent cation transporter.11-13Our group has recently shown that Nramp1 functions as an efflux pump at the phagosomal membrane,11 possibly removing from the phagosomal lumen divalent cations that are either essential to bacterial growth or essential to the activity of bacterial detoxifying enzymes.7 The activity of microbially encoded superoxide dismutases may be essential for survival of bacteria ingested by neutrophils, and it is tempting to speculate that removal of the Mn2+/Fe2+ cofactors that are essential for catalytic activity of such enzymes may be the mechanism by which NRAMP1 contributes to the antimicrobial defenses of the neutrophil.
In macrophages, divalent cation transport by Nramp1 is strictly pH dependent and can be abrogated by folimycin and bafilomycin, potent inhibitors of the vacuolar H+/ATPase, the enzyme responsible for phagosomal acidification.11 This strict requirement of acidic pH for Nramp protein transport has also been established for the mammalian Nramp2 homolog,42 but it is also the hallmark of the eukaryotic and prokaryotic Nramp superfamily (reviewed by Cellier et al6 and Forbes and Gros7). The acidic pH may be required for either protonation of key amino acid residues in the formation of a putative transport site, or the ΔpH may provide a portion or the entire electrochemical gradient required to energize transport. In the present study, immunoblotting of granule populations purified from primary neutrophils indicates that the NRAMP1-positive granule population is also positive for the δ subunit of vacuolar ATPase (Figure 4), an integral, membrane-associated subunit of the enzyme.43This finding suggests that both the pH-dependent NRAMP1 transporter and the enzyme required for the generation of β-pH across the phagosomal membrane (vacuolar ATPase) would be simultaneously delivered to the membrane of the phagosome on recruitment of tertiary granules. This situation would provide a convenient regulatory mechanism for activating NRAMP1 at the desired transport site.
Tertiary granules can also be recruited to fuse with the plasma membrane during degranulation. They contain a pool of membrane glycoproteins such as the fMLP receptor, the FcRIII receptor, and the C3bi receptor that are important for proper adhesion and migration of neutrophils in tissues. Therefore, it may be expected that degranulation of neutrophils may also recruit NRAMP1 to the plasma membrane. Results in Figure 5 indeed verify that NRAMP1 disappears from the tertiary granule fraction on degranulation experimentally induced by PMA treatment. However, this result is not accompanied by appearance of NRAMP1 in the plasma membrane of these cells, which when analyzed in the same experiment remains largely negative for NRAMP1 expression. This situation is similar to the membrane-bound metalloproteinase MT6/MMP-25 that is localized primarily to the membrane of gelatinase granules.39 This situation suggests that NRAMP1 may be unstable at the plasma membrane and is possibly targeted for degradation. Therefore, the activity of NRAMP1 in neutrophils may be limited to possible transport function at the phagosomal membrane on recruitment from tertiary granules.
These findings suggest a possible role of NRAMP1 in neutrophil function and provide a link with the association of NRAMP1 with susceptibility to infectious diseases and inflammatory diseases noted in genetic studies.
We thank Hans Janssen for technical assistance and Nico Ong for assistance with electron microscopy.
Supported by National Institutes of Health grant AI355237 (P.G.) and the Danish Medical Research Council (N.B.). F.C.-H. is supported by a postdoctoral fellowship from Milestone Medica Corporation. P.G. and S.G. are International Research Scholars of the Howard Hughes Medical Institute (HHMI) and Distinguished Scientists of the Canadian Institutes for Health Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Philippe Gros, Department of Biochemistry, McGill University, 3655 Sir William Osler Promenade, Rm 907, Montreal, Quebec, Canada, H3G-1Y6; e-mail: gros@med.mcgill.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal